Monitoring Glycolysis and Respiration Highlights Metabolic Inflexibility of Cryptococcus neoformans
Abstract
:1. Introduction
2. Results
2.1. Assay Optimization for C. neoformans
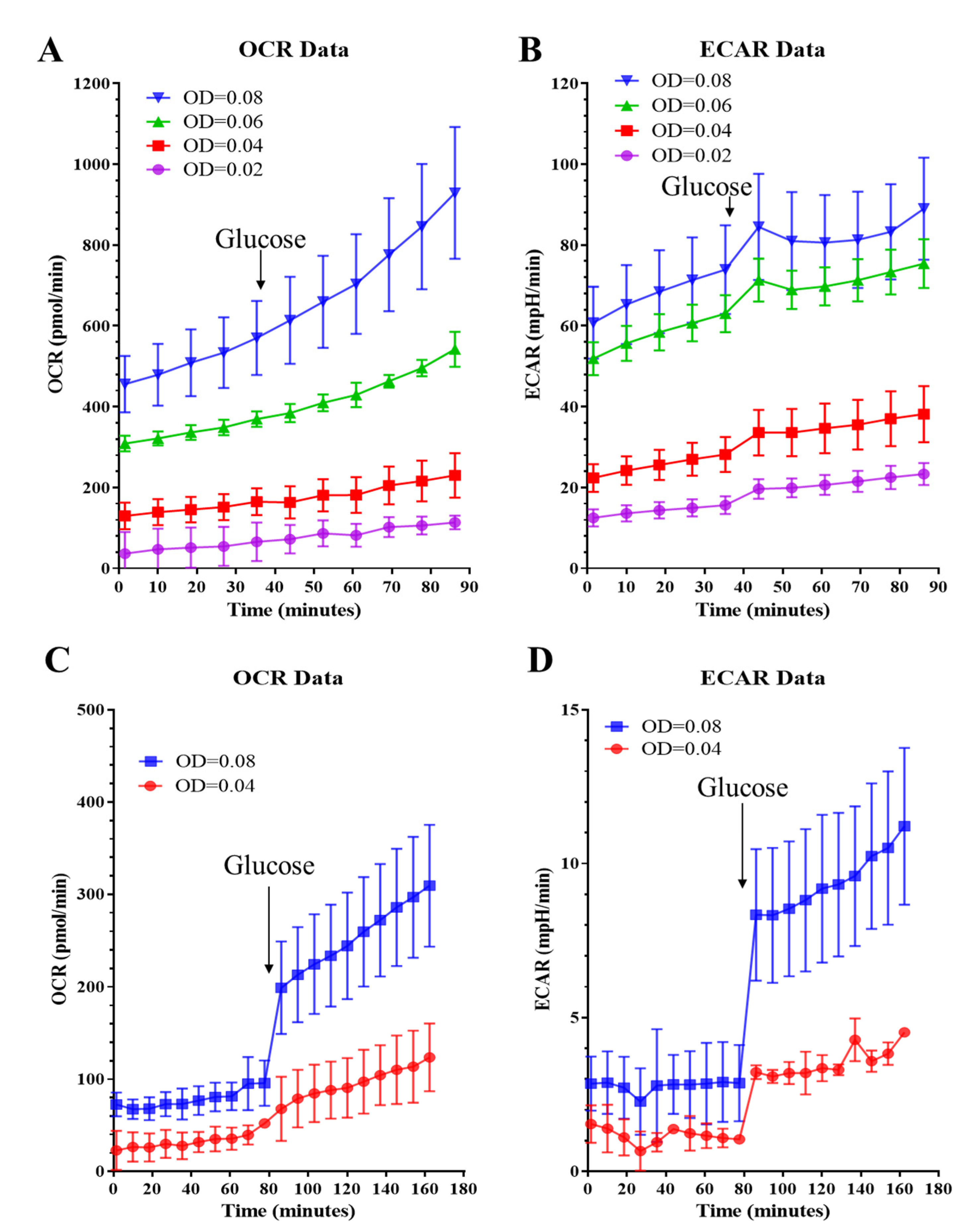
2.1.1. Assay Medium and Cell Seeding Density
2.1.2. Mitochondrial Stress Test
2.1.3. Glycolytic Rate Test
2.2. Measuring Metabolic Activity of C. neoformans and Human Monocytes in Co-Culture
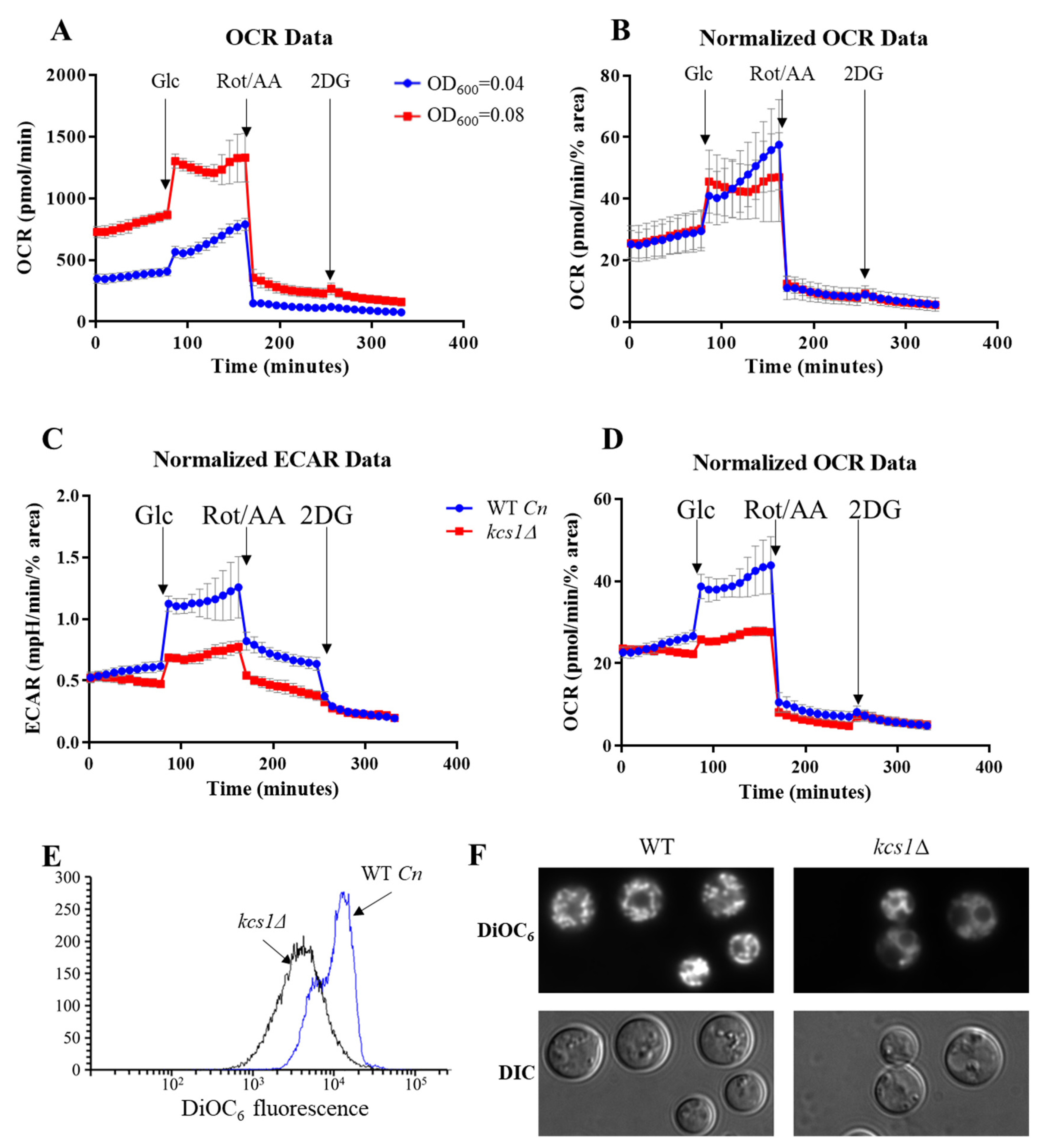
