Lack of Evidence on the Susceptibility of Ticks and Wild Rodent Species to PCV3 Infection
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
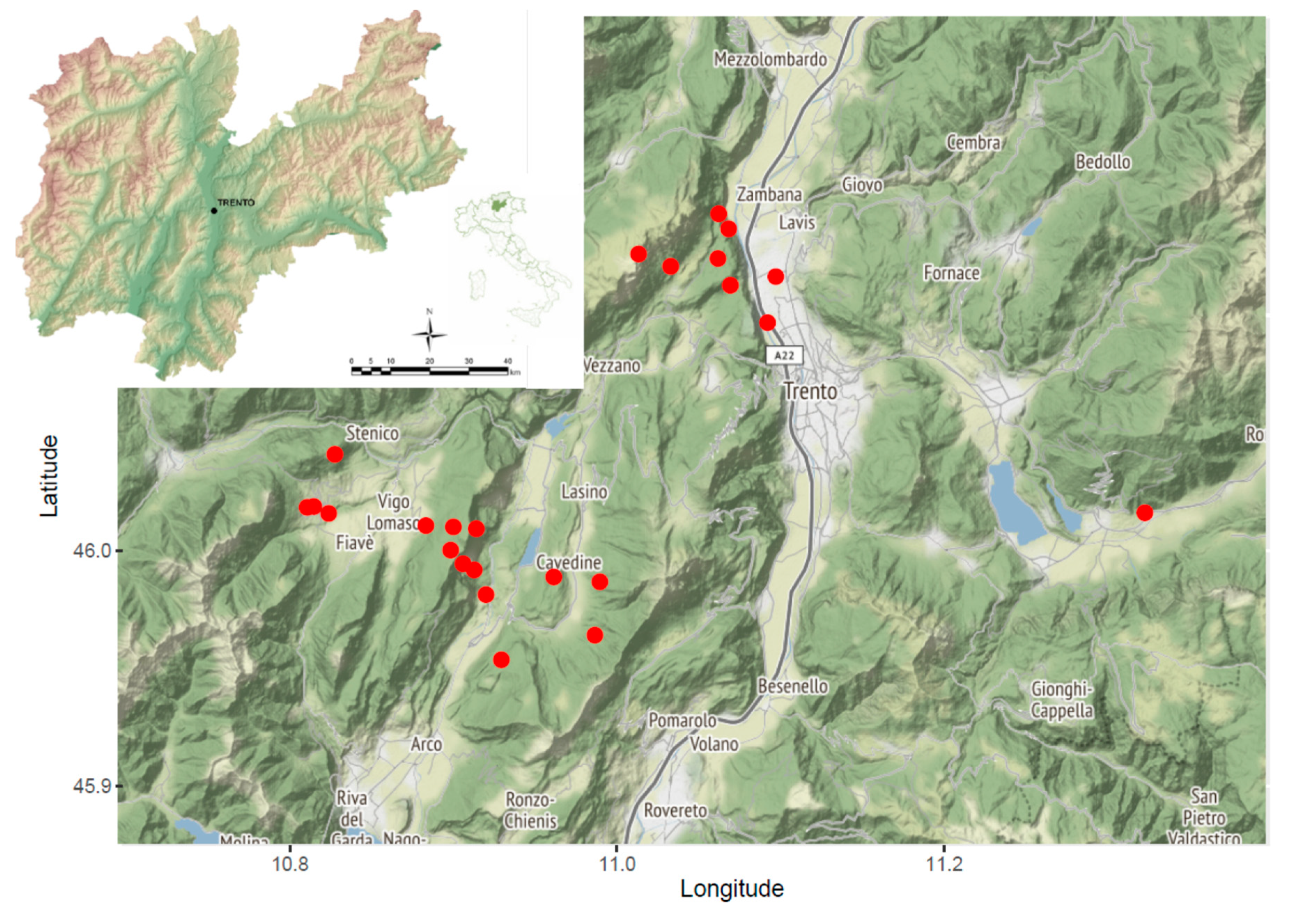
4.1. Sample Collection
4.2. PCV3 Detection
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Klaumann, F.; Correa-Fiz, F.; Franzo, G.; Sibila, M.; Núñez, J.I.; Segalés, J. Current knowledge on Porcine circovirus 3 (PCV3): A novel virus with a yet unknown impact on the swine industry. Front. Vet. Sci. 2018, 5, 1–13. [Google Scholar] [CrossRef]
- Saraiva, G.; Vidigal, P.; Assao, V.; Fajardo, M.; Loreto, A.; Fietto, J.; Bressan, G.; Lobato, Z.; Almeida, M.; Silva-Júnior, A. Retrospective Detection and Genetic Characterization of Porcine circovirus 3 (PCV3) Strains Identified between 2006 and 2007 in Brazil. Viruses 2019, 11, 201. [Google Scholar] [CrossRef] [PubMed]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; He, W.; Correa-Fiz, F.; Li, G.; Legnardi, M.; Su, S.; Segalés, J. A Shift in Porcine Circovirus 3 (PCV3) History Paradigm: Phylodynamic Analyses Reveal an Ancient Origin and Prolonged Undetected Circulation in the Worldwide Swine Population. Adv. Sci. 2019, 6, 1901004. [Google Scholar] [CrossRef]
- Ouyang, T.; Niu, G.; Liu, X.; Zhang, X.; Zhang, Y.; Ren, L. Recent progress on porcine circovirus type 3. Infect. Genet. Evol. 2019, 73, 227–233. [Google Scholar] [CrossRef]
- Fux, R.; Söckler, C.; Link, E.K.; Renken, C.; Krejci, R.; Sutter, G.; Ritzmann, M.; Eddicks, M. Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains. Virol. J. 2018, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Giannitti, F.; Rossow, S.; Marthaler, D.; Knutson, T.P.; Li, L.; Deng, X.; Resende, T.; Vannucci, F.; Delwart, E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol. J. 2016, 13, 1–8. [Google Scholar] [CrossRef]
- Ku, X.; Chen, F.; Li, P.; Wang, Y.; Yu, X.; Fan, S.; Qian, P.; Wu, M.; He, Q. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound. Emerg. Dis. 2017, 64, 703–708. [Google Scholar] [CrossRef]
- Shen, H.; Liu, X.; Zhang, P.; Wang, L.; Liu, Y.; Zhang, L.; Liang, P.; Song, C. Genome characterization of a porcine circovirus type 3 in South China. Transbound. Emerg. Dis. 2018, 65, 264–266. [Google Scholar] [CrossRef]
- Zheng, S.; Wu, X.; Zhang, L.; Xin, C.; Liu, Y.; Shi, J.; Peng, Z.; Xu, S.; Fu, F.; Yu, J.; et al. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound. Emerg. Dis. 2017, 64, 1337–1341. [Google Scholar] [CrossRef]
