Secondary Metabolites in Xylella fastidiosa–Plant Interaction
Abstract
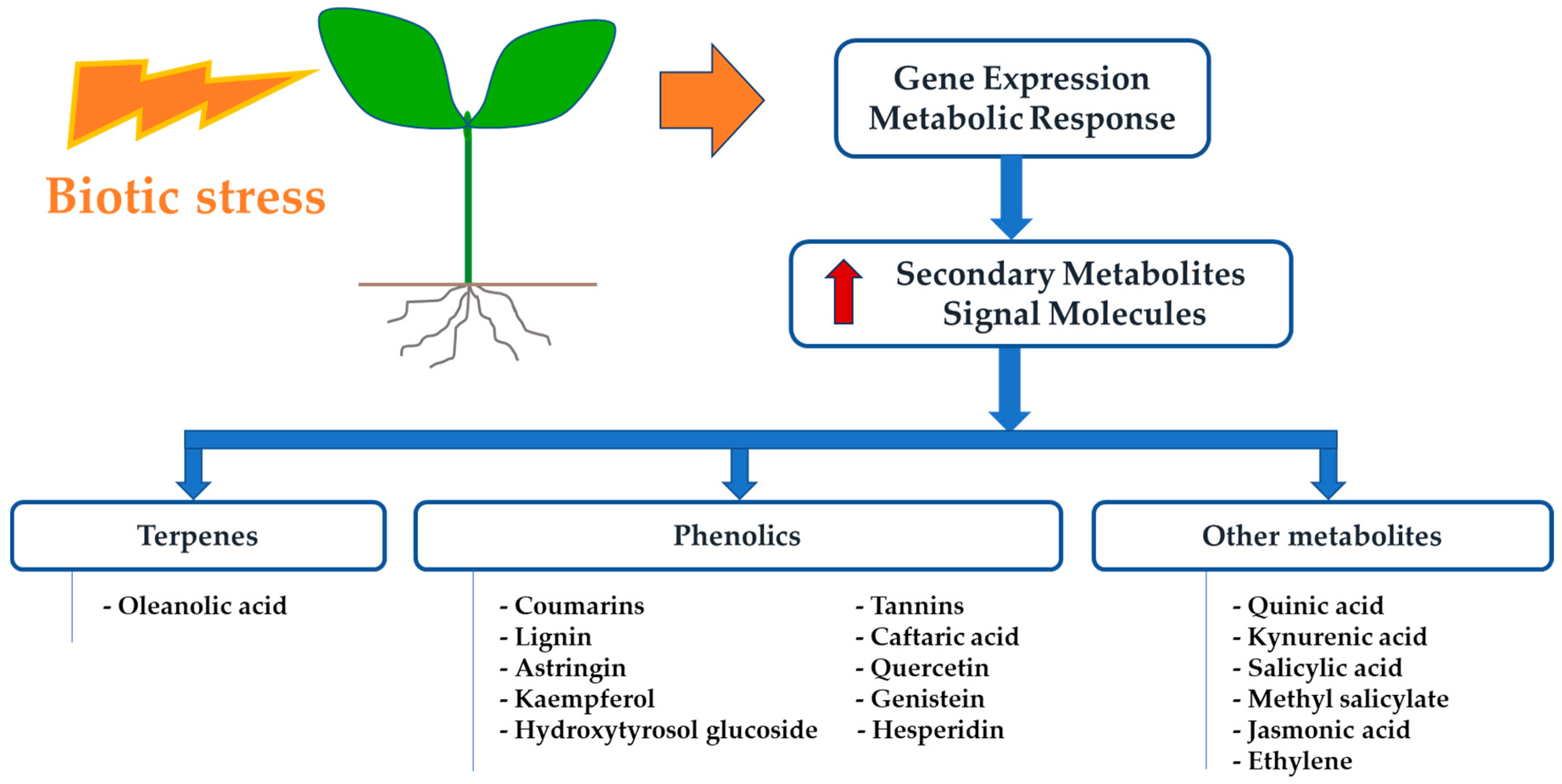
:1. Introduction
2. New Insights on Plant Response to X. fastidiosa
3. Profiling Change of Metabolites in Economic Plants Affected by X. fastidiosa
3.1. Citrus spp.
3.2. Olea europaea L.
3.3. Prunus Dulcis (Mill.) D.A. Webb
3.4. Vitis spp.
| Plant | Evidences | References |
|---|---|---|
| Citrus spp. | Induction of genes involved in phenylpropanoid and flavonoid biosynthesis | [54,55,57,58] |
| Presence of hesperidin in areas where tissues disrupted by X. fastidiosa | [59] | |
| Induction of hesperidin production | [61] | |
| Increase of flavonoids in leaves and coumarins in roots | [62] | |
| Olea europaea L. | Reduction of hydroxytyrosol glucoside and increase of quinic acid | [76] |
| Reduction of hydroxytyrosol glucoside and increase of quinic acid and lignin content | [56] | |
| Higher content of quinic acid in infected leaves | [18] | |
| Increase of flavonoids (such as quercetin, kaempferol and genistein), tannins, oleanolic acid, salycilic, and kynurenic acids | [17] | |
| Prunus dulcis (Mill.) D.A. Webb | Higher concentrations of total phenolic compounds in resistant than susceptible cultivars | [84] |
| Vitis spp. | Increase of catechin, digalloylquinic acid, astringin, multiple catechins, procyanidins, stilbenoids, lignin and condensed tannins | [48] |
| Increase caftaric acid, methyl salicylate and quinic acid | [49,90] | |
| Increase of transcripts for terpene, chalcone, stylbene synthases, erythritol and 2-deoxyerythritol, 1,2-anhydro-myo-inositol, arbutin; glycosidase and tyrosinase | [91] |
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dixon, R. Natural products and plant disease resistance. Nature 2001, 411, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.A. Current trends in the evolutionary ecology of plant defence. Funct. Ecol. 2011, 25, 420–432. [Google Scholar] [CrossRef]
- Schulze, E.-D.; Beck, E.; Buchmann, N.; Clemens, S.; Müller-Hohenstein, K.; Scherer-Lorenzen, M. (Eds.) Plant Ecology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 257–299. [Google Scholar] [CrossRef]
- Bednarek, P.; Osbourn, A. Plant–microbe interactions: Chemical diversity in plant defence. Science 2009, 324, 746–748. [Google Scholar] [CrossRef] [PubMed]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatla, S.C. Secondary matabolites. In Plant Physiology, Development and Metabolism; Bhatla, S.C., Lal, M.A., Eds.; Springer: Singapore, 2018; pp. 1099–1166. [Google Scholar] [CrossRef]
- Zaynab, M.; Fatima, M.; Abbas, S.; Sharif, Y.; Umair, M.; Zafare, M.H.; Bahadarf, K. Role of secondary metabolites in plant defense against pathogens. Microb. Pathog. 2018, 124, 198–202. [Google Scholar] [CrossRef]
- Lemarié, S.; Robert-Seilaniantz, A.; Lariagon, C.; Lemoine, J.; Marnet, N.; Levrel, A.; Jubault, M.; Manzanares-Dauleux, M.; Gravot, A. Camalexin contributes to the partial resistance of Arabidopsis thaliana to the biotrophic soilborne protist Plasmodiophora brassicae. Front. Plant Sci. 2015, 6, 539. [Google Scholar] [CrossRef] [Green Version]
- Cruickshank, I.; Perrin, D.R. Isolation of a phytoalexin from Pisum sativum L. Nature 1960, 187, 799–800. [Google Scholar] [CrossRef]
- Perrin, D.R.; Bottomley, W. Studies on phytoalexins. V. The structure of pisatin from Pisum sativum L. J. Am. Chem. Soc. 1962, 84, 1919–1922. [Google Scholar] [CrossRef]
