A Novel Genotype and First Record of Trypanosoma lainsoni in Argentina
Abstract
:1. Introduction
2. Results
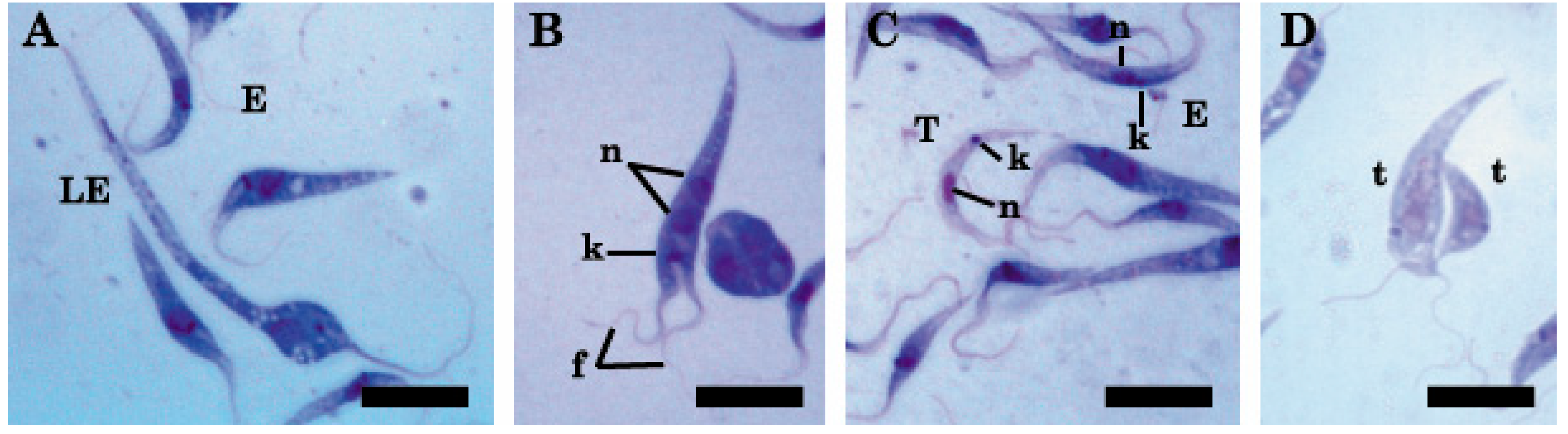
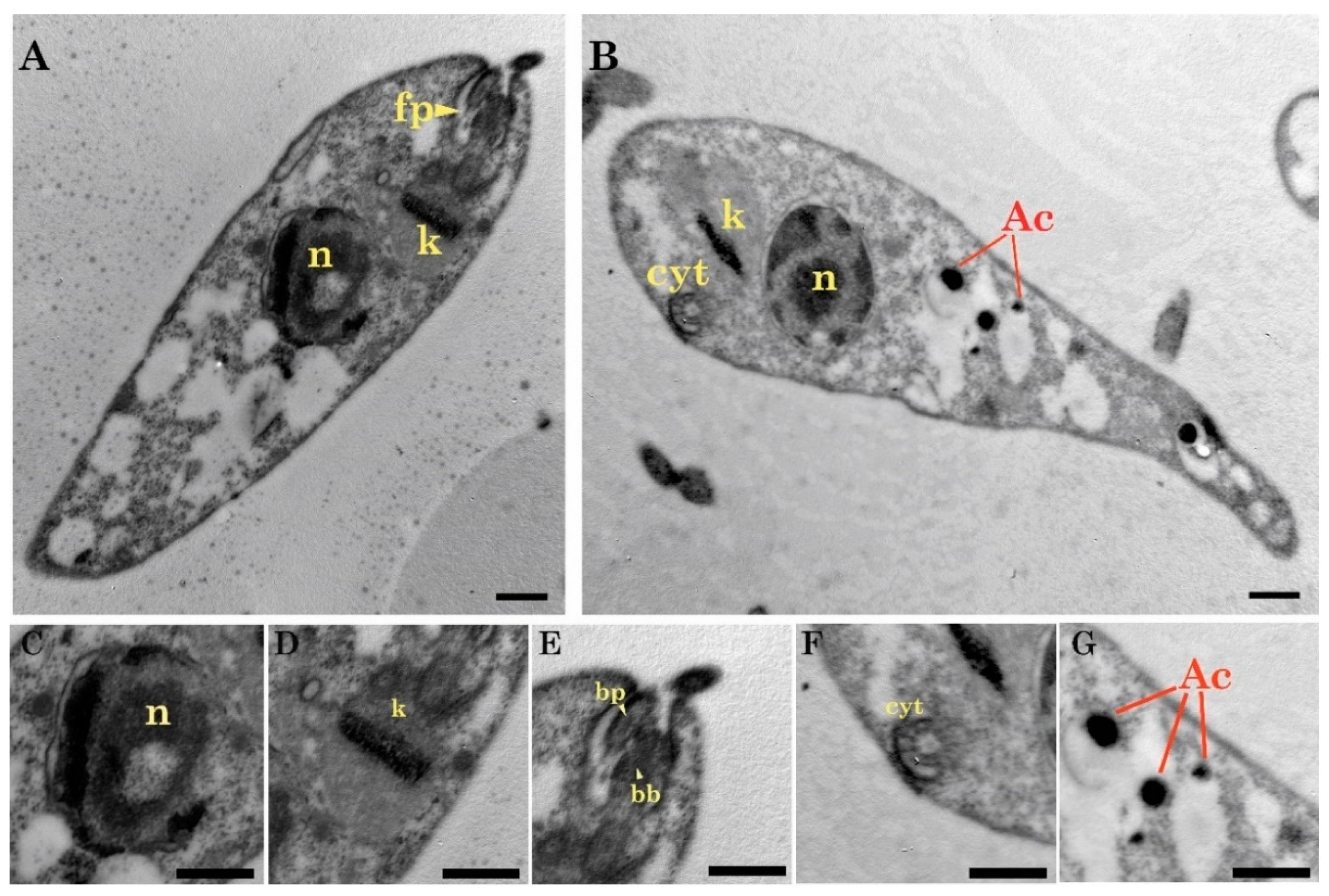
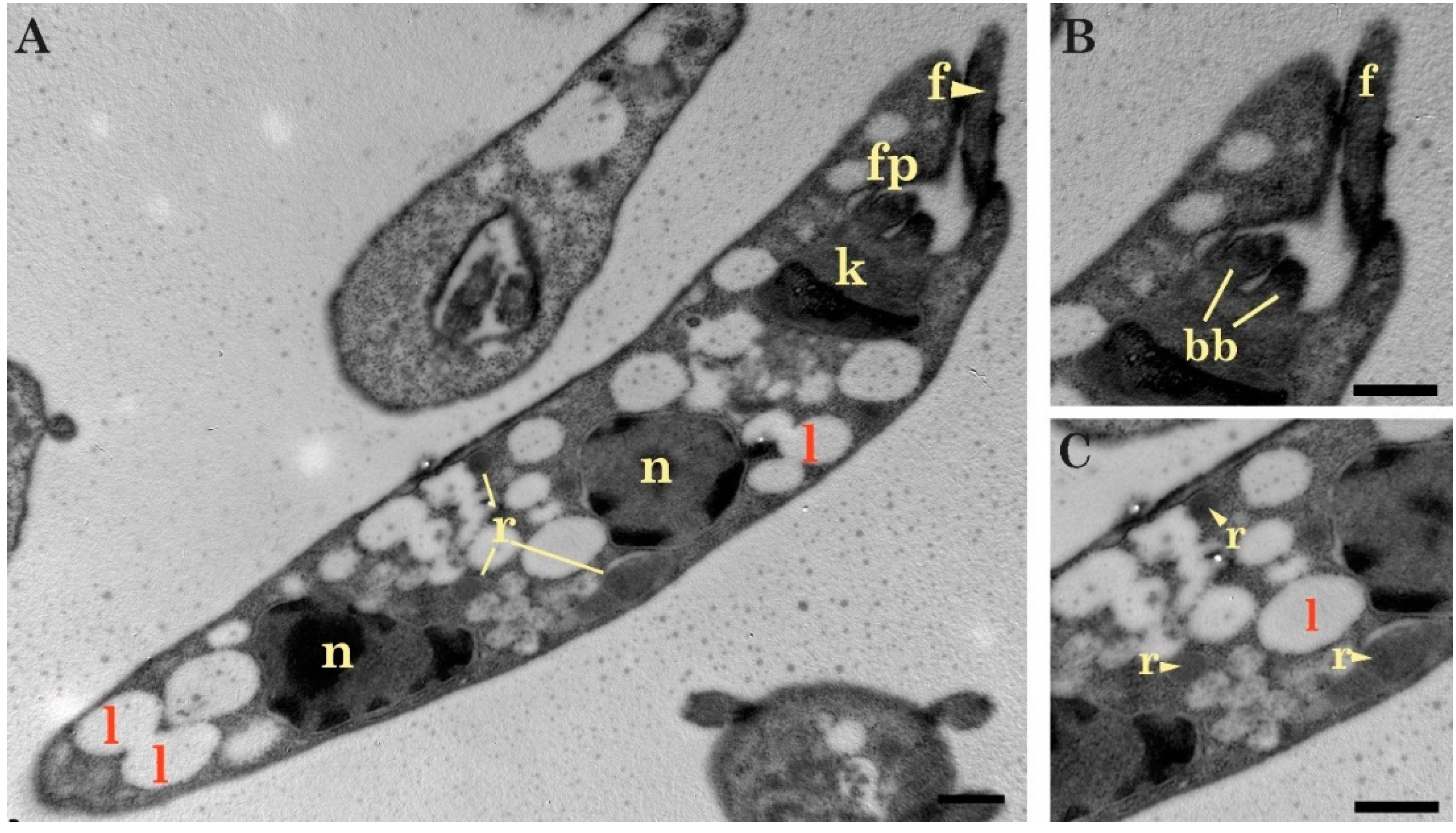
2.1. Light and Transmission Electron Microscopy Classified the Three Isolates to the Genus Trypanosoma (Trypanosomatidae)
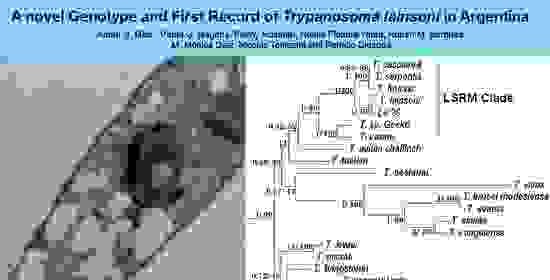
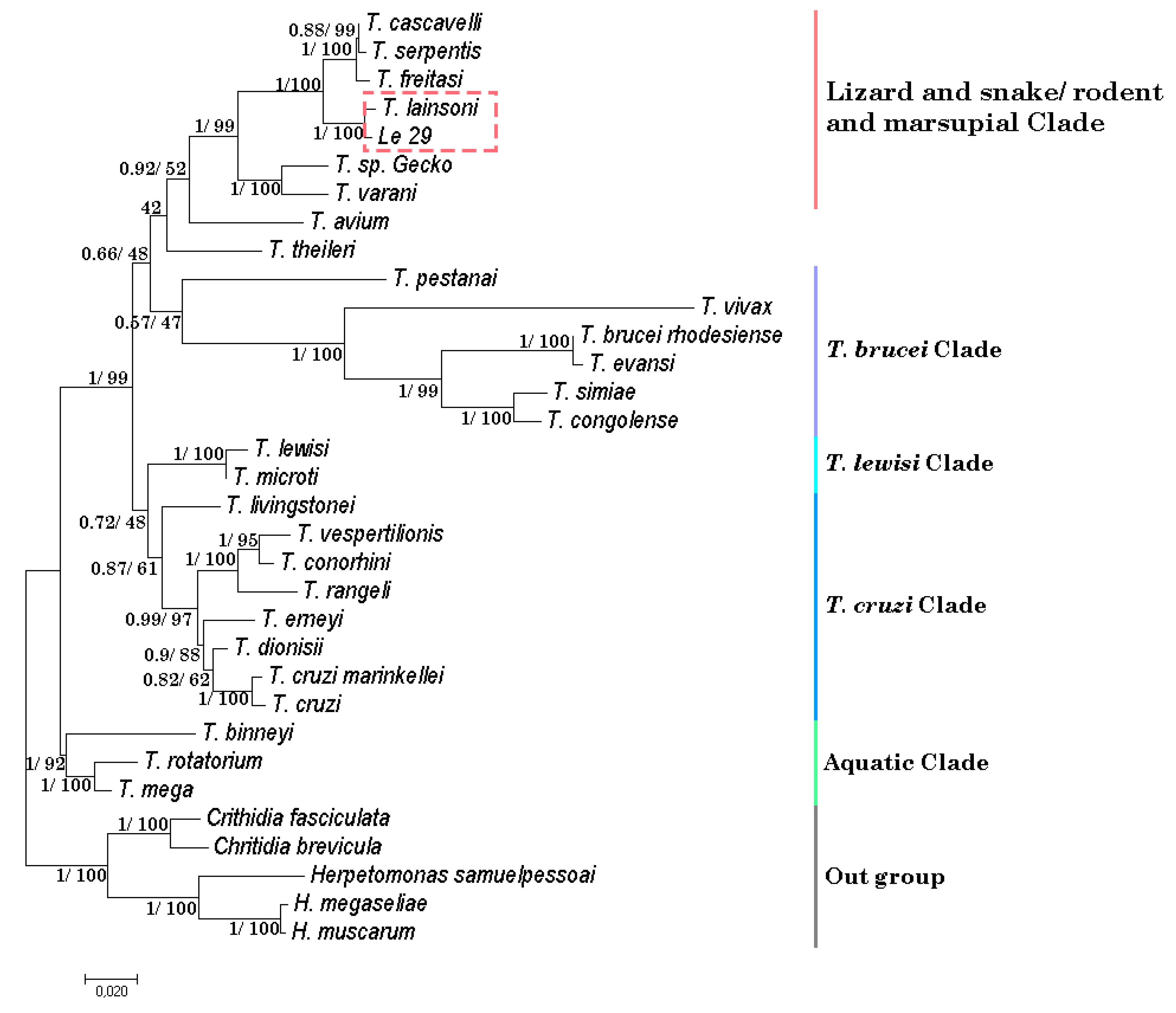
2.2. The Isolated Trypanosomes Belong to Trypanosoma lainsoni and Constitutes a Novel Genotype Different to the Brazilian Isolate
3. Discussion
4. Materials and Methods
4.1. Wild Animal Capture
4.2. Isolation and Culture of Trypanosomes
4.3. Morphological Characterization
Light and Electron Microscopy
4.4. Molecular Characterization
4.4.1. PCR and Sequencing of 18S rDNA and gGAPDH
4.4.2. Data Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vickman, K. The Diversity of the kinetoplastid flagellates. In Biology of the Kinetoplastida; Lumsden, W.H.R., Evans, D.A., Eds.; Academic Press: London, UK; New York, NY, USA; San Francisco, CA, USA, 1976; pp. 1–34. [Google Scholar]
- Svobodová, M.; Zídková, L.; Cepicka, I.; Obornik, M.; Lukes, J.; Votrpka, J. Sergeia podlipaevi gen. Nov., sp. voc. (Tryoanosomatida, Kinetoplastida), parasite of biting midges (Ceratopogonidae, Diptera). Int. J. Syst. Evol. Microbiol. 2007, 57, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Jaskowska, E.; Mayordomo, C.; Preston, G.; Kelly, S. Phytomonas: Tripanosomátidos adaptados a los ambientes vegetales. PLoS Pathog. 2015, 11, e1004484. [Google Scholar]
