Role of the Nitric Oxide Reductase NorVW in the Survival and Virulence of Enterohaemorrhagic Escherichia coli during Infection
Abstract
:1. Introduction
2. Results
2.1. Selection of norV+ EHEC Strains
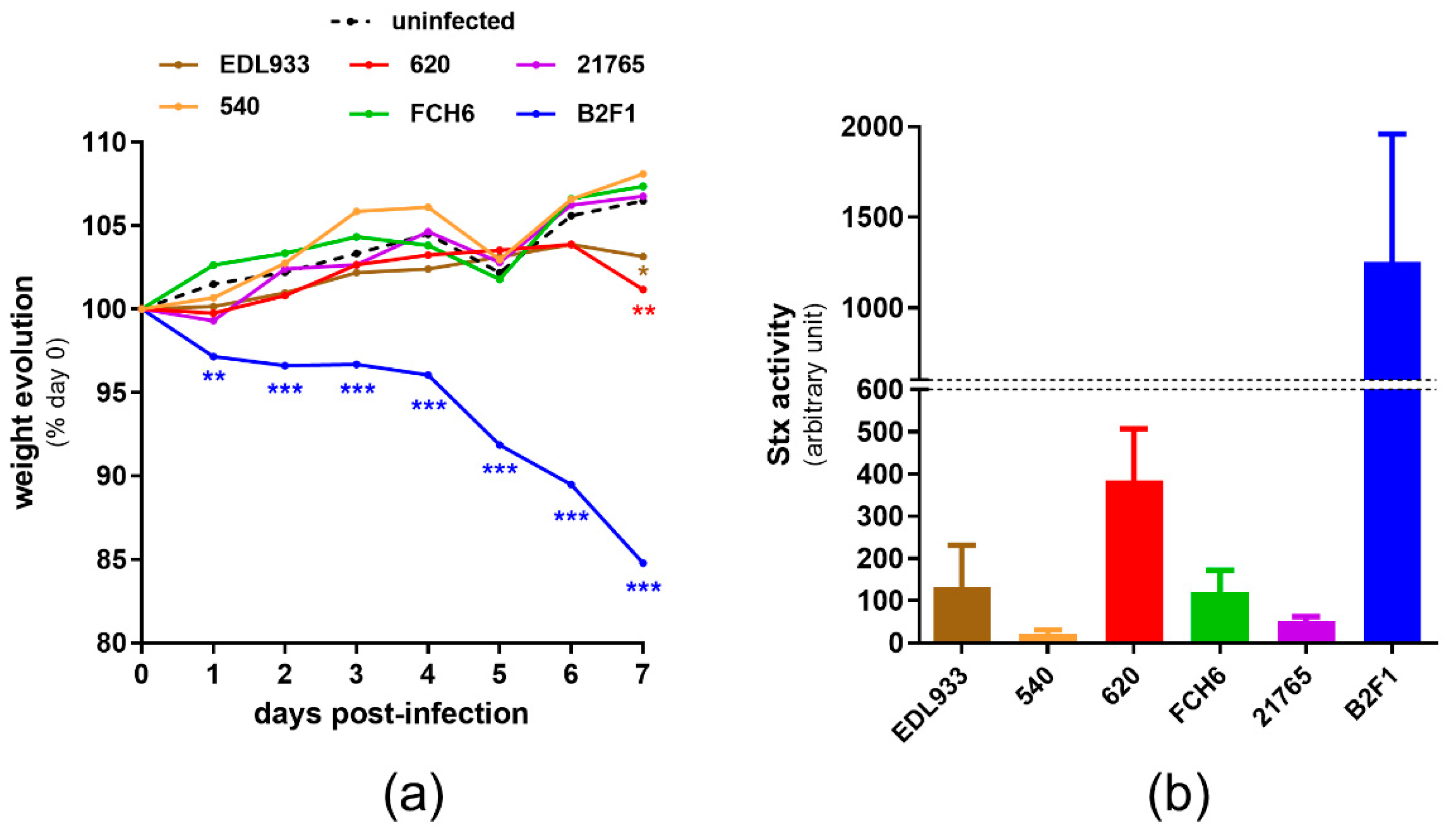
2.2. Virulence of norV+ EHEC Strains in a Mouse Model of Infection
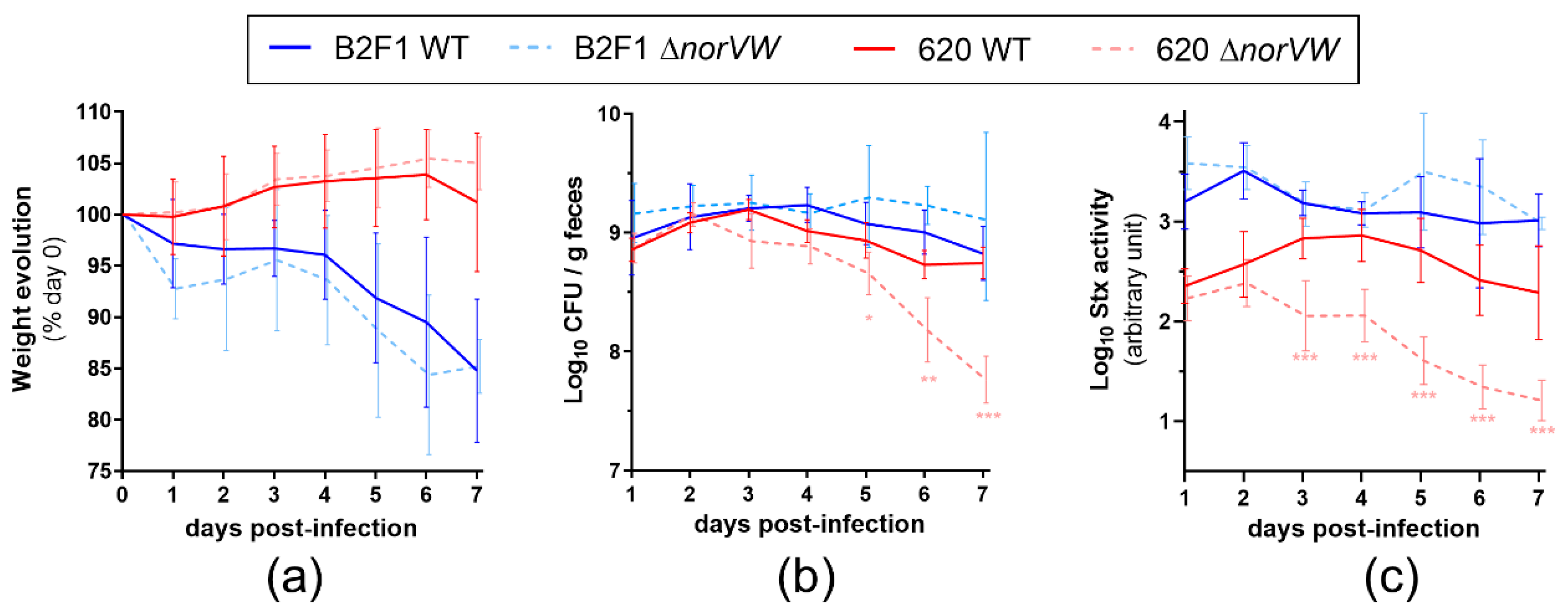
2.3. Role of the norVW Operon in EHEC Virulence
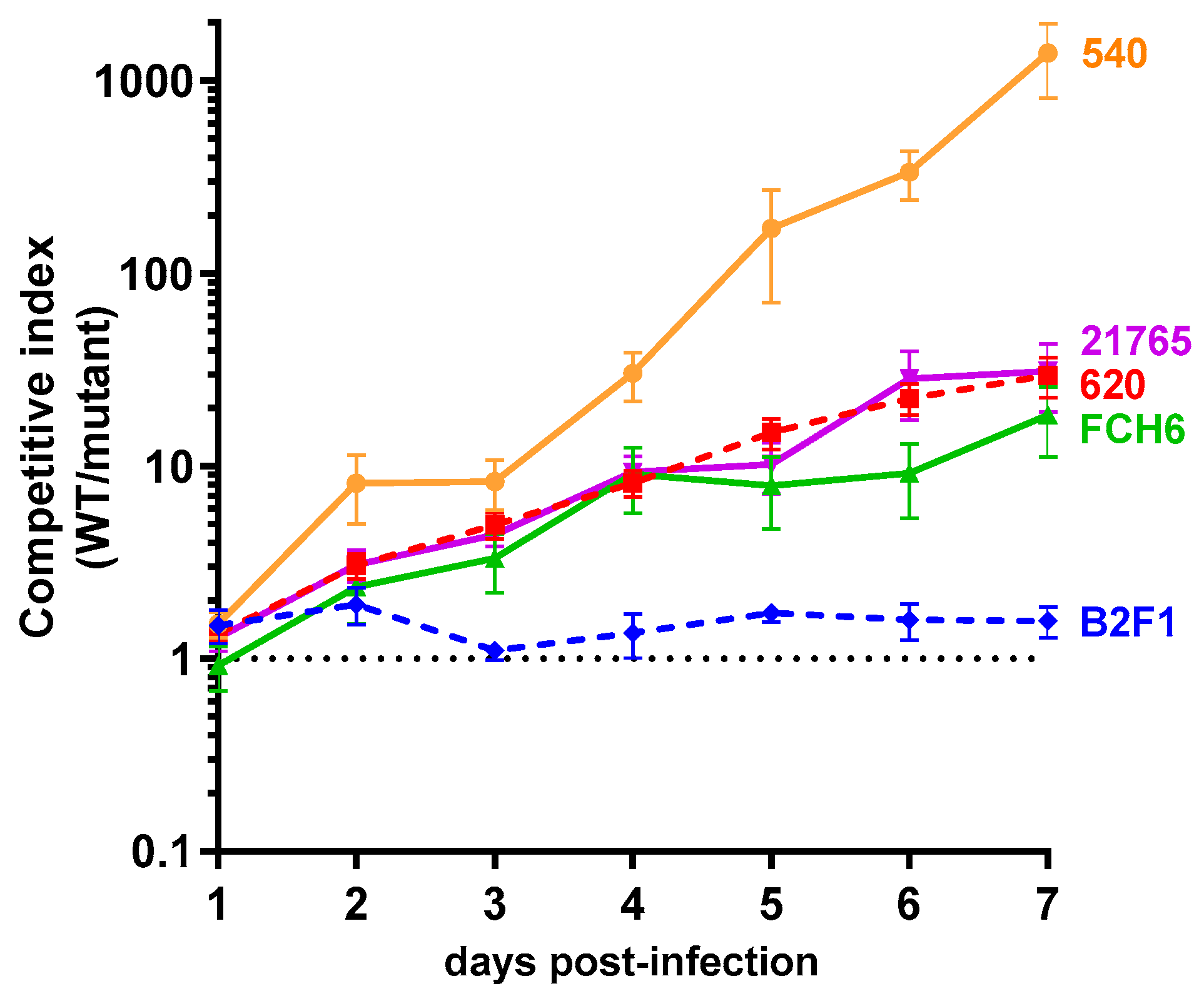
2.4. Role of the norVW Operon in the Fitness of EHEC during Mouse Infection and Influence of NO Synthase Activity from the Host
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Growth Conditions, and Construction of Mutants
4.2. Mouse Infection
4.3. Stx activity Quantification
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hofmann, S.L. Southwestern internal medicine conference: Shiga-like toxins in hemolytic-uremic syndrome and thrombotic thrombocytopenic purpura. Am. J. Med. Sci. 1993, 306, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Karmali, M.A.; Steele, B.T.; Petric, M.; Lim, C. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet 1983, 1, 619–620. [Google Scholar] [CrossRef]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T.; et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Karmali, M.A.; Mascarenhas, M.; Shen, S.; Ziebell, K.; Johnson, S.; Reid-Smith, R.; Isaac-Renton, J.; Clark, C.; Rahn, K.; Kaper, J.B. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 2003, 41, 4930–4940. [Google Scholar] [CrossRef] [Green Version]
- Karmali, M.A.; Gannon, V.; Sargeant, J.M. Verocytotoxin-producing Escherichia coli (VTEC). Vet. Microbiol. 2010, 140, 360–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soysal, N.; Mariani-Kurkdjian, P.; Smail, Y.; Liguori, S.; Gouali, M.; Loukiadis, E.; Fach, P.; Bruyand, M.; Blanco, J.; Bidet, P.; et al. Enterohemorrhagic Escherichia coli Hybrid Pathotype O80:H2 as a New Therapeutic Challenge. Emerg. Infect. Dis. 2016, 22, 1604–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valilis, E.; Ramsey, A.; Sidiq, S.; DuPont, H.L. Non-O157 Shiga toxin-producing Escherichia coli-A poorly appreciated enteric pathogen: Systematic review. Int. J. Infect. Dis. 2018, 76, 82–87. [Google Scholar] [CrossRef] [Green Version]
- Endo, Y.; Tsurugi, K.; Yutsudo, T.; Takeda, Y.; Ogasawara, T.; Igarashi, K. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 1988, 171, 45–50. [Google Scholar] [CrossRef]
- Schuller, S. Shiga toxin interaction with human intestinal epithelium. Toxins 2011, 3, 626–639. [Google Scholar] [CrossRef] [Green Version]
