Multidrug Resistance (MDR) and Collateral Sensitivity in Bacteria, with Special Attention to Genetic and Evolutionary Aspects and to the Perspectives of Antimicrobial Peptides—A Review
Abstract
1. Introduction
2. Multidrug Resistance: Updated Terms and Definitions
2.1. Antibiotic Resistance as a Phenotype
2.2. The Location and Harboring of the Resistance Genes
2.3. Insertion and Excision
2.4. Homolog Recombination
2.5. Intrinsic Resistance (IR)
2.6. Definitions of Antibiotic Polyresistant Strains
2.7. The Updated List of the “ESKAPE” Polyresistant Pathogenic Bacterium Species
3. Resistance Problems Related to Gram-Negative Pathogens
3.1. Β-Lactams vs. ESBL Resistance in Enterobacteriaceae and Klebsiella pneumoniae
3.2. Carbapenems: A Strong Antibiotic (“High Card”) to Beat the “Wedge” ESBL
3.3. Carbapenem Resistance: The Second Strike Back from the Gram-Negative Pathogens (“Wedge” from Nature)
3.4. NDM1
3.5. “Dropped and Rediscovered” Colistin, as a Large Spectral Antibiotic (a “Trump Card”)
3.6. Colistin Resistance: The Third Unexpected Attack (“Wedge”) from Nature
3.7. Efforts to Overcome MDR Problems in Gram-Negative Pathogens in the Absence of Omnipotent (“Jolly Joker”) Antibiotics
4. Resistance Problems Related to Gram-Positive Pathogens
4.1. Methicillin-Resistant Staphylococcus aureus (MRSA)
4.1.1. The Molecular Basis of MRSA
4.1.2. The Accelerated Evolution of MRSA
4.2. Enterococci: The Gram-Positive “Vanguards” of the “MDR Movement”
4.2.1. Comparative Genomics of Enterococci: Species, Strains, Clades Resistance Groups
4.2.2. Clinically Adapted and Non-Clinical Enterococcus strains
4.2.3. MDR Potential of Enterococci
4.2.4. Vancomycin-resistant Enterococcus faecium VRE [298] as a Leading Cause of MDR Hospital Infections
4.2.5. Enterococcus cecorum: An Example for the Recent Speedy Development of Antibiotic Multiresistance in Genus Enterococcus
4.2.6. Vancomycin, Vancomycin Resistance; Daptomycin, Daptomycin Resistance
4.2.7. Daptomycin (DAP)
4.2.8. Daptomycin Resistance DAP (R)
5. The Efforts to Discover Omnipotent (“Jolly Joker”) Antibiotics to Overcome MDR Problems
5.1. Teixobactin: The First Omnipotent (“Jolly Joker”) Antibiotic Active Against Gram-Positive Targets
5.2. Narrow/Spectral Pluripotent Antimicrobial Peptides
5.3. Large Spectral Pluripotent (“Jolly Joker” Candidate) Non-Ribosomal Encoded (NRP) Biosynthetic Antimicrobial Peptides
5.4. Efflux Pump Inhibitors (EPIs)
6. Adaptive Evolution: Trends and Mechanisms in Relation to MDR-Posed Danger
6.1. Resistance vs. Persistence: Darwinian and Lamarckian Approaches
6.2. An Introduction to Experimental Evolution
Resistance Evolution and Mobile Genetic Elements in Enterococci
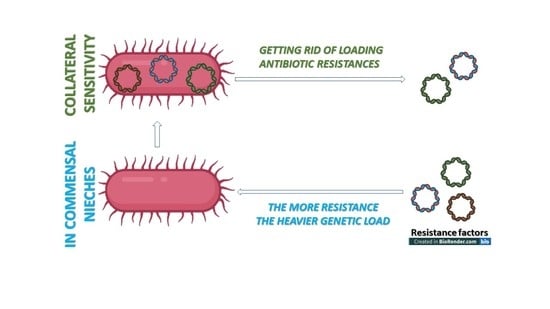
6.3. Resistance Is Not a Positive Selection Marker in a Commensal Niche—The Lesson Learned from Population Genetics and Monitoring Evolutionary Processes