- Seward, E.A.; Votypka, J.; Kment, P.; Lukes, J.; Kelly, S. Description of Phytomonas oxycareni n. sp. from the Salivary Glands of Oxycarenus lavaterae. Protist 2017, 168, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, P.B.; Gibson, W.C.; Stevens, J.R. Patterns of co-evolution between trypanosomes and their hosts deduced from ribosomal RNA and protein-coding gene phylogenies. Mol. Phylogenetics Evol. 2007, 44, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.; Noyes, H.; Gibson, W. The evolution of Trypanosomes infecting humans and primates. Mem. Do Inst. Oswaldo Cruz 1998, 93, 669–676. [Google Scholar] [CrossRef] [Green Version]
- Viola, L.B.; Attias, M.; Takata, C.S.A.; Campaner, M.; De Souza, W.; Camargo, E.P.; Texeira, M.M.G. Phylogenetic analyses based on small subunit rRNA and glycosomal glyceraldehyde-3-phosphate dehydrogenase genes and ultrastructural characterization of two snake Trypanosomes: Trypanosoma serpentis n. sp. from Pseudoboa nigra and Trypanosoma cascavelli from Crotalus durissus terrificus. J. Eukaryot. Microbiol. 2009, 56, 594–602. [Google Scholar]
- Maslov, D.; Podlipaev, S.; Lukes, J. Phylogeny of the Kinetoplastida: Taxonomic Problems and Insights into the evolution of Parasitism. Mem. Do Inst. Oswaldo Cruz. 2001, 96, 397–402. [Google Scholar] [CrossRef] [Green Version]
- Kaufer, A.; Ellis, J.; Stark, D.; Barratt, K. The evolution of trypanosomatid taxonomy. Parasites Vectors 2017, 10, 287. [Google Scholar] [CrossRef]