- Ray, P.E.; Liu, X.H. Pathogenesis of Shiga toxin-induced hemolytic uremic syndrome. Pediatr. Nephrol. 2001, 16, 823–839. [Google Scholar] [CrossRef]
- Melton-Celsa, A.R. Shiga toxin (Stx) classification, structure, and function. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruger, A.; Lucchesi, P.M. Shiga toxins and stx phages: Highly diverse entities. Microbiology 2015, 161, 451–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bielaszewska, M.; Friedrich, A.W.; Aldick, T.; Schurk-Bulgrin, R.; Karch, H. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: Predictor for a severe clinical outcome. Clin. Infect. Dis 2006, 43, 1160–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegler, R.L.; Obrig, T.G.; Pysher, T.J.; Tesh, V.L.; Denkers, N.D.; Taylor, F.B. Response to Shiga toxin 1 and 2 in a baboon model of hemolytic uremic syndrome. Pediatr. Nephrol. 2003, 18, 92–96. [Google Scholar] [CrossRef]
- Schmidt, M.A. LEEways: Tales of EPEC, ATEC and EHEC. Cell. Microbiol. 2010, 12, 1544–1552. [Google Scholar] [CrossRef]
- Kulasekara, B.R.; Jacobs, M.; Zhou, Y.; Wu, Z.; Sims, E.; Saenphimmachak, C.; Rohmer, L.; Ritchie, J.M.; Radey, M.; McKevitt, M.; et al. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect. Immun. 2009, 77, 3713–3721. [Google Scholar] [CrossRef] [Green Version]
- Manning, S.D.; Motiwala, A.S.; Springman, A.C.; Qi, W.; Lacher, D.W.; Ouellette, L.M.; Mladonicky, J.M.; Somsel, P.; Rudrik, J.T.; Dietrich, S.E.; et al. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. USA 2008, 105, 4868–4873. [Google Scholar] [CrossRef] [Green Version]
- Gardner, A.M.; Gessner, C.R.; Gardner, P.R. Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. Role of NorR and sigma54 in the nitric oxide stress response. J. Biol. Chem. 2003, 278, 10081–10086. [Google Scholar] [CrossRef] [Green Version]
- Gardner, A.M.; Helmick, R.A.; Gardner, P.R. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 2002, 277, 8172–8177. [Google Scholar] [CrossRef] [Green Version]
- Hutchings, M.I.; Mandhana, N.; Spiro, S. The NorR protein of Escherichia coli activates expression of the flavorubredoxin gene norV in response to reactive nitrogen species. J. Bacteriol. 2002, 184, 4640–4643. [Google Scholar] [CrossRef] [Green Version]
- Bogdan, C. Nitric oxide synthase in innate and adaptive immunity: An update. Trends Immunol. 2015, 36, 161–178. [Google Scholar] [CrossRef] [PubMed]