6.4. The Loading Capacity of the Bacterial Genome Is Limited—The Lesson Learned from Experimental Evolution
6.4.1. Evolutionary Trends in The Presence of Antibiotics
Resistance, Fitness, and Compensatory Mutations
Collateral Sensitivity
Antibiotic and Antimicrobial Resistance Genes are of Different Mobility Patterns
6.4.2. Evolutionary Trends of Antibiotic Resistance in the Absence of Antibiotic Exposure
6.4.3. Phylogenetic Limitations: Species Specificity of the Antibiotic Resistance Genes
6.5. The CRISPR/Cas Bacterial Immune System: A Molecular Tool to Get Rid of Unwanted Antibiotic Resistance
7. Clonally Evolving Pan-Genomic ESKAPE Pathogens
7.1. Pseudomonas aeruginosa: A Pan-Genomic Hotbed of Multidrug Resistance
7.1.1. A Shortlist of Intrinsic and Acquired Antibiotic Resistance in P. aeruginosa
7.1.2. Pseudomonas Genetics and Genomics
7.1.3. Transcriptomics
7.1.4. Resistance Phenotypes
7.2. Acinetobacter baumannii
7.2.1. Species with a Gradually Expanding (Open) Pangenome
7.2.2. Resistance Mechanisms and Diseases
7.2.3. Genetic Dissection of Colistin Resistance in A. baumannii: Trebosc et al., 2019
7.2.4. Genetics Toolkits to Becoming Multiresistant
7.2.5. Sword of Damocles: Clonal Evolution, Global Spread, and Epidemic Potential of A. baumannii
7.2.6. Genome Plasticity
7.2.7. Will the Epidemic Threat Be Materialized?
8. Concluding Remarks
8.1. Forcasts Based On Evolutionary Data
8.1.1. The Main Question
8.1.2. The “Combat Scenario”
8.1.3. Adaptive Evolution
8.1.4. The “Dialectics” of Resistance and Sensitivity
8.1.5. Collateral Sensitivity: A Biochemically Proven Limiting Factor in MDR Evolution
8.1.6. Counterarguments
8.2. Statistic-Based Conclusions from the Frequencies of MDR-Related Publications in PubMed
8.3. Pharmaceutical Perspectives
8.3.1. Search for Omnipotent (“Jolly Joker”) Antibiotics
8.3.2. The Perspectives of Antimicrobial Peptide (AMP) Molecules
8.4. Closing Remark
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Watkins, R.R.; Bonomo, R.A. Overview: Global and local impact of antibiotic resistance. Infect. Dis. Clin. N. Am. 2016, 30, 313–322. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- El Zowalaty, M.E.; Al Thani, A.A.; Webster, T.J.; El Zowalaty, A.E.; Schweizer, H.P.; Nasrallah, G.K.; Marei, H.E.; Ashour, H.M. Pseudomonas aeruginosa: Arsenal of resistance mechanisms, decades of changing resistance profiles, and future antimicrobial therapies. Future Microbiol. 2015, 10, 1683–1706. 115. [Google Scholar] [CrossRef]
- Imperi, F.; Antunes, L.C.; Blom, J.; Villa, L.; Iacono, M.; Visca, P.; Carattoli, A. The genomics of Acinetobacter baumannii: Insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life 2011, 63, 1068–1074. [Google Scholar] [CrossRef]
- Lean, S.S.; Yeo, C.C. Small, enigmatic plasmids of the nosocomial pathogen, Acinetobacter baumannii: Good, bad, who knows? Front. Microbiol. 2017, 8, 1547. [Google Scholar] [CrossRef] [PubMed]
- Morris, F.C.; Dexter, C.; Kostoulias, X.; Uddin, M.I.; Peleg, A.Y. The mechanisms of disease caused by Acinetobacter baumannii. Front. Microbiol. 2019, 10, 1601. [Google Scholar] [CrossRef]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, B.; Jindal, H.M.; Le, C.F.; Gudimella, R.; Anwar, A.; Razali, R.; Poole-Johnson, J.; Manikam, R.; Sekaran, S.D. Next generation sequencing reveals the antibiotic resistant variants in the genome of Pseudomonas aeruginosa. PLoS ONE 2017, 12, e0182524. [Google Scholar] [CrossRef] [PubMed]
- Jeukens, J.; Freschi, L.; Kukavica-Ibrulj, I.; Emond-Rheault, J.G.; Tucker, N.P.; Levesque, R.C. Genomics of antibiotic-resistance prediction in Pseudomonas aeruginosa. Ann. N. Y. Acad. Sci. 2019, 1435, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.P.; Chain, E. An enzyme from bacteria able to destroy penicillin. Nature 1940, 146, 837. [Google Scholar] [CrossRef]
- Perron, G.G.; Inglis, R.F.; Pennings, P.S.; Cobey, S. Fighting microbial drug resistance: A primer on the role of evolutionary biology in public health. Evol. Appl. 2015, 8, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Khan, A.U. Global economic impact of antibiotic resistance: A review. J. Glob. Antimicrob. Resist. 2019, 19, 313–316. [Google Scholar] [CrossRef]
- Talbot, G.H. What is in the pipeline for Gram-negative pathogens? Expert. Rev. Anti. Infect. Ther. 2008, 6, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Dötsch, A.; Becker, T.; Pommerenke, C.; Magnowska, Z.; Jansch, L.; Haussler, S. Genome-wide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009, 53, 2522–2531. [Google Scholar] [CrossRef]