- Sobolev, V.S. Production of phytoalexins in peanut (Arachis hypogaea) seed elicited by selected microorganisms. J. Agric. Food Chem. 2013, 61, 1850–1858. [Google Scholar] [CrossRef]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for enhanced defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef]
- Wang, N.; Pierson, E.A.; Setubal, J.C.; Xu, J.; Levy, J.G.; Zhang, Y.; Li, J.; Rangel, L.T.; Martins, J.J. The Candidatus Liberibacter-host interface: Insights into pathogenesis mechanisms and disease control. Annu. Rev. Phytopathol. 2017, 55, 451–482. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Nagendran, K.; Rai, A.B.; Singh, B.; Rao, G.P.; Bertaccini, A. Global status of phytoplasma diseases in vegetable crops. Front. Microbiol. 2019, 10, 1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saponari, M.; Giampetruzzi, A.; Loconsole, G.; Boscia, D.; Saldarelli, P. Xylella fastidiosa in olive in Apulia: Where we stand. Phytopathology 2019, 109, 175–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolì, F.; Negro, C.; Nutricati, E.; Vergine, M.; Aprile, A.; Sabella, E.; Damiano, G.; De Bellis, L.; Luvisi, A. Accumulation of azelaic acid in Xylella fastidiosa-infected olive trees: A mobile metabolite for health screening. Phytopathology 2019, 109, 318–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novelli, S.; Gismondi, A.; Di Marco, G.; Canuti, L.; Nanni, V.; Canini, A. Plant defense factors involved in Olea europaea resistance against Xylella fastidiosa infection. J. Plant Res. 2019, 132, 439–455. [Google Scholar] [CrossRef] [PubMed]
- Girelli, C.R.; Angilè, F.; Del Coco, L.; Migoni, D.; Zampella, L.; Marcelletti, S.; Cristella, N.; Marangi, P.; Scortichini, M.; Fanizzi, F.P. 1H-NMR metabolite fingerprinting analysis reveals a disease biomarker and a field treatment response in Xylella fastidiosa subsp. pauca-Infected olive trees. Plants 2019, 8, 115. [Google Scholar] [CrossRef] [Green Version]
- EFSA. Update of the Xylella spp. host plant database—Systematic literature search up to 30 June 2019. EFSA J. 2020, 18, 6114. [Google Scholar] [CrossRef]
- Krugner, R.; Sisterson, M.S.; Chen, J.; Stenger, D.C.; Johnson, M.W. Evaluation of olive as a host of Xylella fastidiosa and associated sharpshooter vectors. Plant Dis. 2014, 98, 1185–1193. [Google Scholar] [CrossRef] [Green Version]
- Roper, M.C.; Greve, L.C.; Labavitch, J.A.; Kirkpatrick, B.C. Detection and visualisation of an exopolysaccharide produced by Xylella fastidiosa in vitro and in planta. Appl. Environ. Microbiol. 2007, 73, 7252–7258. [Google Scholar] [CrossRef] [Green Version]
- Sicard, A.; Zeilinger, A.R.; Vanhove, M.; Schartel, T.E.; Beal, D.J.; Daugherty, M.P.; Almeida, R.P.P. Xylella fastidiosa: Insights into an emerging plant pathogen. Annu. Rev. Phytopathol. 2018, 56, 181–202. [Google Scholar] [CrossRef] [Green Version]
- Purcell, A. Paradigms: Examples from the bacterium Xylella fastidiosa. Ann. Rev. Phytopathol. 2013, 51, 339–356. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.L.; Torres, S.C.Z.; Heredia, M.; Lopes, S.A. Citrus responses to Xylella fastidiosa infection. Plant Dis. 2012, 96, 1245–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, C.M.; Obando, J.J.; Villalobos, W.; Moreira, L.; Rivera, C. First Report of Xylella fastidiosa infecting coffee in Costa Rica. Plant Dis. 2001, 85, 1027. [Google Scholar] [CrossRef] [PubMed]
- Amanifar, N.; Taghavi, M.; Izadpanah, K.; Babaei, G. Isolation and pathogenicity of Xylella fastidiosa from grapevine and almond in Iran. Phytopathol. Mediterr. 2014, 53, 318–327. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization (EPPO). First Report of Xylella fastidiosa in Israel; EPPO Reporting Service: Paris, France, 2019; Volume 6, p. 121.