- Franzo, G.; Grassi, L.; Tucciarone, C.M.; Drigo, M.; Martini, M.; Pasotto, D.; Mondin, A.; Menandro, M.L. A wild circulation: High presence of Porcine circovirus 3 in different mammalian wild hosts and ticks. Transbound. Emerg. Dis. 2019, 66, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Prinz, C.; Stillfried, M.; Neubert, L.K.; Denner, J. Detection of PCV3 in German wild boars. Virol. J. 2019, 16, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Klaumann, F.; Dias-Alves, A.; Cabezón, O.; Mentaberre, G.; Castillo-Contreras, R.; López-Béjar, M.; Casas-Díaz, E.; Sibila, M.; Correa-Fiz, F.; Segalés, J. Porcine circovirus 3 is highly prevalent in serum and tissues and may persistently infect wild boar (Sus scrofa scrofa). Transbound. Emerg. Dis. 2019, 66, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Tucciarone, C.M.; Drigo, M.; Cecchinato, M.; Martini, M.; Mondin, A.; Menandro, M.L. First report of wild boar susceptibility to Porcine circovirus type 3: High prevalence in the Colli Euganei Regional Park (Italy) in the absence of clinical signs. Transbound. Emerg. Dis. 2019, 65, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Z.; Zou, Y.; Zhang, N.; Wang, D.; Tu, D.; Yang, L.; Deng, Z.; Yang, Y.; Jiang, P.; et al. First molecular detection of porcine circovirus type 3 in dogs in China. Virus Genes 2018, 54, 140–144. [Google Scholar] [CrossRef]
- Jiang, S.; Zhou, N.; Li, Y.; An, J.; Chang, T. Detection and sequencing of porcine circovirus 3 in commercially sourced laboratory mice. Vet. Med. Sci. 2019, 5, 176–181. [Google Scholar] [CrossRef]
- Wang, W.; Sun, W.; Cao, L.; Zheng, M.; Zhu, Y.; Li, W.; Liu, C.; Zhuang, X.; Xing, J.; Lu, H.; et al. An epidemiological investigation of porcine circovirus 3 infection in cattle in Shandong province, China. BMC Vet. Res. 2019, 15, 1–8. [Google Scholar] [CrossRef]
- Sun, W.; Wang, W.; Xin, J.; Cao, L.; Zhuang, X.; Zhang, C.; Zhu, Y.; Zhang, H.; Qin, Y.; Du, Q.; et al. An epidemiological investigation of porcine circovirus 3 infection in dogs in the Guangxi Province from 2015 to 2017, China. Virus Res. 2019, 270, 197663. [Google Scholar] [CrossRef]
- Zhai, S.-L.; Lu, S.-S.; Wei, W.-K.; Lv, D.-H.; Wen, X.-H.; Zhai, Q.; Chen, Q.-L.; Sun, Y.-W.; Xi, Y. Reservoirs of Porcine Circoviruses: A Mini Review. Front. Vet. Sci. 2019, 6, 319. [Google Scholar] [CrossRef]