- Cantas, L.; Shah, S.Q.A.; Cavaco, L.M.; Manaia, C.; Walsh, F.; Popowska, M.; Garelick, H.; Bürgmann, H.; Sørum, H. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front. Microbiol. 2013, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- Exner, M.; Bhattacharya, S.; Christiansen, B.; Gebel, J.; Goroncy-Bermes, P.; Hartemann, P.; Heeg, P.; Ilschner, C.; Kramer, A.; Larson, E.; et al. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control 2017, 12. [Google Scholar] [CrossRef]
- Szmolka, A.; Nagy, B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front. Microbiol. 2013, 4, 258. [Google Scholar] [CrossRef]
- Wilk, T.; Szabó, M.; Szmolka, A.; Kiss, J.; Barta, E.; Nagy, T.; Olasz, F.; Nagy, B. Genome sequences of multidrug-resistant Salmonella enterica subsp. enterica serovar infantis strains from broiler chicks in Hungary. Genome Announc. 2016, 4, e01400-16. [Google Scholar] [CrossRef]
- Gebreyes, W.A.; Thakur, S. Multidrug-resistant Salmonella enterica serovar München from pigs and humans and potential interserovar transfer of antimicrobial resistance. Antimicrob. Agents Chemother. 2005, 49, 503–511. [Google Scholar] [CrossRef]
- Endimiani, A.; Hujer, K.M.; Hujer, A.M.; Bertschy, I.; Rossano, A.; Koch, C.; Gerber, V.; Francey, T.; Bonomo, R.A.; Perreten, V. Acinetobacter baumannii isolates from pets and horses in Switzerland: Molecular characterization and clinical data. J. Antimicrob. Chemother. 2011, 66, 2248–2254. [Google Scholar] [CrossRef]
- Moore, A.M.; Patel, S.; Forsberg, K.J.; Wang, B.; Bentley, G.; Razia, Y.; Qin, X.; Tarr, P.I.; Dantas, G. Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes. PLoS ONE 2013, 8, e78822. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.F.; Peterson, A.E.; Julian, K.G.; Greene, W.H.; Price, L.B.; Nelson, K.; Whitener, C.J.; Silbergeld, E.K. Household risk factors for colonization with multidrug-resistant Staphylococcus aureus isolates. PLoS ONE 2013, 8, e54733. [Google Scholar] [CrossRef] [PubMed]
- McManus, B.A.; Coleman, D.C.; Deasy, E.C.; Brennan, G.I.; O’Connell, B.; Monecke, S.; Ehricht, R.; Leggett, B.; Leonard, N.; Shore, A.C. Comparative genotypes, Staphylococcal cassette chromosome mec (SCCmec) genes and antimicrobial resistance amongst Staphylococcus epidermidis and Staphylococcus haemolyticus isolates from infections in humans and companion animals. PLoS ONE 2015, 10, e0138079. [Google Scholar] [CrossRef]
- Rzewuska, M.; Stefanska, I.; Kizerwetter-Swida, M.; Chrobak-Cmiel, D.; Szczygielska, P.; Lesniak, M.; Binek, M. Characterization of extended-spectrum-β-Lactamases produced by Escherichia coli strains isolated from dogs in Poland. Polish J. Microbiol. 2015, 64, 285–288. [Google Scholar] [CrossRef]
- Schwarz, S.; Johnson, A.P. Transferable resistance to colistin: A new but old threat. J. Antimicrob. Chemother. 2016, 71, 2066–2070. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, C.; Gama, L.T.; Belas, A.; Bergström, K.; Beurlet, S.; Briend-Marchal, A.; Broens, E.M.; Costa, M.; Criel, D.; Damborg, P.; et al. European multicenter study on antimicrobial resistance in bacteria isolated from companion animal urinary tract infections. BMC Vet. Res. 2016, 12, 213. [Google Scholar] [CrossRef]
- Fodor, A.; Varga, I.; Hevesi, M.; Máthé-Fodor, A.; Racskó, J.; Hogan, J.A. Anti–microbial peptides of Xenorhabdus origin against multidrug resistant plant pathogens. In A Search for Antibacterial Agents; Bobbarala, V., Ed.; Tech Press: Rijeka, Croatia, 2012; pp. 147–195. [Google Scholar]
- Załuga, J.; Stragier, P.; Baeyen, S.; Haegeman, A.; Van Vaerenbergh, J.; Maes, M.; De Vos, P. Comparative genome analysis of pathogenic and non-pathogenic Clavibacter strains reveals adaptations to their lifestyle. BMC Genomics 2014, 15, 392. [Google Scholar] [CrossRef]
- Förster, H.; McGhee, G.C.; Sundin, G.W.; Adaskaveg, J.E. Characterization of streptomycin resistance in isolates of Erwinia amylovora in California. Phytopathology 2015, 105, 1302–1310. [Google Scholar] [CrossRef]
- Gusberti, M.; Klemm, U.; Meier, M.S.; Maurhofer, M.; Hunger-Glaser, I. Fire blight control: The struggle goes on. A comparison of different fire blight control methods in Switzerland with respect to biosafety, efficacy and durability. Int. J. Environ. Res. Public Health 2015, 12, 11422–11447. [Google Scholar] [CrossRef]
- Fodor, A.; Abate, B.A.; Deák, P.; Fodor, L.; Klein, M.G.; Makrai, M.; Muvevi, J.; Vozik, D. An overview of multi-antibiotic resistance in pathogenic bacteria—From selected genetic and evolutionary aspects—A review. Preprint 2018, 2018080036. [Google Scholar] [CrossRef]