- Ortiz, P.A.; Herakles, A.G.; Lima, L.; da Silva, F.M.; Campaner, M.; Pereira, C.L.; Juttapalapong, S.; Neves, L.; Desquesnes, M.; Camargo, E.P.; et al. Diagnosis and genetic analysis of the worldwide distributed Rattus-borne Trypanosome (Herpetosoma) lewisi and its allied species in blood and fleas of rodents. Infect. Genet. Evol. 2018, 63, 380–390. [Google Scholar] [CrossRef]
- Dario, M.A.; Moratelli, R.; Schwalbl, P.; Jansen, A.M.; Llewellyn, M.S. Small subunit ribosomal metabarcoding reveals extraordinary Trypanosomatid diversity in Brazilian bats. PLoS Negl. Trop. Dis. 2017, 11, e0005790. [Google Scholar] [CrossRef] [Green Version]
- Naiff, R.D.; Barrett, T.V.; Freitas, R.A. Isolation of Trypanosoma freitasi (kinetoplastida: Trypanosomatidae) from Psychodopygus claustrei (Diptera: Psychodidae). Mem. Do Inst. Oswaldo Cruz 1989, 84, 273–275. [Google Scholar] [CrossRef] [Green Version]
- Naiff, R.D.; Barrett, T.V. Trypanosoma (Megatrypanum) lainsoni n. sp. from Mesomys hispidus (Rodentia: Echimyidae) in Brazil: Trypomastigotes described from experimentally infected laboratory mice. Parasite 2013, 20, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fermino, B.R.; Paiva, F.; Viola, L.B.; Rodrigues, C.M.; Garcia, H.A.; Campaner, M.; Takata, C.S.; Sheferaw, D.; Kisakye, J.J.; Kato, A.; et al. Shared species of crocodilian trypanosomes carried by tabanid flies in Africa and South America, including the description of a new species from caimans, Trypanosoma kaiowa n. sp. Parasites Vectors 2019, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Lemos, M.; Morais, D.H.; Carvalho, V.T.; D’agosto, M. First record of Trypanosoma chattoni in Brazil and occurrence of other Trypanosoma species in Brazilian frogs (Anura, Leptodactylidae). J. Parasitol. 2008, 94, 148–151. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, A.T.; Monje, L.D.; Zurvera, D.A.; Beldomenico, P.M. Detection of Trypanosoma evansi infection in wild capybaras from Argentina using smear microscopy and real-time PCR assays. Vet. Parasitol. 2014, 202, 226–233. [Google Scholar] [CrossRef]
- Monzón, C.M.; Mancebo, O.A.; Roux, J.P. Comparison between six parasitological methods for diagnosis of Trypanosoma evansi in the subtropical area of Argentina. Vet. Parasitol. 1990, 36, 141–146. [Google Scholar] [CrossRef]
- Monzón, C.M.; Mancebo, O.A.; Gimenez, J.M.; Russo, A.M. Evolución de la Trypanosomiasis bovina por Trypanosoma vivax en Formosa (Argentina). Años 2007–2012 y su potencial dispersión en el país. Ibero-Latinoam. Parasitol. 2013, 72, 38–44. [Google Scholar]
- Schijman, S.G. Molecular diagnosis of Trypanosoma cruzi. Acta Trop. 2018, 184, 59–66. [Google Scholar] [CrossRef]
- Sant’Anna, C.; Pereira, M.G.; Lemgruber, L.; de Souza, W.; Cunha e Silva, N.L. New insights into the morphology of Trypanosoma cruzi reservosome. Microsc. Res. Tech. 2008, 71, 599–605. [Google Scholar] [CrossRef]
- Ayala, S.C.; Mckay, J.G. Trypanosoma gerrhonoti n. sp., and extrinsic development of lizard trypanosomes in California sandflies. J. Protozool. 1971, 18, 430–433. [Google Scholar] [CrossRef]
- Heoare, C.A. Morphological and taxonomic studies on mammalian trypanosomes X. Revision of the systematics. J. Protozool. 1964, 11, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Ayala, S.C. Two new Trypanosomes from California toads and Lizards. J. Protozool. 1970, 17, 370–373. [Google Scholar] [CrossRef]