- Su, C.C.; Chang, C.J.; Chang, C.M.; Shih, H.T.; Tzeng, K.C.; Jan, F.J.; Kao, C.W.; Deng, W.L. Pierce’s disease of grapevines in Taiwan: Isolation, cultivation and pathogenicity of Xylella fastidiosa. J. Phytopathol. 2013, 161, 389–396. [Google Scholar] [CrossRef]
- Saponari, M.; Boscia, D.; Nigro, F.; Martelli, G.P. Identification of DNA sequences related to Xylella fastidiosa in oleander, almond and olive trees exhibiting leaf scorch symptoms in Apulia (southern Italy). J. Plant Pathol. 2013, 95, 668. [Google Scholar]
- Denancé, N.; Legendre, B.; Briand, M.; Olivier, V.; de Boisseson, C.; Poliakoff, F.; Jacques, M.A. Several subspecies and sequence types are associated with the emergence of Xylella fastidiosa in natural settings in France. Plant Pathol. 2017, 66, 1054–1064. [Google Scholar] [CrossRef] [Green Version]
- Olmo, D.; Nieto, A.; Adrover, F.; Urbano, A.; Beidas, O.; Juan, A.; Marco-Noales, E.; López, M.M.; Navarro, I.; Monterde, A.; et al. First detection of Xylella fastidiosa infecting cherry (Prunus avium) and Polygala myrtifolia plants, in Mallorca Island, Spain. Plant Dis. 2018, 101, 1820. [Google Scholar] [CrossRef]
- Cardinale, M.; Luvisi, A.; Meyer, J.B.; Sabella, E.; De Bellis, L.; Cruz, A.C.; Ampatzidis, Y.; Cherubini, P. Specific fluorescence in situ hybridization (FISH) test to highlight colonisation of xylem vessels by Xylella fastidiosa in naturally infected olive trees (Olea europaea L.). Front. Plant Sci. 2018, 9, 431. [Google Scholar] [CrossRef] [Green Version]
- Kyrkou, I.; Pusa, T.; Ellegaard-Jensen, L.; Sagot, M.F.; Hansen, L.H. Pierce’s disease of grapevines: A review of control strategies and an outline of an epidemiological model. Front. Microbiol. 2018, 9, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Bragard, C.; Dehnen-Schmutz, K.; Di Serio, F.; Gonthier, P.; Jacques, M.A.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; Milonas, P.; et al. Effectiveness of in Planta Control Measures for Xylella Fastidiosa. EFSA J. 2019, 17. [Google Scholar] [CrossRef]
- Azevedo, J.L.; Araújo, W.L.; Lacava, P.T. The diversity of citrus endophytic bacteria and their interaction with Xylella fastidiosa and host plants. Gen. Mol. Biol. 2016, 39, 476–491. [Google Scholar] [CrossRef] [Green Version]
- Deyett, E.; Rolshausen, P.E. Temporal dynamics of the sap microbiome of grapevine under high Pierce’s disease pressure. Front. Plant Sci. 2019, 10, 1246. [Google Scholar] [CrossRef] [PubMed]
- Baptista, P.; Cameirão, C.; Giampetruzzi, A.; Morelli, M.; Kubaa, R.A.; Altamura, G.; D’Attoma, G.; Pereira, J.A.; Neto, T.L.; Saldarelli, P. Understanding the olive microbiome of susceptible and resistant cultivars for sustainable biocontrol. J. Plant Pathol. 2019, 101, 849. [Google Scholar] [CrossRef]
- Vergine, M.; Meyer, J.B.; Cardinale, M.; Sabella, E.; Hartmann, M.; Cherubini, P.; De Bellis, L.; Luvisi, A. The Xylella fastidiosa-Resistant olive cultivar “Leccino” has stable endophytic microbiota during the Olive Quick Decline Syndrome (OQDS). Pathogens 2020, 9, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bordiec, S.; Paquis, S.; Lacroix, H.; Dhondt, S.; Ait, B.E.; Kauffmann, S.; Jeandet, P.; Mazeyrat-Gourbeyre, F.; Clément, C.; Baillieul, F.; et al. Comparative analysis of defence responses induced by the endophytic plant growth-promoting rhizobacterium Burkholderia phytofirmans strain PsJN and the non-host bacterium Pseudomonas syringae pv. pisi in grapevine cell suspensions. J. Exp. Bot. 2011, 62, 595–603. [Google Scholar] [CrossRef] [Green Version]