2.3. Comparing the Metabolic Profiles of C. neoformans WT and the Mutant Strain, Kcs1Δ
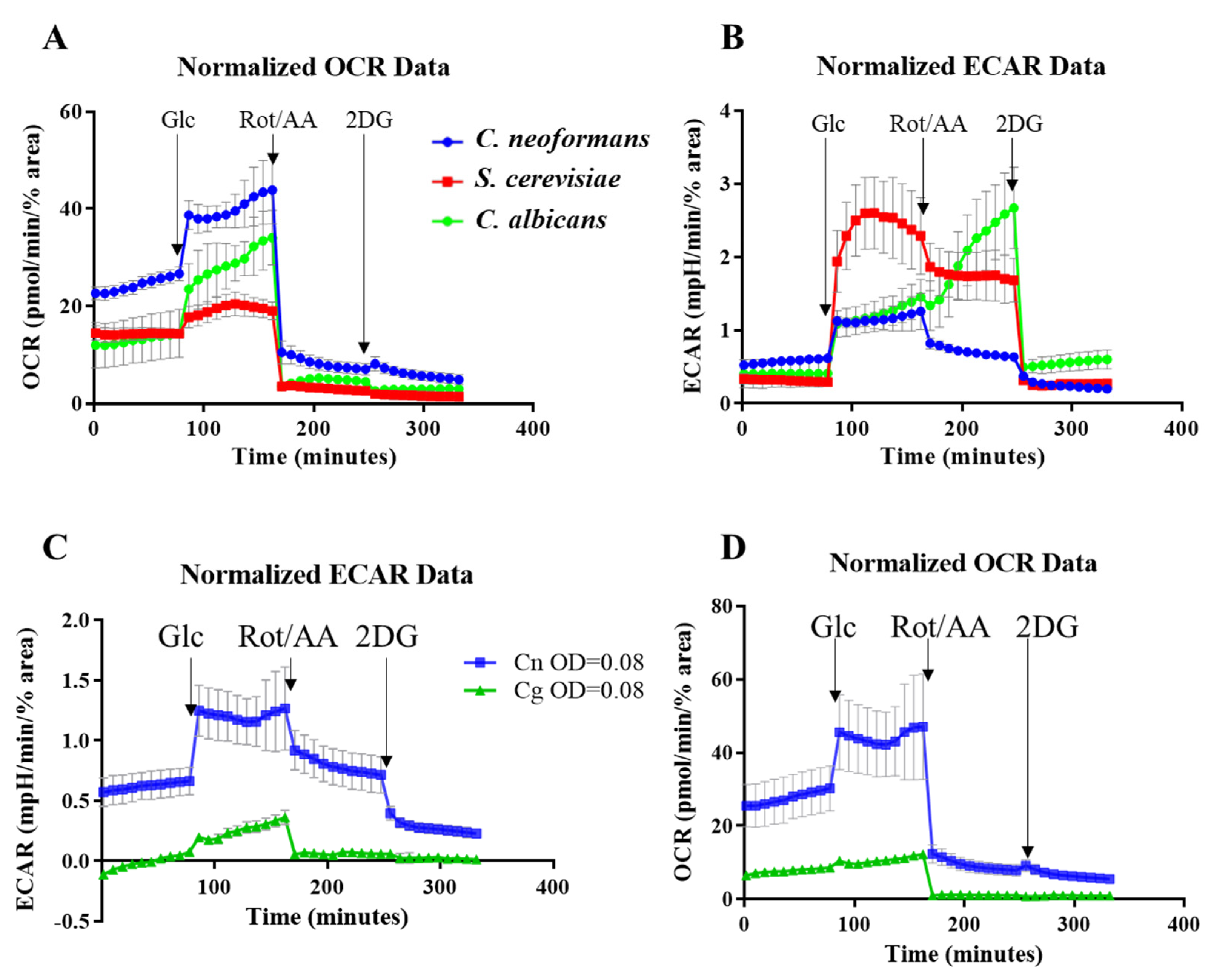
2.4. Comparing the Metabolic Profiles of Three Yeast Species
3. Discussion
4. Materials and Methods
4.1. Fungal Strains, Media and Reagents
4.2. Seahorse XFe24 Analyzer Setup and Optimization for Measuring Metabolism in C. neoformans
4.2.1. Day before the Run
Sensor Cartridge Preparation
Fungal Cell Preparation
4.2.2. Day of the Run
CD14 Monocyte Preparation
Fungal Cell Preparation
Sensor Cartridge Preparation
Assay Setup on Seahorse Analyzer
4.3. Co-Culture Assay
- For host-pathogen co-culture samples, 100 µL of 250,000 CD14+ monocytes in Assay medium (with 10 mM glucose) were combined with 500,000 C. neoformans cells. The cells were mixed and plated on XF tissue culture plate pre-coated with Cell-Tak (Corning) following manufacturer’s instruction.
