Transposon Insertion in the purL Gene Induces Biofilm Depletion in Escherichia coli ATCC 25922
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture of E. coli and Electrocompetent Cells
2.2. Transposon Mutagenesis
2.3. Biofilm Analysis
2.4. Phenotypic Characterization of Mutants
2.4.1. Growth Curves
2.4.2. Swimming Assay
2.4.3. Congo Red Assay
2.4.4. Hemagglutination Assays
2.5. Genotypic Characterization of Mutants
2.5.1. DNA Sequencing
2.5.2. purL Gene Disruption
2.5.3. purL Gene Complementation
2.5.4. Effect of Inosine Addition to the Biofilm Formation Ability of the purL Mutant.
2.6. Proteomic Characterization of Mutants
2.6.1. Protein Isolation
2.6.2. Two-Dimensional SDS-PAGE Analysis
2.6.3. Gel Staining and Protein Detection
2.6.4. Mass-Spectrometry Analysis for Orbitrap
2.6.5. RNA Isolation and cDNA Synthesis
2.6.6. Gene Expression by qRT-PCR
2.7. Data Plotting and Statistical Analysis
3. Results
3.1. Mutant Selection by Biofilm Assays
3.2. Phenotypic Characterization of Mutants
3.2.1. Growth Curves in LB Broth
3.2.2. Swimming, Congo Red and Hemagglutination Assays
3.3. Genotypic Characterization of Mutants
3.3.1. DNA Sequencing
3.3.2. Confirmation of the Role of the purL Gene in Biofilm Formation Using Its Knockout Strain
3.4. Proteomic Characterization of Mutant Tn 263
3.4.1. Comparative Proteomic Analyses of the wt and Tn263 by Two-Dimensional SDS-PAGE
3.4.2. Verification of Biofilm-Related Genes by q-RT-PCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blount, Z.D. The unexhausted potential of E. coli. Elife 2015, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bentley, R.; Meganathan, R. Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol. Rev. 1982, 46, 241–280. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, J.G.; Roth, J.R. Evolution of coenzyme B12 synthesis among enteric bacteria: Evidence for loss and reacquisition of a multigene complex. Genetics 1996, 142, 11–24. [Google Scholar] [PubMed]