- Baccari, C.; Antonova, E.; Lindow, S. Biological control of Pierce’s disease of grape by an endophytic bacterium. Phytopathology 2019, 109, 248–256. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.S.; White, J.F.; Zhang, X.; Hinton, D.M.; Bacon, C.W. Endophyte-mediated adjustments in host morphology and physiology and effects on host fitness traits in grasses. Fungal Ecol. 2012, 5, 322–330. [Google Scholar] [CrossRef]
- De La Fuente, L.; Parker, J.K.; Oliver, J.E.; Granger, S.; Brannen, P.M.; van Santen, E.; Cobine, P.A. The bacterial pathogen Xylella fastidiosa affects the leaf ionome of plant hosts during infection. PLoS ONE 2013, 8, e62945. [Google Scholar] [CrossRef] [Green Version]
- D’Attoma, G.; Morelli, M.; Saldarelli, P.; Saponari, M.; Giampetruzzi, A.; Boscia, D.; Savino, V.N.; La Fuente, L.D.; Cobine, P.A. Ionomic differences between susceptible and resistant olive cultivars infected by Xylella fastidiosa in the outbreak area of Salento, Italy. Pathogens 2019, 8, 272. [Google Scholar] [CrossRef] [Green Version]
- Oliver, J.E.; Sefick, S.A.; Parker, J.K.; Arnold, T.; Cobine, P.A.; De La Fuente, L. Ionome changes in Xylella fastidiosa-infected Nicotiana tabacum correlate with virulence and discriminate between subspecies of bacterial isolates. Mol. Plant Microbe Interact. 2014, 27, 1048–1058. [Google Scholar] [CrossRef] [Green Version]
- Oliver, J.E.; Cobine, P.A.; De La Fuente, L. Xylella fastidiosa isolates from both subsp. multiplex and fastidiosa cause disease on southern highbush blueberry (Vaccinium sp.) under greenhouse conditions. Phytopathology 2015, 105, 855–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.Y.; Bi, J.L.; Morse, J.G.; Toscano, N.C.; Cooksey, D.A. Effect of xylem fluid from susceptible and resistant grapevines on developmental biology of Xylella fastidiosa. Eur. J. Plant Pathol. 2013, 135, 127–135. [Google Scholar] [CrossRef]
- Scala, V.; Pucci, N.; Salustri, M.; Modesti, V.; L’Aurora, A.; Scortichini, M.; Zaccaria, M.; Momeni, B.; Reverberi, M.; Loreti, S. Xylella fastidiosa subsp. pauca and olive produced lipids moderate the switch adhesive versus non-adhesive state and viceversa. PLoS ONE 2020, 15, e0233013. [Google Scholar] [CrossRef]
- Wallis, C.M.; Chen, J. Grapevine phenolic compounds in Xylem Sap and tissues are significantly altered during infection by Xylella fastidiosa. Phytopathology 2012, 102, 816–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallis, C.M.; Wallingford, A.K.; Chen, J. Grapevine rootstock effects on scion sap phenolic levels, resistance to Xylella fastidiosa infection, and progression of Pierce’s disease. Front. Plant Sci. 2013, 4, 502. [Google Scholar] [CrossRef] [Green Version]
- Queiroz-Voltan, R.B.; Paradela Filho, O. Caracterização de estruturas anatômicas de citros infectados com Xylella fastidiosa. Laranja 1999, 20, 55–76. [Google Scholar]
- Rossetti, M.; Garnier, M.; Bove, J.M.; Beretta, M.J.G.; Teixeira, A.R.R.; Quaggio, J.A.; de Negri, J.D. Présence de bactéries dans le xylème d’orangers atteints de chlorose variégée, une nouvelle maladie des agrumes au Brésil. C. R. Acad. Sci. 1990, 310, 345–349. [Google Scholar]