- Fisher, A.C.; Schuster, G.; Cobb, W.J.; James, A.M.; Cooper, S.M.; de León, A.A.; Holman, P.J. Molecular characterization of Trypanosoma (Megatrypanum) spp. infecting cattle (Bos taurus), white-tailed deer (Odocoileus virginianus), and elk (Cervus elaphus canadensis) in the United States. Vet. Parasitol. 2013, 197, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.R.; Leite, R.C.; Doyle, R.L. Tripanosomatides like Trypanosoma theileri in the cattle tick Boophilus microplus. Braz. J. Vet. Parasitol. 2008, 17, 113–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morzaria, S.P.; Latif, A.A.; Jongejan, F.; Walker, A.R. Transmission of a Trypanosoma sp. to cattle by the tick Hyalomma anatolicum anatolicum. Vet. Parasitol. 1986, 19, 13–21. [Google Scholar] [CrossRef]
- Viola, L.B.; Campaner, M.; Takata, C.S.; Ferreira, R.C.; Rodrigues, A.C.; Freitas, R.A.; Duarte, M.R.; Grego, K.F.; Barrett, T.V.; Camargo, E.P.; et al. Phylogeny of snake trypanosomes inferred by SSU rDNA sequences, their possible transmission by phlebotomines, and taxonomic appraisal by molecular, cross-infection and morphological analysis. Parasitology 2008, 135, 595–605. [Google Scholar] [CrossRef]
- Coura, J.R. The main sceneries of Chagas disease transmission. The vectors, blood and oral transmissions—A comprehensive review. Mem. Do Inst. Oswaldo Cruz 2015, 110, 277–282. [Google Scholar] [CrossRef] [Green Version]
- Bisceglia, S.B.C.; Pereira, J.A.; Teta, P.; Quintana, R.D. Food habits of Geoffroy’s cat (Leopardus geoffroyi) in the central Monte desert of Argentina. J. Arid Environ. 2008, 72, 1120–1126. [Google Scholar] [CrossRef]