- Eggesbø, M.; Moen, B.; Peddada, S.; Baird, D.; Rugtveit, J.; Midtvedt, T.; Bushel, P.R.; Sekelja, M.; Rudi, K. Development of gut microbiota in infants not exposed to medical interventions. APMIS 2011, 119, 17–35. [Google Scholar] [CrossRef]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef]
- Soto, S.M. Importance of biofilms in urinary tract infections: New therapeutic approaches. Adv. Biol. 2014, 2014, 1–13. [Google Scholar] [CrossRef]
- Millán-Rodríguez, F.; Palou, J.; Bujons-Tur, A.; Musquera-Felip, M.; Sevilla-Cecilia, C.; Serrallach-Orejas, M.; Baez-Angles, C.; Villavicencio-Mavrich, H. Acute bacterial prostatitis: Two different sub-categories according to a previous manipulation of the lower urinary tract. World J. Urol. 2006, 24, 45–50. [Google Scholar] [CrossRef]
- Beloin, C.; Roux, A.; Ghigo, J.-M. Escherichia coli biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 249–289. [Google Scholar] [CrossRef] [Green Version]
- Wright, K.J.; Seed, P.C.; Hultgren, S.J. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect. Immun. 2005, 73, 7657–7668. [Google Scholar] [CrossRef] [Green Version]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [Green Version]
- Gbejuade, H.O.; Lovering, A.M.; Webb, J.C. The role of microbial biofilms in prosthetic joint infections. Acta Orthop. 2015, 86, 147–158. [Google Scholar] [CrossRef] [PubMed]
- González, J.F.; Hahn, M.M.; Gunn, J.S. Chronic biofilm-based infections: Skewing of the immune response. Pathog. Dis. 2018, 76, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belas, R. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 2014, 22, 517–527. [Google Scholar] [CrossRef]
- Kaplan, J.B. Biofilm dispersal: Mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 2010, 89, 205–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, G.; Sharma, S.; Sharma, P.; Chandola, D.; Dang, S.; Gupta, S.; Gabrani, R. Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 2016, 121, 309–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klemm, P.; Schembri, M. Type 1 fimbriae, curli, and antigen 43: Adhesion, colonization, and biofilm formation. Ecosal Plus 2013, 1, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Beloin, C.; Valle, J.; Latour-Lambert, P.; Faure, P.; Kzreminski, M.; Balestrino, D.; Haagensen, J.A.J.; Molin, S.; Prensier, G.; Arbeille, B.; et al. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 2004, 51, 659–674. [Google Scholar] [CrossRef]
- Mehrotra, A. Bacterial Biofilms. Pediatric Asthmaall. Immunol. 2007, 20, 191–195. [Google Scholar] [CrossRef]
- Prigent-Combaret, C.; Prensier, G.; Le Thi, T.T.; Vidal, O.; Lejeune, P.; Dorel, C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: Role of flagella, curli and colanic acid. Environ. Microbiol. 2000, 2, 450–464. [Google Scholar] [CrossRef]
- Cepas, V.; López, Y.; Gabasa, Y.; Martins, C.B.; Ferreira, J.D.; Correia, M.J.; Santos, L.M.A.; Oliveira, F.; Ramos, V.; Reis, M.; et al. Inhibition of bacterial and fungal biofilm formation by 675 extracts from microalgae and cyanobacteria. Antibiotics 2019, 8, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hultgren, S.J.; Schwan, W.R.; Schaeffer, A.J.; Duncan, J.L. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect. Immun. 1986, 54, 613–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.C.E. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [Green Version]
- Görg, A.; Weiss, W.; Dunn, M.J. Current two-dimensional electrophoresis technology for proteomics. Proteomics 2004, 4, 3665–3685. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Chevallet, M.; Luche, S.; Rabilloud, T. Silver staining of proteins in polyacrylamide gels. Nat. Protoc. 2006, 1, 1852–1858. [Google Scholar] [CrossRef] [Green Version]