- da Silva, F.R.; Vettore, A.L.; Kemper, E.L.; Leite, A.; Arruda, P. Fastidian gum: The Xylella fastidiosa exopolysaccharide possibly involved in bacterial pathogenicity. FEMS Microbiol. Lett. 2001, 203, 165–171. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, F.P.; Stuchi, E.S.; Lourenço, S.A.; Kriss, A.B.; Gottwald, T.R.; Amorim, L. The effect of irrigation on development of citrus variegated chlorosis symptoms. Crop Prot. 2014, 57, 8–14. [Google Scholar] [CrossRef]
- de Souza, A.A.; Takita, M.A.; Coletta-Filho, H.D.; Targon, M.L.P.N.; Carlos, E.F.; Locali-Fabris, E.C.; Amaral, A.M.; Freitas-Astúa, J.; Silva-Pinhati, A.C.O.; Boscariol-Camargo, R.L.; et al. Analysis of expressed sequence tags from Citrus Sinensis L. osbeck infected with Xylella fastidiosa. Genet. Mol. Biol. 2007, 30, 957–964. [Google Scholar] [CrossRef] [Green Version]
- de Souza, A.A.; Takita, M.A.; Coletta-Filho, H.D.; Campos, M.A.; Teixeira, J.E.C.; Targon, M.L.P.N.; Carlos, E.F.; Ravasi, J.F.; Fischer, C.N.; Machado, M.A. Comparative analysis of differentially expressed sequence tags of sweet orange and mandarin infected with Xylella fastidiosa. Genet. Mol. Biol. 2007, 30, 965–971. [Google Scholar] [CrossRef] [Green Version]
- Sabella, E.; Luvisi, A.; Aprile, A.; Negro, C.; Vergine, M.; Nicolì, F.; Miceli, A.; De Bellis, L. Xylella fastidiosa induces differential expression of lignification related-genes and lignin accumulation in tolerant olive trees cv. Leccino. J. Plant Physiol. 2018, 220, 60–68. [Google Scholar] [CrossRef] [PubMed]
- de Souza, A.A.; Takita, M.A.; Amaral, A.M.; Coletta-Filho, H.D.; Machado, M.A. Citrus responses to Xylella fastidiosa infection, the causal agent de citrus variegated chlorosis. Tree Sci. Biotech. 2009, 3, 73–80. [Google Scholar]
- Rodrigues, J.L.M.; Silva-Stenico, M.E.; Gomes, J.E.; Lopes, J.R.S.; Tsai, S.M. Detection and diversity assessment of Xylella fastidiosa in field-collected plant and insect samples by using 16S rRNA and gyrB sequences. Appl. Environ. Microbiol. 2003, 69, 4249–4255. [Google Scholar] [CrossRef] [Green Version]
- Alves, E.; Leite, B.; Pascholati, S.F.; Ishida, M.L.; Andersen, P.C. Citrus Sinensis leaf petiole and blade colonization by Xylella fastidiosa: Details of Xylem Vessel occlusion. Sci. Agric. 2009, 66, 218–224. [Google Scholar] [CrossRef] [Green Version]
- Erickson, L.C. The general physiology of Citrus. In The Citrus Industry: Anatomy, Physiology, Genetics, and Reproduction; Reuther, W., Batchelor, L.D., Webber, H.J., Eds.; University of California Press: Riverside, CA, USA, 1968; pp. 86–122. [Google Scholar]
- Soares, M.S.; da Silva, D.F.; Forim, M.R.; da Silva, M.F.; Fernandes, J.B.; Vieira, P.C.; Silva, D.B.; Lopes, N.P.; de Carvalho, S.A.; de Souza, A.A.; et al. Quantification and localization of hesperidin and rutin in Citrus sinensis Grafted on C. Limonia after Xylella fastidiosa Infection by HPLC-UV and MALDI Imaging Mass Spectrometry. Phytochemistry 2015, 115, 161–170. [Google Scholar] [CrossRef]
- Soares, M.S.; Silva, D.F.; Amaral, J.C.; Silva, M.M.; Forim, M.R.; Rodrigues-Filho, E.; Silva, M.F.; Fernandes, J.B.; Machado, M.A.; de Souza, A.A.; et al. Rapid differentiation of graft Citrus Sinensis with and without Xylella fastidiosa infection by mass spectrometry. Rapid Commun. Mass Spectrom. 2020. [Google Scholar] [CrossRef]
- Haelterman, R.M.; Tolocka, P.A.; Roca, M.E.; Guzmán, F.A.; Fernández, F.D.; Otero, M.L. First presumptive diagnosis of Xylella fastidiosa causing olive scorch in Argentina. J. Plant Pathol. 2015, 97, 393. [Google Scholar]
- Tolocka, P.A.; Mattio, M.F.; Paccioretti, M.A.; Otero, M.L.; Roca, M.E.; Guzmán, F.A.; Haelterman, R.M. Xylella fastidiosa subsp. pauca ST69 in olive in Argentina. J. Plant Pathol. 2017, 99, 803. [Google Scholar] [CrossRef]