- Smits, T.H.M.; Duffy, B.; Blom, J.; Ishimaru, C.A.; Stockwell, V.O.; Pantocin, A. A peptide-derived antibiotic involved in biological control by plant-associated Pantoea species. Arch. Microbiol. 2019, 201, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Aćimović, S.D.; Zeng, Q.; McGhee, G.C.; Sundin, G.W.; Wise, J.C. Control of fire blight (Erwinia amylovora) on apple trees with trunk-injected plant resistance inducers and antibiotics and assessment of induction of pathogenesis-related protein genes. Front. Plant Sci. 2015, 6, 16. [Google Scholar] [CrossRef]
- Stockwell, V.O.; Sundin, G.W.; Jones, A.L. Antibiotic use in plant agriculture. Ann. Rev. Phytopathol. 2002, 40, 443–465. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, V.O.; Duffy, B. Use of antibiotics in plant agriculture. Rev. Sci. Tech. 2012, 31, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Talbot, G.H.; Bradley, J.; Edwards, J.E., Jr.; Gilbert, D.; Scheld, M.; Bartlett, J.G. Bad bugs need drugs: An update on the development pipeline from the antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Diseases 2006, 42, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Plésiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in Enterococci. Expert Rev. Anti. Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef]
- Miller, W.R.; Murray, B.E.; Rice, L.B.; Arias, C.A. Vancomycin-Resistant Enterococci: Therapeutic Challenges in the 21st Century. Infect. Dis. Clin. N. Am. 2016, 30, 415–439. [Google Scholar] [CrossRef]
- Miller, W.R.; Bayer, A.S.; Arias, C.A. Mechanism of action and resistance to daptomycin in Staphylococcus aureus and Enterococci. Cold Spring Harb. Perspect. Med. 2016, 6, a026997. [Google Scholar] [CrossRef]
- Khan, A.; Davlieva, M.; Panesso, D.; Rincon, S.; Miller, W.R.; Diaz, L.; Reyes, J.; Cruz, M.R.; Pemberton, O.; Nguyen, A.H.; et al. Antimicrobial sensing coupled with cell membrane remodeling mediates antibiotic resistance and virulence in Enterococcus faecalis. Proc. Natl. Acad. Sci. USA 2019, 116, 26925–26932. [Google Scholar] [CrossRef] [PubMed]
- Conly, J.M.; Johnston, B.L. Where are all the new antibiotics? The new antibiotic paradox. Can. J. Infect. Dis. Med. Microbiol. 2005, 16, 159–160. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.; Strandwitz, P. Antibiotics right under our nose. Nature Microbiol. 2016, 535, 501–502. [Google Scholar] [CrossRef] [PubMed]
- Stubbings, W.; Labischinski, H. New antibiotics for antibiotic-resistant bacteria. F1000 Biol. Rep. 2009, 1, 40. [Google Scholar] [CrossRef] [PubMed]
- Kosikowska, P.; Lesner, A. Antimicrobial peptides (AMPs) as drug candidates: A patent review (2003–2015). Expert Opin. Ther. Pat. 2016, 26, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.K.; Kim, C.; Seo, C.H.; Park, Y. The therapeutic applications of antimicrobial peptides (AMPs): A patent review. J. Microbiol. 2017, 255, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007, 5, 175–186. [Google Scholar] [CrossRef]
- Pál, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G. The structure and diversity of human, animal and environmental resistomes. Microbiome 2016, 4, 54. [Google Scholar] [CrossRef]
- Bryan, B.E. Genetic modifiers of streptomycin resistance in Pneumococcus. J. Bacteriol. 1961, 82, 461–470. [Google Scholar] [CrossRef]
- Hershberg, R. Antibiotic-independent adaptive effects of antibiotic resistance mutations. Trends Genet. 2017, 33, 521–528. [Google Scholar] [CrossRef]
- Georghiou, S.B.; Seifert, M.; Catanzaro, D.G.; Garfein, R.S.; Rodwell, T.C. Increased tuberculosis patient mortality associated with Mycobacterium tuberculosis mutations conferring resistance to second-line antituberculous drugs. J. Clin. Microbiol. 2017, 55, 1928–1937. [Google Scholar] [CrossRef]
- Feuerriegel, S.; Cox, H.S.; Zarkua, N.; Karimovich, H.A.; Braker, K.; Rüsch-Gerdes, S.; Niemann, S. Sequence analyses of just four genes to detect extensively drug-resistant Mycobacterium tuberculosis strains in multidrug-resistant tuberculosis patients undergoing treatment. Antimicrob. Agents Chemother. 2009, 53, 3353–3356. [Google Scholar] [CrossRef] [PubMed]
- Georghiou, S.B.; Magana, M.; Garfein, R.S.; Catanzaro, D.G.; Catanzaro, A.; Rodwell, T.C. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: A systematic review. PLoS ONE 2012, 7, e33275. [Google Scholar] [CrossRef] [PubMed]