- Branchu, P.; Matrat, S.; Vareille, M.; Garrivier, A.; Durand, A.; Crepin, S.; Harel, J.; Jubelin, G.; Gobert, A.P. NsrR, GadE, and GadX interplay in repressing expression of the Escherichia coli O157:H7 LEE pathogenicity island in response to nitric oxide. PLoS Pathog. 2014, 10, e1003874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gobert, A.P.; Vareille, M.; Glasser, A.L.; Hindre, T.; de Sablet, T.; Martin, C. Shiga toxin produced by enterohemorrhagic Escherichia coli inhibits PI3K/NF-kappaB signaling pathway in globotriaosylceramide-3-negative human intestinal epithelial cells. J. Immunol. 2007, 178, 8168–8174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichimura, K.; Shimizu, T.; Matsumoto, A.; Hirai, S.; Yokoyama, E.; Takeuchi, H.; Yahiro, K.; Noda, M. Nitric oxide-enhanced Shiga toxin production was regulated by Fur and RecA in enterohemorrhagic Escherichia coli O157. Microbiologyopen 2017, 6. [Google Scholar] [CrossRef]
- Naili, I.; Gardette, M.; Garrivier, A.; Daniel, J.; Desvaux, M.; Pizza, M.; Gobert, A.; Marchal, T.; Loukiadis, E.; Jubelin, G. Interplay between enterohaemorrhagic Escherichia coli and nitric oxide during the infectious process. Emerg. Microbes Infect. 2020, 9, 1065–1076. [Google Scholar] [CrossRef]
- Shimizu, T.; Tsutsuki, H.; Matsumoto, A.; Nakaya, H.; Noda, M. The nitric oxide reductase of enterohaemorrhagic Escherichia coli plays an important role for the survival within macrophages. Mol. Microbiol. 2012, 85, 492–512. [Google Scholar] [CrossRef]
- Espie, E.; Vaillant, V.; Mariani-Kurkdjian, P.; Grimont, F.; Martin-Schaller, R.; De Valk, H.; Vernozy-Rozand, C. Escherichia coli O157 outbreak associated with fresh unpasteurized goats’ cheese. Epidemiol. Infect. 2006, 134, 143–146. [Google Scholar] [CrossRef]
- Douellou, T.; Delannoy, S.; Ganet, S.; Mariani-Kurkdjian, P.; Fach, P.; Loukiadis, E.; Montel, M.; Thevenot-Sergentet, D. Shiga toxin-producing Escherichia coli strains isolated from dairy products-Genetic diversity and virulence gene profiles. Int. J. Food Microbiol. 2016, 232, 52–62. [Google Scholar] [CrossRef]
- King, L.A.; Loukiadis, E.; Mariani-Kurkdjian, P.; Haeghebaert, S.; Weill, F.X.; Baliere, C.; Ganet, S.; Gouali, M.; Vaillant, V.; Pihier, N.; et al. Foodborne transmission of sorbitol-fermenting Escherichia coli O157:H7 via ground beef: An outbreak in northern France, 2011. Clin. Microbiol. Infect. 2014, 20, O1136–O1144. [Google Scholar] [CrossRef] [Green Version]