- Gutiérrez Hernández, O.; García, L.V. Incidencia de Xylella fastidiosa en las Islas Baleares y distribución potencial en la península ibérica. Investig. Geogr. 2018, 69, 55–72. [Google Scholar] [CrossRef] [Green Version]
- European and Mediterranean Plant Protection Organization (EPPO). Update on the Situation of Xylella fastidiosa in Spain; EPPO Reporting Service: Paris, France, 2019; in press.
- Della Coletta-Filho, H.; Francisco, C.S.; Spotti-Lopes, J.R.; De Oliveira, A.F.; De Oliveira da Silva, L.F. First report of olive leaf scorch in Brazil associated with Xylella fastidiosa subs. pauca. Phytopathol. Mediterr. 2016, 55, 130–135. [Google Scholar]
- Wong, F.; Cooksey, D.A.; Costa, H.S. Documentation and characterisation of Xylella fastidiosa strains in landscape hosts. In Proceedings of the California Department of Food and Agriculture Symposium, Pierce’s Disease Research, Coronado, CA, USA, 7–10 December 2004; pp. 238–241. [Google Scholar]
- Maggiore, G.; Semeraro, T.; Aretano, R.; De Bellis, L.; Luvisi, A. GIS analysis of land-use change in threatened landscapes by Xylella fastidiosa. Sustainability 2019, 11, 253. [Google Scholar] [CrossRef] [Green Version]
- Semeraro, T.; Gatto, E.; Buccolieri, R.; Vergine, M.; Gao, Z.; De Bellis, L.; Luvisi, A. Changes in olive urban forests infected by Xylella fastidiosa: Impact on microclimate and social health. Int. J. Environ. Res. Public Health 2019, 16, 2642. [Google Scholar] [CrossRef] [Green Version]
- Martelli, G.P. The current status of the quick decline syndrome of olive in Southern Italy. Phytoparasitica 2016, 44, 1–10. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; Morelli, M.; Saponari, M.; Loconsole, G.; Chiumenti, M.; Boscia, D.; Savino, V.N.; Martelli, G.P.; Saldarelli, P. Transcriptome profiling of two olive cultivars in response to infection by the CoDiRO strain of Xylella fastidiosa subsp. pauca. BMC Genom. 2016, 17, 475. [Google Scholar] [CrossRef] [Green Version]
- Saponari, M.; Boscia, D.; Altamura, G.; Loconsole, G.; Zicca, S.; D’Attoma, G.; Morelli, M.; Palmisano, F.; Saponari, A.; Tavano, D.; et al. Isolation and pathogenicity of Xylella fastidiosa associated to the olive quick decline syndrome in Southern Italy. Sci. Rep. 2017, 7, 17723. [Google Scholar] [CrossRef]
- Sabella, E.; Aprile, A.; Genga, A.; Siciliano, T.; Nutricati, E.; Nicolì, F.; Vergine, M.; Negro, C.; De Bellis, L.; Luvisi, A. Xylem cavitation susceptibility and refilling mechanisms in olive trees infected by Xylella fastidiosa. Sci. Rep. 2019, 9, 9602. [Google Scholar] [CrossRef] [Green Version]
- Kover, P.X.; Schaal, B.A. Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions. Proc. Natl. Acad. Sci. USA 2002, 99, 11270–11274. [Google Scholar] [CrossRef] [Green Version]
- Luvisi, A.; Aprile, A.; Sabella, E.; Vergine, M.; Nicolì, F.; Nutricati, E.; Miceli, A.; Negro, C.; De Bellis, L. Xylella fastidiosa subsp. pauca (CoDiRO strain) infection in four olive (Olea europaea L.) cultivars: Profile of phenolic compounds in leaves and progression of leaf scorch symptoms. Phytopathol. Mediterr. 2017, 56, 259−273. [Google Scholar] [CrossRef]
- Sanborn, R.R.; Mircetitch, S.M.; Nyland, G.; Moller, W.J. Golden death—A new leaf scorch threat to almond growers. Calif. Agric. 1974, 28, 4–5. [Google Scholar]