- Myneedu, V.P.; Singhal, R.; Khayyam, K.U.; Sharma, P.P.; Bhalla, M.; Behera, D.; Sarin, R. First and second line drug resistance among treatment naïve pulmonary tuberculosis patients in a district under Revised National Tuberculosis Control Programme (RNTCP) in New Delhi. J. Epidemiol. Glob. Health 2015, 5, 365–373. [Google Scholar] [CrossRef]
- Dookie, N.; Rambaran, S.; Padayatchi, N.; Mahomed, S.; Naidoo, K. Evolution of drug resistance in Mycobacterium tuberculosis: A review on the molecular determinants of resistance and implications for personalized care. J. Antimicrob. Chemother. 2018, 73, 1138–1151. [Google Scholar] [CrossRef] [PubMed]
- Tomasz, A.; Albino, A.; Zanati, E. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 1970, 227, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Tiniakow, G.G.; Terentieva, E.L. Cubitus interruptus, a new genovariation of the fourth chromosome of Drosophila melanogaster. Genetics 1933, 18, 117–120. [Google Scholar] [PubMed]
- Thompson, J.N., Jr. A test of the influence of isoallelic variation upon a quantitative character. Heredity 1975, 35, 401–406. [Google Scholar] [CrossRef]
- Villee, C.A.; Lavin, G.I. The production of phenocopies in Drosophila using visible light and a photodynamic dye. Anat. Rec. 1946, 96, 565. [Google Scholar] [CrossRef]
- Ivanovics, G.; Lantos, J. Phenocopy of resistance to phage W in Bacillus anthracis. Acta Microbiol. Acad. Sci. Hung. 1962, 9, 237–246. [Google Scholar]
- Ruppé, E.; Cherkaoui, A.; Lazarevic, V.; Emonet, S.; Schrenzel, J.O. Establishing genotype-to-phenotype relationships in bacteria causing hospital-acquired pneumonia: A prelude to the application of clinical metagenomics. Antibiotics 2017, 6, E30. [Google Scholar] [CrossRef]
- Arps, P.J.; Winkler, M.E. Structural analysis of the Escherichia coli K-12 hisT operon by using a kanamycin resistance cassette. J. Bacteriol. 1987, 169, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Domingues, S.; Da Silva, G.J.; Nielsen, K.M. Global dissemination patterns of common gene cassette arrays in class 1 integrons. Microbiology 2015, 161, 1313–1337. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, D.; Peters, B.M.; Li, L.; Li, B.; Xu, Z.; Shirliff, M.E. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb. Pathol. 2016, 101, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Rosdahl, V.T. Localisation of the penicillinase gene in naturally occurring Staphylococcus aureus strains. Acta Pathol. Microbiol. Scand. Ser. B Microbiol. 1985, 93, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Rosdahl, V.T.; Rosendal, K. Resistance to cadmium, arsenate and mercury among Danish strains of Staphylococcus aureus isolated from cases of bacteriaemia, 1957–1974. J. Med. Microbiol. 1980, 13, 383–391. [Google Scholar] [CrossRef]
- Si, H.; Zhang, W.J.; Chu, S.; Wang, X.M.; Dai, L.; Hua, X.; Dong, Z.; Schwarz, S.; Liu, S. Novel plasmid-borne multidrug resistance gene cluster including lsa (E) from a linezolid-resistant Enterococcus faecium isolate of swine origin. Antimicrob. Agents Chemother. 2015, 59, 7113–7116. [Google Scholar] [CrossRef]
- San Millan, A. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 2018, 26, 978–985. [Google Scholar] [CrossRef]
- Wein, T.; Hülter, N.F.; Mizrahi, I.; Dagan, T. Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat. Commun. 2019, 10, 2595. [Google Scholar] [CrossRef]
- Tauch, A.; Schlüter, A.; Bischoff, N.; Goesmann, A.; Meyer, F.; Pühler, A. The 79,370-bp conjugative plasmid pB4 consists of an IncP-1beta backbone loaded with a chromate resistance transposon, the strA-strB streptomycin resistance gene pair, the oxacillinase gene bla(NPS-1), and a tripartite antibiotic efflux system of the resistance-nodulation-division family. Mol. Genet. Genom. 2003, 268, 570–584. [Google Scholar]
- Poey, M.E.; Azpiroz, M.F.; Laviña, M. On sulfonamide resistance, sul genes, class 1 integrons and their horizontal transfer in Escherichia coli. Microb. Pathog. 2019, 135, 103611. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Ogata, C.; Sato, S. Episome-mediated transfer of drug resistance in Enterobacteriaceae VIII. J. Bacteriol. 1964, 88, 922–928. [Google Scholar] [CrossRef]
- Soufi, L.; Abbassi, M.S.; Sáenz, Y.; Vinué, L.; Somalo, S.; Zarazaga, M.; Abbas, A.; Dbaya, R.; Khanfir, L.; Ben Hassen, A.; et al. Prevalence and diversity of integrons and associated resistance genes in Escherichia coli isolates from poultry meat in Tunisia. Foodborne Pathog. Dis. 2009, 6, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Salyers, A.A.; Shoemaker, N.B. Resistance gene transfer in anaerobes: New insights, new problems. Clin. Infect. Dis. 1996, 23, S36–S43. [Google Scholar] [CrossRef] [PubMed]