- Ha, Z.; Li, J.F.; Xie, C.Z.; Li, C.H.; Zhou, H.N.; Zhang, Y.; Hao, P.F.; Nan, F.L.; Zhang, J.Y.; Han, J.C.; et al. First detection and genomic characterization of porcine circovirus 3 in mosquitoes from pig farms in China. Vet. Microbiol. 2020, 240, 1–7. [Google Scholar] [CrossRef]
- Czyżewska-Dors, E.; Núñez, J.I.; Saporiti, V.; Huerta, E.; Riutord, C.; Cabezón, O.; Segalés, J.; Sibila, M. Detection of Porcine Circovirus 3 in Wildlife Species in Spain. Pathogens 2020, 9, 341. [Google Scholar] [CrossRef] [PubMed]
- Baráková, I.; Derdáková, M.; Selyemová, D.; Chvostáč, M.; Špitalská, E.; Rosso, F.; Collini, M.; Rosà, R.; Tagliapietra, V.; Girardi, M.; et al. Tick-borne pathogens and their reservoir hosts in northern Italy. Ticks Tick Borne Dis. 2018, 9, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Segalés, J.; Klaumann, F.; Legnardi, M.; Mweu, M.M.; Mahmmod, Y.S. Diagnostic accuracy of two DNA-based molecular assays for detection of porcine circovirus 3 in swine population using Bayesian latent class analysis. Lett. Appl. Microbiol. 2019, 69, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Lőrincz, M.; Cságola, A.; Biksi, I.; Szeredi, L.; Dan, Á.; Tuboly, T. Detection of Porcine circovirus in rodents—Short communication. Acta Vet. Hung. 2010, 58, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.L.; Chen, S.N.; Liu, W.; Li, X.P.; Deng, S.F.; Wen, X.H.; Luo, M.L.; Lv, D.H.; Wei, W.K.; Chen, R.A. Molecular detection and genome characterization of porcine circovirus type 2 in rats captured on commercial swine farms. Arch. Virol. 2016, 161, 3237–3244. [Google Scholar] [CrossRef]
- Pinheiro, A.L.B.C.; Bulos, L.H.S.; Onofre, T.S.; de Paula Gabardo, M.; de Carvalho, O.V.; Fausto, M.C.; Guedes, R.M.C.; de Almeida, M.R.; Silva, A. Verification of natural infection of peridomestic rodents by PCV2 on commercial swine farms. Res. Vet. Sci. 2013, 94, 764–768. [Google Scholar] [CrossRef]
- Ciebiera, O.; Jerzak, L.; Nowak-Chmura, M.; Bocheński, M. Ticks (Acari: Ixodida) on birds (Aves) migrating through the Polish Baltic coast. Exp. Appl. Acarol. 2019, 77, 241–251. [Google Scholar] [CrossRef]
- Mendoza-Roldan, A.J.; Colella, V.; Paolo Lia, R.; Nguyen, V.L.; Barros-Battesti, D.M.; Iatta, R.; Dantas-Torres, F.; Otranto, D. Borrelia burgdorferi (sensu lato) in ectoparasites and reptiles in southern Italy. Parasit. Vectors 2019, 12, 1–9. [Google Scholar] [CrossRef]