- For cryptococcal control samples, 100 µL of Assay medium were added to cryptococcal cells. The cells were resuspended and plated on XF tissue culture plate.
- For monocyte control samples, 100 µL of 250,000 CD14+ monocytes in Assay medium were plated on XF tissue culture plate.
- The plate was incubated at 37 °C, 5% CO2 for 30 min.
- To improve the attachment and provide even dispersal of the cells, the plate was centrifuged at 100× g for 1 min, deceleration 3. Then the plate was turned 180° and similarly centrifuged.
- 400 µL Assay medium were slowly added to each well.
- The plate was incubated at 37 °C without CO2 enrichment until ready to insert to the Analyzer (~1.5 h from the start of co-incubation).
- Rot/AA 0.5 µM and 2DG 50 mM were injected into the plate wells during the course of the Assay.
4.4. Data Normalization for Fungal Cells
- Process → Sharpen (improves separation between cells)
- Process → Binary → Convert to Mask (creates binary black and white image)
- Select representative area of the micrograph using buttons in the main ImageJ menu, or use the entire image by default
- Analyze → Analyze Particles (select Size as 3-infinity [in pixels]; tick “Summarize” option). If “%Area” is not displayed in the “Summary” window, tick “Area” option in the Analyze → Set Measurements window
4.5. DiOC6 Staining
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rajasingham, R.; Smith, R.M.; Park, B.J.; Jarvis, J.N.; Govender, N.P.; Chiller, T.M.; Denning, D.W.; Loyse, A.; Boulware, D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017, 17, 873–881. [Google Scholar] [CrossRef] [Green Version]
- Coelho, C.; Bocca, A.L.; Casadevall, A. The tools for virulence of Cryptococcus neoformans. Adv. Appl. Microbiol. 2014, 87, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Charlier, C.; Nielsen, K.; Daou, S.; Brigitte, M.; Chretien, F.; Dromer, F. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect. Immun. 2009, 77, 120–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufman-Francis, K.; Djordjevic, J.T.; Juillard, P.G.; Lev, S.; Desmarini, D.; Grau, G.E.R.; Sorrell, T.C. The early innate immune response to, and phagocyte-dependent entry of, Cryptococcus neoformans map to the perivascular space of cortical post-capillary venules in neurocryptococcosis. Am. J. Pathol. 2018, 188, 1653–1665. [Google Scholar] [CrossRef]
- Sorrell, T.C.; Juillard, P.G.; Djordjevic, J.T.; Kaufman-Francis, K.; Dietmann, A.; Milonig, A.; Combes, V.; Grau, G.E. Cryptococcal transmigration across a model brain blood-barrier: Evidence of the Trojan horse mechanism and differences between Cryptococcus neoformans var. grubii strain H99 and Cryptococcus gattii strain R265. Microbes Infect. Inst. Pasteur 2015. [Google Scholar] [CrossRef]
- Denham, S.T.; Brown, J.C.S. Mechanisms of pulmonary escape and dissemination by Cryptococcus neoformans. J. Fungi 2018, 4, 25. [Google Scholar] [CrossRef] [Green Version]
- Odds, F.C.; De Backer, T.; Dams, G.; Vranckx, L.; Woestenborghs, F. Oxygen as limiting nutrient for growth of Cryptococcus neoformans. J. Clin. Microbiol. 1995, 33, 995–997. [Google Scholar] [CrossRef] [Green Version]
- Ingavale, S.S.; Chang, Y.C.; Lee, H.; McClelland, C.M.; Leong, M.L.; Kwon-Chung, K.J. Importance of mitochondria in survival of Cryptococcus neoformans under low oxygen conditions and tolerance to cobalt chloride. PLoS Pathog. 2008, 4, e1000155. [Google Scholar] [CrossRef] [Green Version]
- Chun, C.D.; Liu, O.W.; Madhani, H.D. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2007, 3, e22. [Google Scholar] [CrossRef] [Green Version]