- Marcelino, I.; Colomé-Calls, N.; Holzmuller, P.; Lisacek, F.; Reynaud, Y.; Canals, F.; Vachiéry, N. Sweet and sour ehrlichia: Glycoproteomics and phosphoproteomics reveal new players in Ehrlichia ruminantium physiology and pathogenesis. Front. Microbiol. 2019, 10, 1–18. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility, 29th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Jandu, N.; Ho, N.K.L.; Donato, K.A.; Karmali, M.A.; Mascarenhas, M.; Duffy, S.P.; Tailor, C.; Sherman, P.M. Enterohemorrhagic Escherichia coli O157:H7 gene expression profiling in response to growth in the presence of host epithelia. PLoS ONE 2009, 4, e4889. [Google Scholar] [CrossRef]
- Reid, S.D.; Herbelin, C.J.; Bumbaugh, A.C.; Selander, R.K.; Whittam, T.S. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 2000, 406, 64–67. [Google Scholar] [CrossRef]
- Nhu, N.T.K.; Phan, M.D.; Peters, K.M.; Lo, A.W.; Forde, B.M.; Chong, T.M.; Yin, W.F.; Chan, K.G.; Chromek, M.; Brauner, A.; et al. Discovery of new genes involved in curli production by a uropathogenic Escherichia coli strain from the highly virulent O45:K1:H7 lineage. MBio 2018, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Serra, D.O.; Richter, A.M.; Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 2013, 195, 5540–5554. [Google Scholar] [CrossRef] [Green Version]
- Kozmin, S.G.; Wang, J.; Schaaper, R.M. Role for CysJ flavin reductase in molybdenum cofactor-dependent resistance of Escherichia coli to 6-N-Hydroxylaminopurine. J. Bacteriol. 2010, 192, 2026–2033. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Lee, J.; Lee, J.; Park, C. Characterization of the Escherichia coli YajL, YhbO and ElbB glyoxalases. FEMS Microbiol. Lett. 2016, 363, fnv239. [Google Scholar] [CrossRef] [Green Version]
- Burnett, B.J.; Altman, R.B.; Ferrao, R.; Alejo, J.L.; Kaur, N.; Kanji, J.; Blanchard, S.C. Elongation factor Ts directly facilitates the formation and disassembly of the Escherichia coli elongation factor Tu·GTP·Aminoacyl-tRNA ternary complex. J. Biol. Chem. 2013, 288, 13917–13928. [Google Scholar] [CrossRef] [Green Version]