- Butaye, P.; Cloeckaert, A.; Schwarz, S. Mobile genes coding for efflux-mediated antimicrobial resistance in Gram-positive and Gram-negative bacteria. Int. J. Antimicrob. Agents 2003, 22, 205–210. [Google Scholar] [CrossRef]
- Poirel, L.; Bonnin, R.A.; Nordmann, P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life 2011, 63, 1061–1067. [Google Scholar] [CrossRef]
- Wendlandt, S.; Shen, J.; Kadlec, K.; Wang, Y.; Li, B.; Zhang, W.J.; Feßler, A.T.; Wu, C.; Schwarz, S. Multidrug resistance genes in staphylococci from animals that confer resistance to critically and highly important antimicrobial agents in human medicine. Trends Microbiol. 2015, 23, 44–54. [Google Scholar] [CrossRef]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- García-Solache, M.; Lebreton, F.; McLaughlin, R.E.; Whiteaker, J.D.; Gilmore, M.S.; Rice, L.B. Homologous recombination within large chromosomal regions facilitates acquisition of β-lactam and vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 2016, 60, 5777–5786. [Google Scholar] [CrossRef]
- Shin, J.E.; Lin, C.; Lim, H.N. Horizontal transfer of DNA methylation patterns into bacterial chromosomes. Nucleic Acids Res. 2016, 44, 4460–4471. [Google Scholar] [CrossRef]
- Heuer, H.; Smalla, K. Horizontal gene transfer between bacteria. Environ. Biosafety Res. 2007, 6, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Broaders, E.; Gahan, G.M.; Marchesi, J.R. Mobile genetic elements of the human gastrointestinal tract. Potential for spread of antibiotic resistance genes. Gut Microbes 2013, 4, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, R.; Rolinson, G.N. Characteristics of methicillin-resistant Staphylococci. J. Bacteriol. 1964, 87, 887–899. [Google Scholar] [CrossRef]
- Arzanlou, M.; Chai, W.C.; Venter, H. Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem. 2017, 61, 49–59. [Google Scholar] [PubMed]
- Dalbadie-McFarland, G.; Neitzel, J.J.; Richards, J.H. Active-site mutants of beta-lactamase: Use of an inactive double mutant to study requirements for catalysis. Biochemistry 1986, 25, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.H.; Labgold, M.R.; Richards, J.H. Altering enzymatic activity: Recruitment of carboxypeptidase activity into an RTEM beta-lactamase/penicillin-binding protein 5 chimera. Proc. Natl. Acad Sci. USA 1990, 87, 2823–2827. [Google Scholar] [CrossRef]
- Yan, Y.H.; Li, G.; Li, G.B. Principles and current strategies targeting metallo-β-lactamase mediated antibacterial resistance. Med. Res. Rev 2020, in press. [Google Scholar] [CrossRef]
- Baltz, R.H. Spontaneous and induced mutations to rifampicin, streptomycin and spectinomycin resistances in actinomycetes: Mutagenic mechanisms and applications for strain improvement. J. Antibiot. 2014, 67, 619–624. [Google Scholar] [CrossRef]
- Behmard, E.; Najafi, A.; Ahmadi, A. Understanding the resistance mechanism of penicillin binding protein 1a mutant against cefotaxime using molecular dynamic simulation. J. Biomol. Struct. Dyn. 2019, 37, 741–749. [Google Scholar] [CrossRef]
- Poole, K. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 2001, 3, 255–264. [Google Scholar]
- Poole, K. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 2004, 10, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Blanco, P.; Alcalde-Rico, M.; Corona, F.; Reales-Calderón, J.A.; Sánchez, M.B.; Martínez, J.L. Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resist. Update 2016, 28, 13–27. [Google Scholar] [CrossRef]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef]
- Juhas, M. Horizontal gene transfer in human pathogens. Crit. Rev. Microbiol. 2015, 41, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef]
- Fullmer, M.S.; Ouellette, M.; Louyakis, A.S.; Papke, R.T.; Gogarten, J.P. The Patchy Distribution of Restriction—Modification System Genes and the Conservation of Orphan Methyltransferases in Halobacteria. Genes 2019, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Lin, J. Factors influencing horizontal gene transfer in the intestine. Anim. Health Res. Rev. 2017, 18, 153–159. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Van Schaik, W.; Top, J.; Riley, D.R.; Boekhorst, J.; Vrijenhoek, J.E.; Schapendonk, C.M.; Hendrickx, A.P.; Nijman, I.J.; Bonten, M.J.; Tettelin, H.; et al. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genom. 2010, 11, 239. [Google Scholar] [CrossRef]
- Coombs, J.M. Potential for horizontal gene transfer in microbial communities of the terrestrial subsurface. Methods Mol. Biol. 2009, 532, 413–433. [Google Scholar] [PubMed]