- Kollien, A.H.; Schaub, G.A. The development of Trypanosoma cruzi in triatominae. Parasitol. Today 2000, 16, 381–387. [Google Scholar] [CrossRef]
- Hayes, P.M.; Lawton, S.P.; Smit, N.J.; Gibson, W.C.; Davies, A.J. Morphological and molecular characterization of a marine fish trypanosome from South Africa, including its development in a leech vector. Parasites Vectors 2014, 7, 50. [Google Scholar] [CrossRef] [Green Version]
- Lemos, M.; Fermino, B.; Simas-Rodrigues, C.; Hoffmann, L.; Silva, R.; Camargo, E.; Texeira, M.; Sauto-Padro, T. Phylogenetic and morphological characterization of trypanosomes from Brazilian armoured carfishes and leeches reveal high species diversity, mixed infections and a new fish trypanosome specie. Parasites Vectors 2015, 8, 573–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. Mega 7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Soc. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomasini, N.; Lauthier, J.J.; Llewellyn, M.S.; Diosque, P. MLSTest: Novel software for multi-locus sequence data analysis in eukaryotic organisms. Infect. Genet. Evol. 2013, 20, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 2001, 294, 2310–2314. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar]
- Hasegawa, M.; Kishino, H.; Yano, T.A. Dating of the human ape splitting by a molecular clock of mitochondrial-DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]

| Form | Body Average Length | Free Average Flagellum | Total Average Length | Average Width |
|---|---|---|---|---|
| Trypomastigote | 18.15 ± 6.26 | 8.67 ± 3.02 | 28.13 ± 6.74 | 1.5 ± 0.47 |