- Stricker, J.; Erickson, H.P. In Vivo characterization of Escherichia coli ftsZ mutants: Effects on Z-Ring structure and function. J. Bacteriol. 2003, 185, 4796–4805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonntag, I.; Schwarz, H.; Hirota, Y.; Henning, U. Cell envelope and shape of Escherichia coli: Multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J. Bacteriol. 1978, 136, 280–285. [Google Scholar] [CrossRef] [Green Version]
- Franklin, M.J.; Chang, C.; Akiyama, T.; Bothner, B. New technologies for studying biofilms. Microbiol. Spectr. 2015, 3, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Green, S.M.; Malik, T.; Giles, I.G.; Drabble, W.T. The purB gene of Escherichia coli K-12 is located in an operon. Microbiology 1996, 142, 3219–3230. [Google Scholar] [CrossRef] [Green Version]
- Anand, R.; Hoskins, A.A.; Stubbe, J.; Ealick, S.E. Domain organization of Salmonella typhimurium formylglycinamide ribonucleotide amidotransferase revealed by X-ray crystallography. Biochemistry 2004, 43, 10328–10342. [Google Scholar] [CrossRef]
- Zhang, Y.; Morar, M.; Ealick, S.E. Structural biology of the purine biosynthetic pathway. Cell. Mol. Life Sci. 2008, 65, 3699–3724. [Google Scholar] [CrossRef] [Green Version]
- Soto, S.M.; Smithson, A.; Martinez, J.A.; Horcajada, J.P.; Mensa, J.; Vila, J. Biofilm formation in uropathogenic Escherichia coli strains: Relationship with prostatitis, urovirulence factors and antimicrobial resistance. J. Urol. 2007, 177, 365–368. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef]
- Vidal, O.; Longin, R.; Prigent-Combaret, C.; Dorel, C.; Hooreman, M.; Lejeune, P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: Involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998, 180, 2442–2449. [Google Scholar] [CrossRef] [Green Version]
- Cepas, V.; López, Y.; Muñoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef] [Green Version]
- Gualdi, L.; Tagliabue, L.; Bertagnoli, S.; Ieranò, T.; De Castro, C.; Landini, P. Cellulose modulates biofilm formation by counteracting curli-mediated colonization of solid surfaces in Escherichia coli. Microbiology 2008, 154, 2017–2024. [Google Scholar] [CrossRef] [Green Version]