- Tyrrell, C.; .Burgess, C.M.; Brennan, F.P.; Walsh, F. Antibiotic resistance in grass and soil. Biochem Soc. Trans. 2019, 47, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Giordano, C.; Barnini, S.; Tsioutis, C.; Chlebowicz, M.A.; Scoulica, E.V.; Gikas, A.; Rossen, J.W.; Friedrich, A.W.; Bathoorn, E. Expansion of KPC-producing Klebsiella pneumoniae with various mgrB mutations giving rise to colistin resistance: The role of ISL3 on plasmids. Int. J. Antimicrob. Agents 2018, 51, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Scoulica, E.V.; Neonakis, I.K.; Gikas, A.I.; Tselentis, Y.J. Spread of bla (VIM-1)-producing E. coli in a university hospital in Greece. Genetic analysis of the integron carrying the bla (VIM-1) metallo-beta-lactamase gene. Diagn. Microbiol. Infect. Dis. 2004, 48, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Gray, T.A.; Derbyshire, K.M. Blending genomes: Distributive conjugal transfer in mycobacteria, a sexier form of HGT. Mol. Microbiol. 2018, 108, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti. Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef]
- Schwaber, M.J.; Navon-Venezia, S.; Schwartz, D.; Carmeli, Y. High levels of antimicrobial coresistance among extended-spectrum-β-lactamase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2005, 49, 2137–2139. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: “No ESKAPE”. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESCAPE! An Update from the Infectious Diseases Society of America. Clin. Infec. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Rice, L.B. Progress and challenges in implementing the research on ESKAPE pathogens. Infect. Control. Hosp. Epidemiol. 2010, 1, S7–S10. [Google Scholar] [CrossRef]
- Schwaber, M.J.; Carmeli, Y. An ongoing national intervention to contain the spread of carbapenem-resistant Enterobacteriaceae. Clin. Infect. Dis. 2014, 58, 697–703. 426. [Google Scholar] [CrossRef] [PubMed]
- Sandiumenge, A.; Lisboa, T.; Gomez, F.; Hernandez, P.; Canadell, L.; Rello, J. Effect of antibiotic diversity on ventilator-associated pneumonia caused by ESKAPE organisms. Chest 2011, 140, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Penes, N.O.; Muntean, A.A.; Moisoiu, A.; Muntean, M.M.; Chirca, A.; Bogdan, M.A.; Popa, M.I. An overview of resistance profiles ESKAPE pathogens from 2010–2015 in a tertiary respiratory center in Romania. Rom. J. Morphol. Embryol. 2017, 58, 909–922. [Google Scholar] [PubMed]
- Azzopardi, E.A.; Azzopardi, S.M.; Boyce, D.E.; Dickson, W.A. Emerging gram-negative infections in burn wounds. J. Burn Care Res. 2011, 32, 570–576. [Google Scholar] [CrossRef]
- Navidinia, M. The clinical importance of emerging ESKAPE pathogens in nosocomial infections. J. Paramed. Sci. 2016, 7, 2008–4978. [Google Scholar]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Bodro, M.; Gudiol, C.; Garcia-Vidal, C.; Tubau, F.; Contra, A.; Boix, L.; Domingo-Domenech, E.; Calvo, M.; Carratalà, J. Epidemiology, antibiotic therapy and outcomes of bacteremia caused by drug-resistant ESKAPE pathogens in cancer patients. Support Care Cancer 2014, 22, 603–610. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Horne, D.; Tomasz, A. Tolerant response of Streptococcus sanguis to beta-lactams and other cell wall inhibitors. Antimicrob. Agents Chemother. 1977, 11, 888–896. [Google Scholar] [CrossRef][Green Version]
- Tomasz, A. Penicillin-binding proteins and the antibacterial effectiveness of beta-lactam antibiotics. Rev. Infect. Dis. 1986, 8, S260–S278. [Google Scholar] [CrossRef]
- Quinn, J.P. Clinical significance of extended-spectrum beta-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 1994, 13, S39–S42. [Google Scholar] [CrossRef] [PubMed]
- McDanel, J.; Schweizer, M.; Crabb, V.; Nelson, R.; Samore, M.; Khader, K.; Blevins, A.E.; Diekema, D.; Chiang, H.Y.; Nair, R.; et al. Incidence of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella infections in the United States: A systematic literature review. Infect. Control. Hosp. Epidemiol. 2017, 38, 1209–1215. [Google Scholar] [CrossRef] [PubMed]
- Flokas, M.E.; Alevizakos, M.; Shehadeh, F.; Andreatos, N.; Mylonakis, E. Extended-spectrum β-lactamase-producing Enterobacteriaceae colonisation in long-term care facilities: A systematic review and meta-analysis. Int. J. Antimicrob. Agents 2017, 50, 649–656. [Google Scholar] [CrossRef]