- Grahl, N.; Shepardson, K.M.; Chung, D.; Cramer, R.A. Hypoxia and fungal pathogenesis: To air or not to air? Eukaryot. Cell 2012, 11, 560–570. [Google Scholar] [CrossRef] [Green Version]
- Eltzschig, H.K.; Carmeliet, P. Hypoxia and inflammation. N. Engl. J. Med. 2011, 364, 656–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nizet, V.; Johnson, R.S. Interdependence of hypoxic and innate immune responses. Nat. Rev. Immunol. 2009, 9, 609–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, G.; Cheng, P.Y.; Sham, A.; Perfect, J.R.; Kronstad, J.W. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol. Microbiol. 2008, 69, 1456–1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panepinto, J.; Liu, L.; Ramos, J.; Zhu, X.; Valyi-Nagy, T.; Eksi, S.; Fu, J.; Jaffe, H.A.; Wickes, B.; Williamson, P.R. The DEAD-box RNA helicase Vad1 regulates multiple virulence-associated genes in Cryptococcus neoformans. J. Clin. Investig. 2005, 115, 632–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.S.; Betancourt-Quiroz, M.; Price, J.L.; Toffaletti, D.L.; Vora, H.; Hu, G.; Kronstad, J.W.; Perfect, J.R. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio 2011, 2, e00103-11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lev, S.; Li, C.; Desmarini, D.; Saiardi, A.; Fewings, N.L.; Schibeci, S.D.; Sharma, R.; Sorrell, T.C.; Djordjevic, J.T. Fungal inositol pyrophosphate IP7 is crucial for metabolic adaptation to the host environment and pathogenicity. mBio 2015, 6, e00531-15. [Google Scholar] [CrossRef] [Green Version]
- Mookerjee, S.A.; Goncalves, R.L.; Gerencser, A.A.; Nicholls, D.G.; Brand, M.D. The contributions of respiration and glycolysis to extracellular acid production. Biochim. Biophys. Acta 2015, 1847, 171–181. [Google Scholar] [CrossRef] [Green Version]
- Mookerjee, S.A.; Brand, M.D. Measurement and analysis of extracellular acid production to determine glycolytic rate. J. Vis. Exp. 2015, e53464. [Google Scholar] [CrossRef] [Green Version]
- Kidd, S.E.; Hagen, F.; Tscharke, R.L.; Huynh, M.; Bartlett, K.H.; Fyfe, M.; Macdougall, L.; Boekhout, T.; Kwon-Chung, K.J.; Meyer, W. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. USA 2004, 101, 17258–17263. [Google Scholar] [CrossRef] [Green Version]
- Hagen, F.; Khayhan, K.; Theelen, B.; Kolecka, A.; Polacheck, I.; Sionov, E.; Falk, R.; Parnmen, S.; Lumbsch, H.T.; Boekhout, T. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol. 2015, 78, 16–48. [Google Scholar] [CrossRef] [Green Version]
- Trevijano-Contador, N.; Rossi, S.A.; Alves, E.; Landin-Ferreiroa, S.; Zaragoza, O. Capsule enlargement in Cryptococcus neoformans is dependent on mitochondrial activity. Front. Microbiol. 2017, 8, 1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, J.; Meyer, J.D.; Lodge, J.K. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 2000, 7, 125–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhya, R.; Campbell, L.T.; Donlin, M.J.; Aurora, R.; Lodge, J.K. Global transcriptome profile of Cryptococcus neoformans during exposure to hydrogen peroxide induced oxidative stress. PLoS ONE 2013, 8, e55110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obando, D.; Koda, Y.; Pantarat, N.; Lev, S.; Zuo, X.; Bijosono Oei, J.; Widmer, F.; Djordjevic, J.T.; Sorrell, T.C.; Jolliffe, K.A. Synthesis and evaluation of a series of bis(pentylpyridinium) compounds as antifungal agents. ChemMedChem 2018, 13, 1421–1436. [Google Scholar] [CrossRef]
- Chen, W.; Gueron, M. The inhibition of bovine heart hexokinase by 2-deoxy-D-glucose-6-phosphate: Characterization by 31P NMR and metabolic implications. Biochimie 1992, 74, 867–873. [Google Scholar] [CrossRef]