- Garavaglia, M.; Rossi, E.; Landini, P. The pyrimidine nucleotide biosynthetic pathway modulates production of biofilm determinants in Escherichia coli. PLoS ONE 2012, 7, e31252. [Google Scholar] [CrossRef] [Green Version]
- Shaffer, C.L.; Zhang, E.W.; Dudley, A.G.; Dixon, B.R.E.A.; Guckes, K.R.; Breland, E.J.; Floyd, K.A.; Casella, D.P.; Algood, H.M.S.; Clayton, D.B.; et al. Purine biosynthesis metabolically constrains intracellular survival of uropathogenic Escherichia Coli. Infect. Immun. 2017, 85, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Prüß, B.M.; Besemann, C.; Denton, A.; Wolfe, A.J. A complex transcription network controls the early stages of biofilm development by Escherichia Coli. J. Bacteriol. 2006, 188, 3731–3739. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, S.; Arita-Morioka, K.; Terao, A.; Yamanaka, K.; Ogura, T.; Mizunoe, Y. Multitasking of Hsp70 chaperone in the biogenesis of bacterial functional amyloids. Commun. Biol. 2018, 1, 52. [Google Scholar] [CrossRef]
- Gannesen, A.V.; Zdorovenko, E.L.; Botchkova, E.A.; Hardouin, J.; Massier, S.; Kopitsyn, D.S.; Gorbachevskii, M.V.; Kadykova, A.A.; Shashkov, A.S.; Zhurina, M.V.; et al. Composition of the biofilm matrix of Cutibacterium acnes acneic strain RT5. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Prüß, B.M.; Verma, K.; Samanta, P.; Sule, P.; Kumar, S.; Wu, J.; Christianson, D.; Horne, S.M.; Stafslien, S.J.; Wolfe, A.J.; et al. Environmental and genetic factors that contribute to Escherichia coli K-12 biofilm formation. Arch. Microbiol. 2010, 192, 715–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gervais, F.G.; Phoenix, P.; Drapeau, G.R. The rcsB gene, a positive regulator of colanic acid biosynthesis in Escherichia coli, is also an activator of ftsZ expression. J. Bacteriol. 1992, 174, 3964–3971. [Google Scholar] [CrossRef] [Green Version]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 2015, 3, 1191–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Wood, T.K. Protein acetylation in prokaryotes increases stress resistance. Biochem. Biophys. Res. Commun. 