- Sisterson, M.S.; Chen, J.; Viveros, M.A.; Civerolo, E.L.; Ledbetter, C.; Groves, R.L. Effects of almond leaf scorch disease on almond yield: Implications for management. Plant Dis. 2008, 92, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.P.P.; Purcell, A.H. Biological traits of Xylella fastidiosa strains for grapes and almonds. Appl. Environ. Microbiol. 2003, 69, 7447–7452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loconsole, G.; Saponari, M.; Boscia, D.; D’Attoma, G.; Morelli, M.; Martelli, G.; Almeida, R.P.P. Intercepted isolates of Xylella fastidiosa in Europe reveal novel genetic diversity. Eur. J. Plant Pathol. 2016, 146, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Giampetruzzi, A.; Velasco-Amo, M.P.; Marco-Noales, E.; Montes-Borrego, M.; Román-Écija, M.; Navarro, I.; Monterde, A.; Barbé, S.; Almeida, R.P.P.; Saldarelli, P.; et al. Draft genome resources of two strains (“ESVL” and “IVIA5901”) of Xylella fastidiosa associated with almond leaf scorch disease in Alicante, Spain. Phytopathology 2019, 109, 219–221. [Google Scholar] [CrossRef] [Green Version]
- Rogers, E.E.; Ledbetter, C. Susceptibility to Xylella fastidiosa in a first-generation hybrid from a non-traditional peach-almond cross. HortScience 2015, 50, 337–340. [Google Scholar] [CrossRef] [Green Version]
- Krugner, R.; Ledbetter, C.A. Rootstock effects on almond leaf scorch disease incidence and severity. Plant Dis. 2016, 100, 1617–1621. [Google Scholar] [CrossRef]
- Wilhelm, M.; Brodbeck, B.V.; Andersen, P.C.; Kasun, G.W.; Kirkpatrick, B.C. Analysis of xylem fluid components in almond cultivars differing in resistance to almond leaf scorch disease. Plant Dis. 2011, 95, 166–172. [Google Scholar] [CrossRef]
- Goheen, A.C.; Nyland, G.; Lowe, S.K. Association of a rickettsia-like organism with Pierce’s disease of grapevines and alfalfa dwarf and heat therapy of the disease in grapevines. Phytopathology 1973, 63, 341–345. [Google Scholar] [CrossRef]
- Hopkins, D.L.; Mollenhauer, H.H. Rickettsia-like bacterium associated with Pierce’s disease of grapes. Science 1973, 179, 298–300. [Google Scholar] [CrossRef]
- Hopkins, D.L. Diseases caused by leafhopper-borne rickettsia-like bacteria. Annu. Rev. Phytopathol. 1977, 17, 277–294. [Google Scholar] [CrossRef]
- Galvez, L.C.; Korus, K.; Fernandez, J.; Behn, J.L.; Banjara, N. The Threat of Pierce’s Disease to Midwest Wine and Table Grapes; American Phytopathological Society: St. Paul, MN, USA, 2010. [Google Scholar]
- Sanscartier, C.A.; Arora, A.K.; Tulgetske, G.M.; Miller, T.A. Glassy-winged sharpshooter population survey and Xylella fastidiosa detection. Undergrad. Res. J. 2012, 6, 31. [Google Scholar]
- Wallis, C.M.; Wallingford, A.K.; Chen, J. Effects of cultivar, phenology, and Xylella fastidiosa infection on grapevine Xylem Sap and tissue phenolic content. Physiol. Mol. Plant Pathol. 2013, 84, 28–35. [Google Scholar] [CrossRef]
- Zaini, P.A.; Nascimento, R.; Gouran, H.; Cantu, D.; Chakraborty, S.; Phu, M.; Goulart, L.R.; Dandekar, A.M. Molecular profiling of Pierce’s disease outlines the response circuitry of Vitis vinifera to Xylella fastidiosa infection. Front. Plant Sci. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- European Commission. Regulation (EC) No 1107/2009 of the European Parliament and of the Council of 21 October 2009 concerning the placing of plant protection products on the market and repealing Council Directives 79/117/EEC and 91/414/EEC. Off. J. Eur. Union 2009, 309, 1–50. [Google Scholar]
- Cantrell, C.L.; Dayan, F.E.; Duke, S.O. Natural products as source for new pesticides. J. Nat. Prod. 2012, 75, 1231–1242. [Google Scholar] [CrossRef]