- Rosà, R.; Tagliapietra, V.; Manica, M.; Arnoldi, D.; Hauffe, H.C.; Rossi, C.; Rosso, F.; Henttonen, H.; Rizzoli, A. Changes in host densities and co-feeding pattern efficiently predict tick-borne encephalitis hazard in an endemic focus in northern Italy. Int. J. Parasitol. 2019, 49, 779–787. [Google Scholar] [CrossRef]
- Franzo, G.; Legnardi, M.; Centelleghe, C.; Tucciarone, C.M.; Cecchinato, M.; Cortey, M.; Segalés, J.; Drigo, M. Development and validation of direct PCR and quantitative PCR assays for the rapid, sensitive, and economical detection of porcine circovirus 3. J. Vet. Diagn. Investig. 2018, 30, 538–544. [Google Scholar] [CrossRef]
- Mangold, A.J.; Bargues, M.D.; Mas-Coma, S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). Parasitol. Res. 1998, 84, 478–484. [Google Scholar] [CrossRef] [PubMed]
| Number of Captures | A. flavicollis | A. sylvaticus | M. glareolus | Total |
|---|---|---|---|---|
| 1 | 51 | 1 | 11 | 63 |
| 2 | 13 | 10 | 23 | |
| 3 | 8 | 1 | 2 | 11 |
| 4 | 5 | 3 | 8 | |
| 5 | 5 | 5 | ||
| 6 | 4 | 4 | ||
| 7 | 1 | 1 | ||
| 9 | 3 | 3 | ||
| Total samples | 90 | 2 | 26 | 118 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grassi, L.; Tagliapietra, V.; Rizzoli, A.; Martini, M.; Drigo, M.; Franzo, G.; Menandro, M.L. Lack of Evidence on the Susceptibility of Ticks and Wild Rodent Species to PCV3 Infection. Pathogens 2020, 9, 682. https://doi.org/10.3390/pathogens9090682
Grassi L, Tagliapietra V, Rizzoli A, Martini M, Drigo M, Franzo G, Menandro ML. Lack of Evidence on the Susceptibility of Ticks and Wild Rodent Species to PCV3 Infection. Pathogens. 2020; 9(9):682. https://doi.org/10.3390/pathogens9090682
Chicago/Turabian StyleGrassi, Laura, Valentina Tagliapietra, Annapaola Rizzoli, Marco Martini, Michele Drigo, Giovanni Franzo, and Maria Luisa Menandro. 2020. "Lack of Evidence on the Susceptibility of Ticks and Wild Rodent Species to PCV3 Infection" Pathogens 9, no. 9: 682. https://doi.org/10.3390/pathogens9090682
APA StyleGrassi, L., Tagliapietra, V., Rizzoli, A., Martini, M., Drigo, M., Franzo, G., & Menandro, M. L. (2020). Lack of Evidence on the Susceptibility of Ticks and Wild Rodent Species to PCV3 Infection. Pathogens, 9(9), 682. https://doi.org/10.3390/pathogens9090682