2011, 410, 846–851. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.-B.; Chen, M.; Zhang, Y.; Wang, M.; Wang, Y.; Huang, Q.; Wang, X.; Wang, G. The phosphotransferase system gene ptsI in the endophytic bacterium Bacillus cereus is required for biofilm formation, colonization, and biocontrol against wheat sharp eyespot. FEMS Microbiol. Lett. 2014, 354, 142–152. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Lv, X.; Lu, J.; Yu, S.; Jin, Y.; Hu, J.; Zuo, J.; Mi, R.; Huang, Y.; Qi, K.; et al. The role of the ptsI gene on AI-2 internalization and pathogenesis of avian pathogenic Escherichia coli. Microb. Pathog. 2017, 113, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.S.; Santos, A.J.M.; Bejerano-Sagie, M.; Correia, P.B.; Marques, J.C.; Xavier, K.B. Phosphoenolpyruvate phosphotransferase system regulates detection and processing of the quorum sensing signal autoinducer-2. Mol. Microbiol. 2012, 84, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Barrios, A.F.G.; Zuo, R.; Ren, D.; Wood, T.K. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 2006, 93, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Leonardo, M.R.; Dailly, Y.; Clark, D.P. Role of NAD in regulating the adhE gene of Escherichia coli. J. Bacteriol. 1996, 178, 6013–6018. [Google Scholar] [CrossRef] [Green Version]
- Colón-González, M.; Méndez-Ortiz, M.M.; Membrillo-Hernández, J. Anaerobic growth does not support biofilm formation in Escherichia coli K-12. Res. Microbiol. 2004, 155, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J.; Langborg, A.; Minhas, H.S. Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem. J. 1999, 344, 109. [Google Scholar] [CrossRef] [PubMed]
- Aristarkhov, A.; Mikulskis, A.; Belasco, J.G.; Lin, E.C.C. Translation of the adhE transcript to produce ethanol dehydrogenase requires RNase III cleavage in Escherichia coli. J. Bacteriol. 1996, 178, 4327–4332. [Google Scholar] [CrossRef] [Green Version]