- Kerangart, S.; Douellou, T.; Delannoy, S.; Fach, P.; Beutin, L.; Sergentet-Thevenot, D.; Cournoyer, B.; Loukiadis, E. Variable tellurite resistance profiles of clinically-relevant Shiga toxin-producing Escherichia coli (STEC) influence their recovery from foodstuffs. Food Microbiol. 2016, 59, 32–42. [Google Scholar] [CrossRef]
- Bonanno, L.; Loukiadis, E.; Mariani-Kurkdjian, P.; Oswald, E.; Garnier, L.; Michel, V.; Auvray, F. Diversity of Shiga Toxin-Producing Escherichia coli (STEC) O26:H11 Strains Examined via stx Subtypes and Insertion Sites of Stx and EspK Bacteriophages. Appl. Environ. Microbiol. 2015, 81, 3712–3721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teel, L.D.; Melton-Celsa, A.R.; Schmitt, C.K.; O’Brien, A.D. One of two copies of the gene for the activatable shiga toxin type 2d in Escherichia coli O91:H21 strain B2F1 is associated with an inducible bacteriophage. Infect. Immun. 2002, 70, 4282–4291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, H.; Terai, A.; Kurazono, H.; Takeda, Y.; Nishibuchi, M. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 1990, 8, 47–60. [Google Scholar] [CrossRef]
- Galia, W.; Mariani-Kurkdjian, P.; Loukiadis, E.; Blanquet-Diot, S.; Leriche, F.; Brugere, H.; Shima, A.; Oswald, E.; Cournoyer, B.; Thevenot-Sergentet, D. Genome sequence and annotation of a human infection isolate of Escherichia coli O26:H11 involved in a raw milk cheese outbreak. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [Green Version]
- Lindgren, S.W.; Melton, A.R.; O’Brien, A.D. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect. Immun. 1993, 61, 3832–3842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinones, B.; Massey, S.; Friedman, M.; Swimley, M.S.; Teter, K. Novel cell-based method to detect Shiga toxin 2 from Escherichia coli O157:H7 and inhibitors of toxin activity. Appl. Environ. Microbiol. 2009, 75, 1410–1416. [Google Scholar] [CrossRef] [Green Version]
- Gardette, M.; Le Hello, S.; Mariani-Kurkdjian, P.; Fabre, L.; Gravey, F.; Garrivier, A.; Loukiadis, E.; Jubelin, G. Identification and prevalence of in vivo-induced genes in enterohaemorrhagic Escherichia coli. Virulence 2019, 10, 180–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John, M.; Kudva, I.T.; Griffin, R.W.; Dodson, A.W.; McManus, B.; Krastins, B.; Sarracino, D.; Progulske-Fox, A.; Hillman, J.D.; Handfield, M.; et al. Use of in vivo-induced antigen technology for identification of Escherichia coli O157:H7 proteins expressed during human infection. Infect. Immun. 2005, 73, 2665–2679. [Google Scholar] [CrossRef] [Green Version]