- Wick, A.N.; Drury, D.R.; Nakada, H.I.; Wolfe, J.B. Localization of the primary metabolic block produced by 2-deoxyglucose. J. Biol. Chem. 1957, 224, 963–969. [Google Scholar]
- Viola, A.; Munari, F.; Sanchez-Rodriguez, R.; Scolaro, T.; Castegna, A. The metabolic signature of macrophage responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.K.S.; Al-Sharea, A.; Shihata, W.A.; Bertuzzo Veiga, C.; Cooney, O.D.; Fleetwood, A.J.; Flynn, M.C.; Claeson, E.; Palmer, C.S.; Lancaster, G.I.; et al. Glycolysis is required for LPS-induced activation and adhesion of human CD14(+)CD16(−) monocytes. Front. Immunol. 2019, 10, 2054. [Google Scholar] [CrossRef]
- Voelz, K.; Lammas, D.A.; May, R.C. Cytokine signaling regulates the outcome of intracellular macrophage parasitism by Cryptococcus neoformans. Infect. Immun. 2009, 77, 3450–3457. [Google Scholar] [CrossRef] [Green Version]
- Kelly, B.; O’Neill, L.A. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015, 25, 771–784. [Google Scholar] [CrossRef] [Green Version]
- Raulien, N.; Friedrich, K.; Strobel, S.; Rubner, S.; Baumann, S.; von Bergen, M.; Korner, A.; Krueger, M.; Rossol, M.; Wagner, U. Fatty acid oxidation compensates for lipopolysaccharide-induced warburg effect in glucose-deprived monocytes. Front. Immunol. 2017, 8, 609. [Google Scholar] [CrossRef] [PubMed]
- Tucey, T.M.; Verma, J.; Harrison, P.F.; Snelgrove, S.L.; Lo, T.L.; Scherer, A.K.; Barugahare, A.A.; Powell, D.R.; Wheeler, R.T.; Hickey, M.J.; et al. Glucose homeostasis is important for immune cell viability during candida challenge and host survival of systemic fungal infection. Cell Metab. 2018, 27, 988–1006.e7. [Google Scholar] [CrossRef] [PubMed]
- Fleetwood, A.J.; Lee, M.K.S.; Singleton, W.; Achuthan, A.; Lee, M.C.; O’Brien-Simpson, N.M.; Cook, A.D.; Murphy, A.J.; Dashper, S.G.; Reynolds, E.C.; et al. Metabolic remodeling, inflammasome activation, and pyroptosis in macrophages stimulated by Porphyromonas gingivalis and its outer membrane vesicles. Front. Cell. Infect. Microbiol. 2017, 7, 351. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Coelho, C.; Alanio, A. Mechanisms of Cryptococcus neoformans-mediated host damage. Front. Immunol. 2018, 9, 855. [Google Scholar] [CrossRef] [PubMed]
- Coelho, C.; Souza, A.C.; Derengowski Lda, S.; de Leon-Rodriguez, C.; Wang, B.; Leon-Rivera, R.; Bocca, A.L.; Goncalves, T.; Casadevall, A. Macrophage mitochondrial and stress response to ingestion of Cryptococcus neoformans. J. Immunol. 2015, 194, 2345–2357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, L.M.; Dixon, E.F.; May, R.C. The fungal pathogen Cryptococcus neoformans manipulates macrophage phagosome maturation. Cell. Microbiol. 2015, 17, 702–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, S.A.; May, R.C. Cryptococcus interactions with macrophages: Evasion and manipulation of the phagosome by a fungal pathogen. Cell. Microbiol. 2013, 15, 403–411. [Google Scholar] [CrossRef]
- Ma, H.; Croudace, J.E.; Lammas, D.A.; May, R.C. Expulsion of live pathogenic yeast by macrophages. Curr. Biol. 2006, 16, 2156–2160. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Lev, S.; Saiardi, A.; Desmarini, D.; Sorrell, T.C.; Djordjevic, J.T. Identification of a major IP5 kinase in Cryptococcus neoformans confirms that PP-IP5/IP7, not IP6, is essential for virulence. Sci. Rep. 2016, 6, 23927. [Google Scholar] [CrossRef]
- Perry, S.W.; Norman, J.P.; Barbieri, J.; Brown, E.B.; Gelbard, H.A. Mitochondrial membrane potential probes and the proton gradient: A practical usage guide. Biotechniques 2011, 50, 98–115. [Google Scholar] [CrossRef]