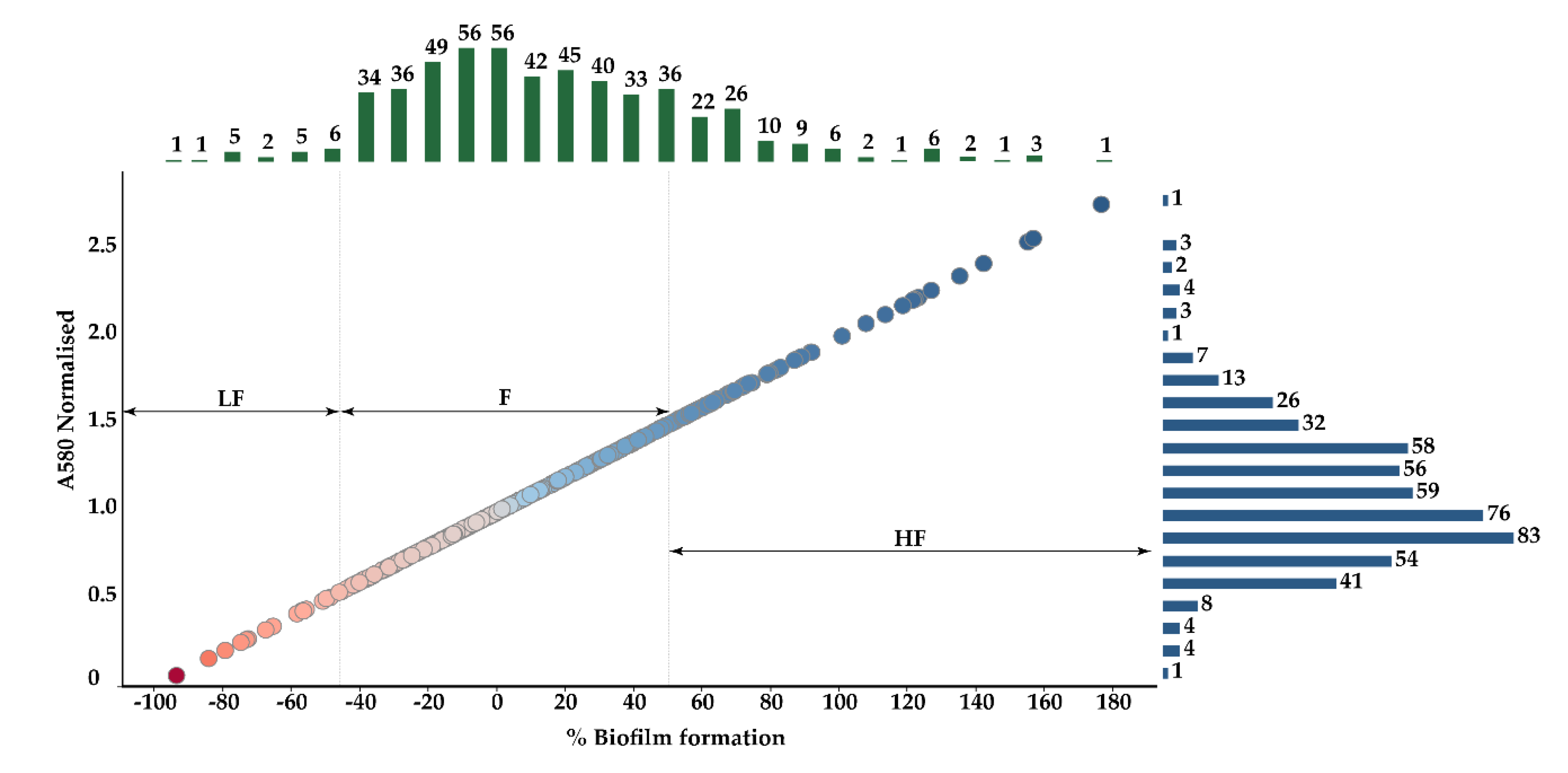
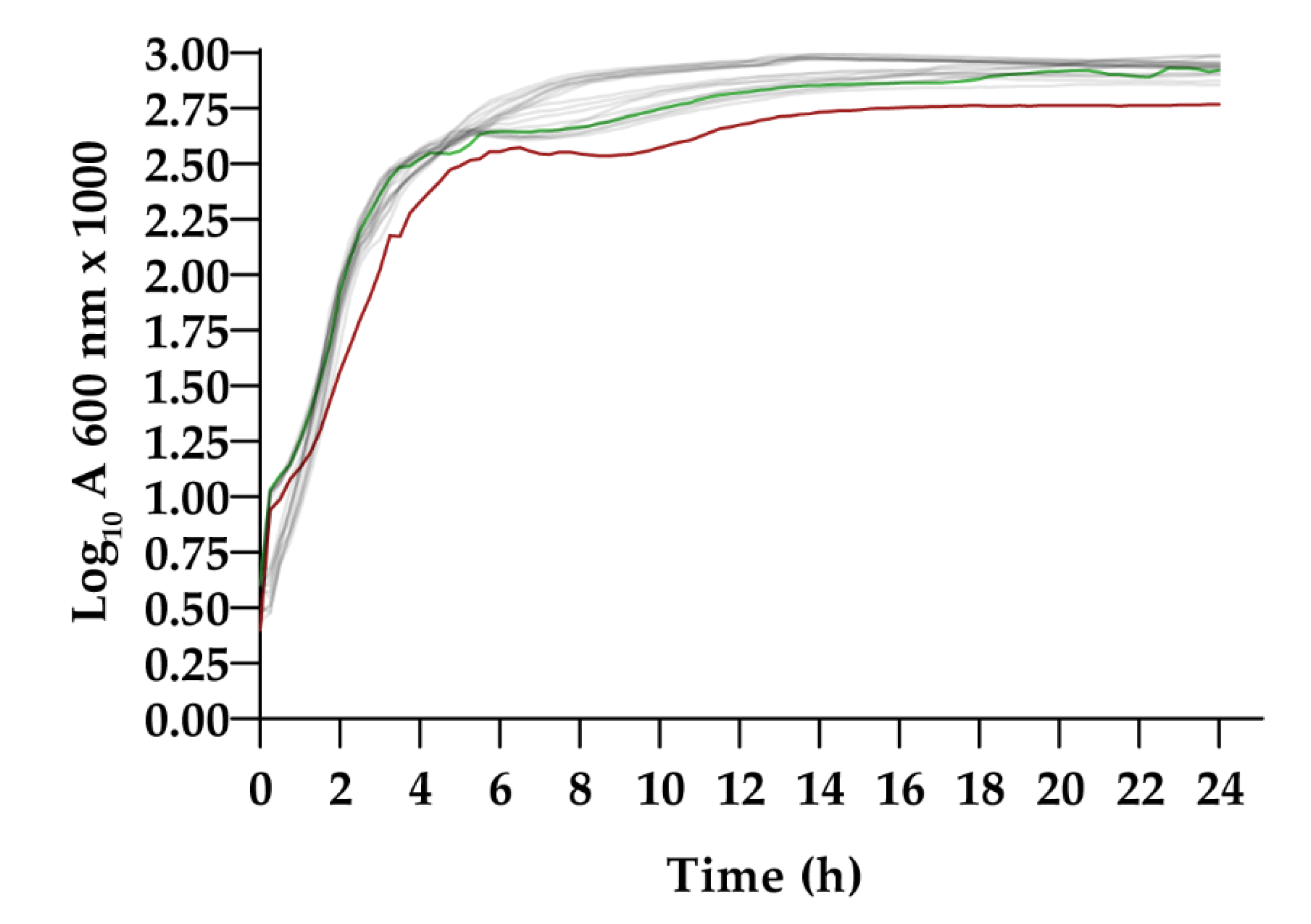
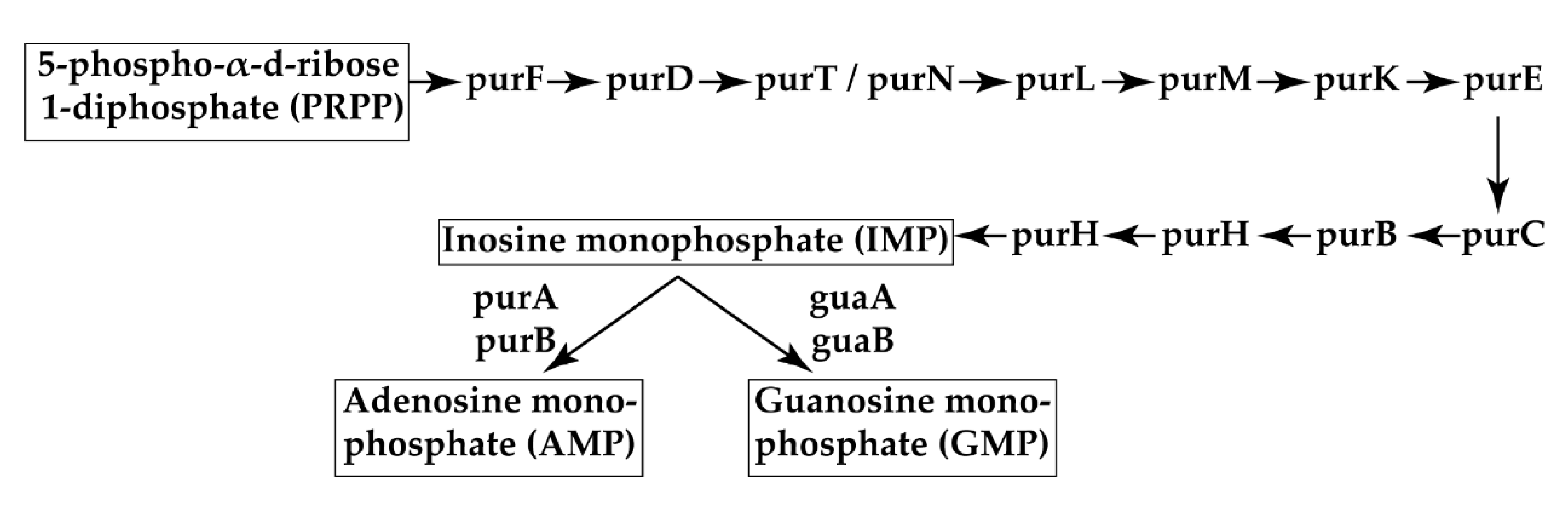



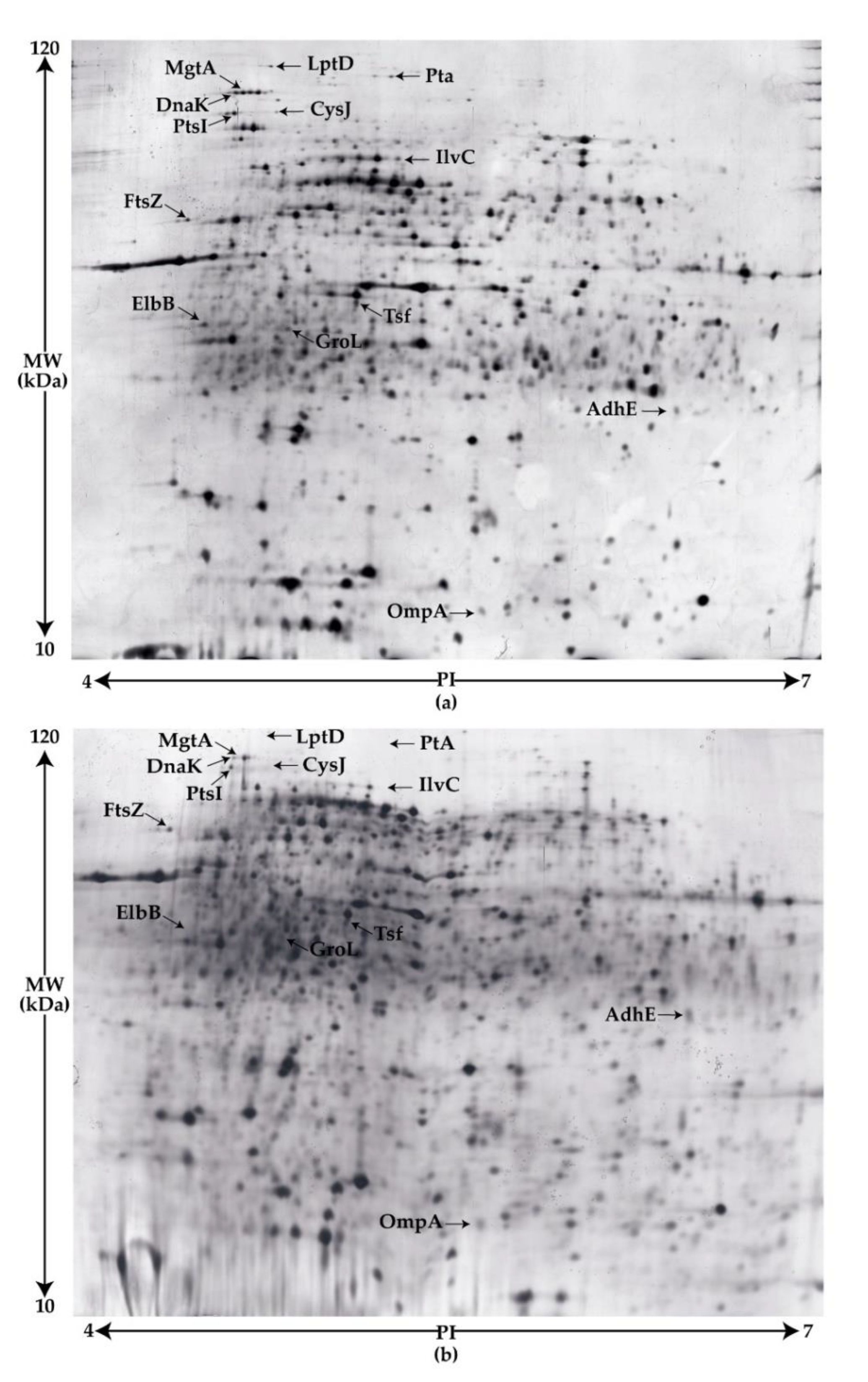

| Sample | Swimming | Curli Production | Hemagglutination (Log2) |
|---|---|---|---|
| Tn5 | + | + | 10 |
| Tn29 | + | + | 10 |
| Tn42 | + | + | 10 |
| Tn82 | + | + | 10 |
| Tn90 | + | + | 10 |
| Tn119 | + | + | 10 |
| Tn143 | + | + | 10 |
| Tn 249 | + | + | 10 |
| Tn 251 | + | + | 10 |
| Tn 262 | + | + | 10 |
| Tn263 | + | - | 10 |
| Tn 337 | + | + | 10 |
| Tn 373 | + | + | 10 |
| Tn 406 | + | + | 10 |
| Tn 420 | + | + | 10 |
| Tn 425 | + | + | 10 |
| Tn 457 | + | + | 10 |
| Tn 463 | + | - | 10 |
| Tn 467 | + | + | 10 |
| Tn 474 | + | + | 10 |
| Log10 CFU/mL ± SD | Live/Dead | ||||
|---|---|---|---|---|---|
| Biofilm | Planktonic | Integrated Density | Live (% ± SD) | Dead (% ± SD) | |
| wild type | 9.09 ± 0.24 | 8.99 ± 0.05 | 2.03 × 107 ± 4.86 × 106 | 85.40 ± 6.30 | 14.60 ± 6.30 |
| Tn263 | 6.40 ± 0.51 | 6.96 ± 0.01 | 9.69 × 106 ± 2.79 × 106 | 86.32 ± 4.01 | 13.68 ± 4.01 |
| ΔpurL::cat | 6.22 ± 0.44 | 7.02 ± 0.02 | 1.11 × 107 ± 2.34 × 106 | 80.38 ± 6.88 | 19.62 ± 6.88 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cepas, V.; Ballén, V.; Gabasa, Y.; Ramírez, M.; López, Y.; Soto, S.M. Transposon Insertion in the purL Gene Induces Biofilm Depletion in Escherichia coli ATCC 25922. Pathogens 2020, 9, 774. https://doi.org/10.3390/pathogens9090774
Cepas V, Ballén V, Gabasa Y, Ramírez M, López Y, Soto SM. Transposon Insertion in the purL Gene Induces Biofilm Depletion in Escherichia coli ATCC 25922. Pathogens. 2020; 9(9):774. https://doi.org/10.3390/pathogens9090774
Chicago/Turabian StyleCepas, Virginio, Victoria Ballén, Yaiza Gabasa, Miriam Ramírez, Yuly López, and Sara Mª Soto. 2020. "Transposon Insertion in the purL Gene Induces Biofilm Depletion in Escherichia coli ATCC 25922" Pathogens 9, no. 9: 774. https://doi.org/10.3390/pathogens9090774