- Vareille, M.; de Sablet, T.; Hindre, T.; Martin, C.; Gobert, A.P. Nitric oxide inhibits Shiga-toxin synthesis by enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. USA 2007, 104, 10199–10204. [Google Scholar] [CrossRef] [Green Version]
- Hausladen, A.; Gow, A.; Stamler, J.S. Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc. Natl. Acad. Sci. USA 2001, 98, 10108–10112. [Google Scholar] [CrossRef] [Green Version]
- Poock, S.R.; Leach, E.R.; Moir, J.W.; Cole, J.A.; Richardson, D.J. Respiratory detoxification of nitric oxide by the cytochrome c nitrite reductase of Escherichia coli. J. Biol. Chem. 2002, 277, 23664–23669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Vine, C.E.; Balasiny, B.K.; Rizk, J.; Bradley, C.L.; Tinajero-Trejo, M.; Poole, R.K.; Bergaust, L.L.; Bakken, L.R.; Cole, J.A. The roles of the hybrid cluster protein, Hcp and its reductase, Hcr, in high affinity nitric oxide reduction that protects anaerobic cultures of Escherichia coli against nitrosative stress. Mol. Microbiol. 2016, 100, 877–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Justino, M.C.; Vicente, J.B.; Teixeira, M.; Saraiva, L.M. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 2005, 280, 2636–2643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, J.L.; Brynildsen, M.P. Discovery and dissection of metabolic oscillations in the microaerobic nitric oxide response network of Escherichia coli. Proc. Natl. Acad. Sci. USA 2016, 113, E1757–E1766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, T.; Matsumoto, A.; Noda, M. Cooperative roles of nitric oxide-metabolizing enzymes to counteract nitrosative stress in enterohemorrhagic Escherichia coli. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, H.H.; Liu, Y.; Zhang, M.Q.; Spiro, S. Genome-wide analysis of the response to nitric oxide in uropathogenic Escherichia coli CFT073. Microbial Genom. 2015, 1, e000031. [Google Scholar] [CrossRef] [Green Version]
- Robinson, C.M.; Sinclair, J.F.; Smith, M.J.; O’Brien, A.D. Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc. Natl. Acad. Sci. USA 2006, 103, 9667–9672. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Yin, X.; Feng, Y.; Chambers, J.R.; Guo, A.; Gong, J.; Zhu, J.; Gyles, C.L. Verotoxin 2 enhances adherence of enterohemorrhagic Escherichia coli O157:H7 to intestinal epithelial cells and expression of β1-integrin by IPEC-J2 cells. Appl. Environ. Microbiol. 2010, 76, 4461–4468. [Google Scholar] [CrossRef] [Green Version]
- Bang, I.S.; Liu, L.; Vazquez-Torres, A.; Crouch, M.L.; Stamler, J.S.; Fang, F.C. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J. Biol. Chem. 2006, 281, 28039–28047. [Google Scholar] [CrossRef] [Green Version]