| Epimastigote | 21.88 ± 3.28 | 11.43 ± 3.81 | 33.61 ± 6.08 | 2.3 ± 0.8 |
| Long Epimastigote | 44.25 ± 7.22 | 17.13 ± 6.32 | 61.85 ± 13.51 | 3.84 ± 1.55 |
| Transitional form | 13.98 ± 6.76 | 12.85 ± 3.52 | 27.26 ± 9.69 | 4.19 ± 1.34 |
| Spheromastigote | 6.12 ± 1.7 | 16.27 ± 5.19 | 23.01 ± 5.64 | 4.29 ± 1.8 |
| Partial Sequence | |
| Le29 | MT363779 |
| Ca37 | MT363780 |
| Ca47 | MT363781 |
| Complete Sequence | |
| Le29 | MT373343/MT373412 |
| Ca37 | MT363778/MT373413 |
| Ca47 | MT363777/MT373414 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz, A.G.; Ragone, P.G.; Rusman, F.; Floridia-Yapur, N.; Barquez, R.M.; Díaz, M.M.; Tomasini, N.; Diosque, P. A Novel Genotype and First Record of Trypanosoma lainsoni in Argentina. Pathogens 2020, 9, 731. https://doi.org/10.3390/pathogens9090731
Díaz AG, Ragone PG, Rusman F, Floridia-Yapur N, Barquez RM, Díaz MM, Tomasini N, Diosque P. A Novel Genotype and First Record of Trypanosoma lainsoni in Argentina. Pathogens. 2020; 9(9):731. https://doi.org/10.3390/pathogens9090731
Chicago/Turabian StyleDíaz, Anahí G., Paula G. Ragone, Fanny Rusman, Noelia Floridia-Yapur, Rubén M. Barquez, M. Mónica Díaz, Nicolás Tomasini, and Patricio Diosque. 2020. "A Novel Genotype and First Record of Trypanosoma lainsoni in Argentina" Pathogens 9, no. 9: 731. https://doi.org/10.3390/pathogens9090731
APA StyleDíaz, A. G., Ragone, P. G., Rusman, F., Floridia-Yapur, N., Barquez, R. M., Díaz, M. M., Tomasini, N., & Diosque, P. (2020). A Novel Genotype and First Record of Trypanosoma lainsoni in Argentina. Pathogens, 9(9), 731. https://doi.org/10.3390/pathogens9090731