- Kennedy, G.G.; Barbour, J.D. Resistance variation in natural and managed systems. In Plant Resistance to Herbivores and Pathogens; Fritz, R.S., Slimms, E.L., Eds.; University of Chicago Press: Chicago, IL, USA, 1992. [Google Scholar]
- Bosso, L.; Di Febbraro, M.; Cristinzio, G.; Zoina, A.; Russo, D. Shedding light on the effects of climate change on the potential distribution of Xylella fastidiosa in the Mediterranean basin. Biol. Invasions 2016, 18, 1759–1768. [Google Scholar] [CrossRef]
- Luvisi, A.; Nicolì, F.; De Bellis, L. Sustainable management of plant quarantine pests: The case of Olive Quick Decline Syndrome. Sustainability 2017, 9, 659. [Google Scholar] [CrossRef] [Green Version]
- Mouden, S.; Klinkhamer, P.G.L.; Choi, Y.H.; Leiss, K.A. Towards eco-friendly crop protection: Natural deep eutectic solvents and defensive secondary metabolites. Phytochem. Rev. 2017, 16, 935–951. [Google Scholar] [CrossRef] [Green Version]
- Gandikota, M.; De Kochko, A.; Chen, L.; Ithal, N.; Fauquet, C.; Reddy, A.R. Development of transgenic rice plants expressing maize anthocyanin genes and increased blast resistance. Mol. Breed. 2001, 7, 73–83. [Google Scholar] [CrossRef]
- Shadle, G.L.; Wesley, S.V.; Korth, K.L.; Chen, F.; Lamb, C.; Dixon, R.A. Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of L-phenylalanine ammonia-lyase. Phytochemistry 2003, 64, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Way, H.M.; Kazan, K.; Mitter, N.; Goulter, K.C.; Birch, R.G.; Manners, J.M. Constitutive expression of a phenylalanine ammonia-lyase gene from Stylosanthes humilis in transgenic tobacco leads to enhanced disease resistance but impaired plant growth. Physiol. Mol. Plant Pathol. 2002, 60, 275–282. [Google Scholar] [CrossRef]
- Sunitha, S.; Rock, C.D. CRISPR/Cas9-mediated targeted mutagenesis of TAS4 and MYBA7 loci in grapevine rootstock 101-14. Transgen. Res. 2020, 29, 355–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindow, S.; Newman, K.; Chatterjee, S.; Baccari, C.; Lavarone, A.T.; Ionescu, M. Production of Xylella fastidiosa diffusible signal factor in transgenic grape causes pathogen confusion and reduction in severity of Pierce’s disease. Mol. Plant Microbe Interact. 2014, 27, 244–254. [Google Scholar] [CrossRef] [Green Version]
- Caserta, R.; Souza-Neto, R.R.; Takita, M.A.; Lindow, S.E.; De Souza, A.A. Ectopic expression of Xylella fastidiosa rpfF conferring production of diffusible signal factor in transgenic tobacco and citrus alters pathogen behavior and reduces disease severity. Mol. Plant Microbe Interact. 2017, 30, 866–875. [Google Scholar] [CrossRef] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vergine, M.; Nicolì, F.; Sabella, E.; Aprile, A.; De Bellis, L.; Luvisi, A. Secondary Metabolites in Xylella fastidiosa–Plant Interaction. Pathogens 2020, 9, 675. https://doi.org/10.3390/pathogens9090675
Vergine M, Nicolì F, Sabella E, Aprile A, De Bellis L, Luvisi A. Secondary Metabolites in Xylella fastidiosa–Plant Interaction. Pathogens. 2020; 9(9):675. https://doi.org/10.3390/pathogens9090675
Chicago/Turabian StyleVergine, Marzia, Francesca Nicolì, Erika Sabella, Alessio Aprile, Luigi De Bellis, and Andrea Luvisi. 2020. "Secondary Metabolites in Xylella fastidiosa–Plant Interaction" Pathogens 9, no. 9: 675. https://doi.org/10.3390/pathogens9090675
APA StyleVergine, M., Nicolì, F., Sabella, E., Aprile, A., De Bellis, L., & Luvisi, A. (2020). Secondary Metabolites in Xylella fastidiosa–Plant Interaction. Pathogens, 9(9), 675. https://doi.org/10.3390/pathogens9090675