- Jones-Carson, J.; Husain, M.; Liu, L.; Orlicky, D.J.; Vazquez-Torres, A. Cytochrome bd-dependent bioenergetics and antinitrosative defenses in Salmonella Pathogenesis. mBio 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Karlinsey, J.E.; Bang, I.S.; Becker, L.A.; Frawley, E.R.; Porwollik, S.; Robbins, H.F.; Thomas, V.C.; Urbano, R.; McClelland, M.; Fang, F.C. The NsrR regulon in nitrosative stress resistance of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2012, 85, 1179–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, A.R.; Dunman, P.M.; Fang, F.C. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 2006, 61, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Sebbane, F.; Lemaitre, N.; Sturdevant, D.E.; Rebeil, R.; Virtaneva, K.; Porcella, S.F.; Hinnebusch, B.J. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc. Natl. Acad. Sci. USA 2006, 103, 11766–11771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svensson, L.; Poljakovic, M.; Save, S.; Gilberthorpe, N.; Schon, T.; Strid, S.; Corker, H.; Poole, R.K.; Persson, K. Role of flavohemoglobin in combating nitrosative stress in uropathogenic Escherichia coli: Implications for urinary tract infection. Microb. Pathog. 2010, 49, 59–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arai, H.; Iiyama, K. Role of nitric oxide-detoxifying enzymes in the virulence of Pseudomonas aeruginosa against the silkworm, Bombyx mori. Biosci. Biotechnol. Biochem. 2013, 77, 198–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Dong, Y.; Wang, N.; Ma, S.; Lu, C.; Liu, Y. Diverse effects of nitric oxide reductase NorV on Aeromonas hydrophila virulence-associated traits under aerobic and anaerobic conditions. Vet. Res. 2019, 50, 67. [Google Scholar] [CrossRef] [Green Version]
- Burton, N.A.; Schurmann, N.; Casse, O.; Steeb, A.K.; Claudi, B.; Zankl, J.; Schmidt, A.; Bumann, D. Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe 2014, 15, 72–83. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, T.; Hirai, S.; Yokoyama, E.; Ichimura, K.; Noda, M. An evolutionary analysis of nitric oxide reductase gene norV in enterohemorrhagic Escherichia coli O157. Infect. Genet. Evol. 2015, 33, 176–181. [Google Scholar] [CrossRef]
- Boerlin, P.; McEwen, S.A.; Boerlin-Petzold, F.; Wilson, J.B.; Johnson, R.P.; Gyles, C.L. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 1999, 37, 497–503. [Google Scholar] [CrossRef] [Green Version]