- Wright, L.; Bubb, W.; Davidson, J.; Santangelo, R.; Krockenberger, M.; Himmelreich, U.; Sorrell, T. Metabolites released by Cryptococcus neoformans var. neoformans and var. gattii differentially affect human neutrophil function. Microbes Infect. Inst. Pasteur 2002, 4, 1427–1438. [Google Scholar] [CrossRef]
- Bubb, W.A.; Wright, L.C.; Cagney, M.; Santangelo, R.T.; Sorrell, T.C.; Kuchel, P.W. Heteronuclear NMR studies of metabolites produced by Cryptococcus neoformans in culture media: Identification of possible virulence factors. Magn. Reson. Med. 1999, 42, 442–453. [Google Scholar] [CrossRef]
- Glenn, K.; Ingram-Smith, C.; Smith, K.S. Biochemical and kinetic characterization of xylulose 5-phosphate/fructose 6-phosphate phosphoketolase 2 (Xfp2) from Cryptococcus neoformans. Eukaryot. Cell 2014, 13, 657–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, P.A.; Ravi, S.; Chacko, B.; Johnson, M.S.; Darley-Usmar, V.M. A review of the mitochondrial and glycolytic metabolism in human platelets and leukocytes: Implications for their use as bioenergetic biomarkers. Redox Biol. 2014, 2, 206–210. [Google Scholar] [CrossRef] [Green Version]
- Dominguez-Andres, J.; Arts, R.J.W.; Ter Horst, R.; Gresnigt, M.S.; Smeekens, S.P.; Ratter, J.M.; Lachmandas, E.; Boutens, L.; van de Veerdonk, F.L.; Joosten, L.A.B.; et al. Rewiring monocyte glucose metabolism via C-type lectin signaling protects against disseminated candidiasis. PLoS Pathog. 2017, 13, e1006632. [Google Scholar] [CrossRef]
- Thapa, B.; Lee, K. Metabolic influence on macrophage polarization and pathogenesis. BMB Rep. 2019, 52, 360–372. [Google Scholar] [CrossRef]
- Dugo, L.; Belluomo, M.G.; Fanali, C.; Russo, M.; Cacciola, F.; Maccarrone, M.; Sardanelli, A.M. Effect of cocoa polyphenolic extract on macrophage polarization from proinflammatory M1 to Anti-inflammatory M2 state. Oxid. Med. Cell. Longev. 2017, 2017, 6293740. [Google Scholar] [CrossRef]
- Calahorra, M.; Sanchez, N.S.; Pena, A. Characterization of glycolytic metabolism and ion transport of Candida albicans. Yeast 2012, 29, 357–370. [Google Scholar] [CrossRef]
- Pfeiffer, T.; Morley, A. An evolutionary perspective on the Crabtree effect. Front. Mol. Biosci. 2014, 1, 17. [Google Scholar] [CrossRef]
- Kim, J.H.; Haff, R.P.; Faria, N.C.; Martins Mde, L.; Chan, K.L.; Campbell, B.C. Targeting the mitochondrial respiratory chain of Cryptococcus through antifungal chemosensitization: A model for control of non-fermentative pathogens. Molecules 2013, 18, 8873–8894. [Google Scholar] [CrossRef]
- Lev, S.; Kaufman-Francis, K.; Desmarini, D.; Juillard, P.G.; Li, C.; Stifter, S.A.; Feng, C.G.; Sorrell, T.C.; Grau, G.E.; Bahn, Y.S.; et al. Pho4 is essential for dissemination of Cryptococcus neoformans to the Host brain by promoting phosphate uptake and growth at alkaline pH. mSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lev, S.; Li, C.; Desmarini, D.; Liuwantara, D.; Sorrell, T.C.; Hawthorne, W.J.; Djordjevic, J.T. Monitoring Glycolysis and Respiration Highlights Metabolic Inflexibility of Cryptococcus neoformans. Pathogens 2020, 9, 684. https://doi.org/10.3390/pathogens9090684
Lev S, Li C, Desmarini D, Liuwantara D, Sorrell TC, Hawthorne WJ, Djordjevic JT. Monitoring Glycolysis and Respiration Highlights Metabolic Inflexibility of Cryptococcus neoformans. Pathogens. 2020; 9(9):684. https://doi.org/10.3390/pathogens9090684
Chicago/Turabian StyleLev, Sophie, Cecilia Li, Desmarini Desmarini, David Liuwantara, Tania C. Sorrell, Wayne J. Hawthorne, and Julianne T. Djordjevic. 2020. "Monitoring Glycolysis and Respiration Highlights Metabolic Inflexibility of Cryptococcus neoformans" Pathogens 9, no. 9: 684. https://doi.org/10.3390/pathogens9090684