- Ostroff, S.M.; Tarr, P.I.; Neill, M.A.; Lewis, J.H.; Hargrett-Bean, N.; Kobayashi, J.M. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J. Infect. Dis. 1989, 160, 994–998. [Google Scholar] [CrossRef]
- Gill, A.E.; Amyes, S.G. The contribution of a novel ribosomal S12 mutation to aminoglycoside resistance of Escherichia coli mutants. J. Chemother. 2004, 16, 347–349. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinones, B.; Swimley, M.S. Use of a Vero cell-based fluorescent assay to assess relative toxicities of Shiga toxin 2 subtypes from Escherichia coli. Methods Mol. Biol. 2011, 739, 61–71. [Google Scholar] [CrossRef] [PubMed]
- King, L.A.; Mailles, A.; Mariani-Kurkdjian, P.; Vernozy-Rozand, C.; Montet, M.P.; Grimont, F.; Pihier, N.; Devalk, H.; Perret, F.; Bingen, E.; et al. Community-wide outbreak of Escherichia coli O157:H7 associated with consumption of frozen beef burgers. Epidemiol. Infect. 2009, 137, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Makino, K.; Ohnishi, M.; Kurokawa, K.; Ishii, K.; Yokoyama, K.; Han, C.G.; Ohtsubo, E.; Nakayama, K.; Murata, T.; et al. Complete Genome Sequence of Enterohemorrhagic Escherichia coli O157:H7 and Genomic Comparison with a Laboratory Strain K-12. DNA Res. 2001, 8, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neto, M.; Delannoy, S.; Auvray, F.; Oswald, E.; Fach, P.; Loukiadis, E. Public health significance of E. coli 026 isolated from foodstuffs: Toward genetic predictors of their virulence. Zoonoses Public Health 2012, 59, 19–90. [Google Scholar] [CrossRef]
- Ogura, Y.; Gotoh, Y.; Itoh, T.; Sato, M.P.; Seto, K.; Yoshino, S.; Isobe, J.; Etoh, Y.; Kurogi, M.; Kimata, K.; et al. Population structure of Escherichia coli O26: H11 with recent and repeated stx2 acquisition in multiple lineages. Microb. Genom. 2017, 3, e000141. [Google Scholar] [CrossRef]
- Mariani-Kurkdjian, P.; Denamur, E.; Milon, A.; Picard, B.; Cave, H.; Lambert-Zechovsky, N.; Loirat, C.; Goullet, P.; Sansonetti, P.J.; Elion, J. Identification of a Clone of Escherichia coli 0103:H2 as a Potential Agent of Hemolytic-Uremic Syndrome in France. J. Clin. Microbiol. 1993, 31, 296–301. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.L.; Köhler, B.; Oswald, E.; Beutin, L.; Karch, H.; Morabito, S.; Caprioli, A.; Suerbaum, S.; Schmidt, H. Genetic Diversity of Intimin Genes of Attaching and Effacing Escherichia coli Strains. J. Clin. Microbiol. 2002, 40, 4486–4492. [Google Scholar] [CrossRef] [Green Version]
- Bibbal, D.; Loukiadis, E.; Kérourédan, M.; Ferré, F.; Dilasser, F.; Peytavin De Garam, C.; Cartier, P.; Oswald, E.; Gay, E.; Auvray, F.; et al. Prevalence of Carriage of Shiga Toxin-Producing Escherichia coli Serotypes O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 among Slaughtered Adult Cattle in France. Appl. Environ. Microbiol. 2015, 81, 1397–1405. [Google Scholar] [CrossRef] [Green Version]

| Serotype | Strain | norV | eaeb | stx1 | stx2 |
|---|---|---|---|---|---|
| O157:H7 | 620 | + | + | – | + |
| 540 | + | + | + | + | |
| FCH6 | + | + | – | + | |
| RD9 | - | + | + | + | |
| EDL 933 | - | + | + | + | |
| Sakaï | - | + | + | + | |
| O26:H11 | 21765 | + | + | – | + |
| 37.40 | + | + | + | – | |
| 279/8 | + | + | + | – | |
| 11368 | + | + | + | – | |
| O103:H2 | PMK5 | + | + | + | – |
| 590 | + | + | + | – | |
| 2503 | + | + | + | – | |
| 03.35 | + | + | + | – | |
| 2455-1 | + | + | + | + | |
| O111:H8 | CL37 | + | + | – | – |
| J43 | + | + | + | – | |
| 622-4 | + | + | + | + | |
| O145:H28 | 2513-21 | + | + | – | + |
| 991 | + | + | + | – | |
| 1036 | + | + | – | + | |
| O121:H19 | 12652 | + | + | – | + |
| 12805 | + | + | – | + | |
| S3075 | + | + | – | + | |
| O45:H2 | 12047 | + | + | + | – |
| O91:H21 | 13199 | + | – | + | + |
| B2F1 | + | – | – | + | |
| 13694 | + | – | – | + | |
| O113:H21 | 13341 | + | – | – | + |
| 14032 | + | – | – | + | |
| O113:H4 | 13137 | + | – | + | – |
| O80:H2 | 38009 | + | + | – | + |
| RDEx444 | + | + | – | + | |
| 40963 | + | + | – | + |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gardette, M.; Daniel, J.; Loukiadis, E.; Jubelin, G. Role of the Nitric Oxide Reductase NorVW in the Survival and Virulence of Enterohaemorrhagic Escherichia coli during Infection. Pathogens 2020, 9, 683. https://doi.org/10.3390/pathogens9090683
Gardette M, Daniel J, Loukiadis E, Jubelin G. Role of the Nitric Oxide Reductase NorVW in the Survival and Virulence of Enterohaemorrhagic Escherichia coli during Infection. Pathogens. 2020; 9(9):683. https://doi.org/10.3390/pathogens9090683
Chicago/Turabian StyleGardette, Marion, Julien Daniel, Estelle Loukiadis, and Grégory Jubelin. 2020. "Role of the Nitric Oxide Reductase NorVW in the Survival and Virulence of Enterohaemorrhagic Escherichia coli during Infection" Pathogens 9, no. 9: 683. https://doi.org/10.3390/pathogens9090683
APA StyleGardette, M., Daniel, J., Loukiadis, E., & Jubelin, G. (2020). Role of the Nitric Oxide Reductase NorVW in the Survival and Virulence of Enterohaemorrhagic Escherichia coli during Infection. Pathogens, 9(9), 683. https://doi.org/10.3390/pathogens9090683