- Dame-Korevaar, A.; Fischer, E.A.J.; van der Goot, J.; Stegeman, A.; Mevius, D. Transmission routes of ESBL/pAmpC producing bacteria in the broiler production pyramid, a literature review. Prev. Vet. Med. 2019, 162, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D. Multiresistant Enterobacteriaceae: New threat of an old problem. Expert Rev. Anti. Infect. Ther. 2008, 6, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Sacco, E.; Cortes, M.; Josseaume, N.; Bouchier, C.; Dubée, V.; Hugonnet, J.E.; Mainardi, J.L.; Rice, L.B.; Arthur, M. Mutation landscape of acquired cross-resistance to glycopeptide and β-lactam antibiotics in Enterococcus faecium. Antimicrob. Agents Chemother. 2015, 59, 5306–5315. [Google Scholar] [CrossRef][Green Version]
- Tiwari, S.; Jamal, S.B.; Hassan, S.S.; Carvalho, P.V.; Almeida, S.; Barh, D.; Ghosh, P.; Silva, A.; Castro, T.L.; Azevedo, V. Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: An overview. Front. Microbiol. 2017, 8, 1878. [Google Scholar] [CrossRef]
- Spengler, G.; Kincses, A.; Gajdács, M.; Amaral, L. New roads leading to old destinations: Efflux pumps as targets to reverse multidrug resistance in bacteria. Molecules 2017, 22, E468. [Google Scholar] [CrossRef]
- Schmutzhard, E.; Williams, K.J.; Vukmirovits, G.; Chmelik, V.; Pfausler, B.; Featherstone, A. The Meropenem Meningitis Study Group 1995: A randomized comparison of meropenem with cefotaxime or ceftriaxone for the treatment of bacterial meningitis in adults. J. Antimicrobiol. Chemother. 1995, 36, 85–97. [Google Scholar] [CrossRef]
- Almeida, M.V.A.; Cangussú, Í.M.; Carvalho, A.L.S.; Brito, I.L.P.; Costa, R.A. Drug resistance, AmpC-β-lactamase and extended-spectrum β-lactamase-producing Enterobacteriaceae isolated from fish and shrimp. Rev. Inst. Med. Trop. Sao Paulo 2017, 59, e70. [Google Scholar] [CrossRef]
- Brewer, N.S.; Hellinger, W.C. The monobactams. Mayo Clin. Proc. 1991, 66, 1152–1157. [Google Scholar] [CrossRef]
- Jain, P.; Roy, S.; Viswanathan, R.; Basu, S.; Singh, A.K.; Dutta, S. Concurrent and transferable resistance to extended-spectrum cephalosporins, monobactam and fluoroquinolone in a Salmonella enterica serovar Worthington blood isolate from a neonate in Kolkata, India. Int. J. Antimicrob. Agents 2013, 41, 494–495. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, C.; MacGowan, A.P. A review of the pharmacokinetics and pharmacodynamics of aztreonam. J. Antimicrob. Chemother. 2016, 71, 2704–2712. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.J.; Wilkowske, C.J. The penicillins. Mayo Clin. Proc. 1987, 62, 806–820. [Google Scholar] [CrossRef]
- Adnan, S.; Paterson, D.L.; Lipman, J.; Roberts, J.A. Ampicillin/sulbactam: Its potential use in treating infections in critically ill patients. Int. J. Antimicrob. Agents 2013, 42, 384–389. [Google Scholar] [CrossRef]
- Chen, C.W.; Ming, C.C.; Ma, C.J.; Shan, Y.S.; Yeh, Y.S.; Wang, J.Y. Prospective, randomized, study of ampicillin-sulbactam versus moxifloxacin monotherapy for the treatment of community-acquired complicated intra-abdominal infections. Surg. Infect. 2013, 4, 389–396. [Google Scholar] [CrossRef]
- Housman, S.T.; Hagihara, M.; Nicolau, D.P.; Kuti, J.L. In vitro pharmacodynamics of human-simulated exposures of ampicillin/sulbactam, doripenem and tigecycline alone and in combination against multidrug-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 2013, 8, 2296–2304. [Google Scholar] [CrossRef][Green Version]
- Yokoyama, Y.; Matsumoto, K.; Ikawa, K.; Watanabe, E.; Yamamoto, H.; Imoto, Y.; Morikawa, N.; Takeda, Y. The pharmacokinetics of ampicillin-sulbactam in anuric patients: Dosing optimization for prophylaxis during cardiovascular surgery. Int. J. Clin. Pharm. 2016, 38, 771–775. [Google Scholar] [CrossRef]
- Hoogkamp-Korstanje, J.A.; Westerdaal, N. Activity and synergy of ureido penicillins and aminoglycosides against Pseudomonas aeruginosa. Infection 1982, 10, S257–S261. [Google Scholar] [CrossRef]
- Giamarellou, H.; Antoniadou, A. Antipseudomonal antibiotics. Med. Clin. N. Am. 2001, 85, 19–42. [Google Scholar] [CrossRef]
- Butterfield, J.M.; Lodise, T.P.; Beegle, S.; Rosen, J.; Farkas, J.; Pai, M.P. Pharmacokinetics and pharmacodynamics of extended-infusion piperacillin/tazobactam in adult patients with cystic fibrosis-related acute pulmonary exacerbations. J. Antimicrob. Chemother. 2014, 9, 176–180. [Google Scholar] [CrossRef][Green Version]
- Lee, J.; Oh, C.E.; Choi, E.H.; Lee, H.J. The impact of the increased use of piperacillin/ tazobactam on the selection of antibiotic resistance among invasive Escherichia coli and Klebsiella pneumoniae isolates. Int. J. Infect. Dis. 2013, 17, e638–e643. [Google Scholar] [CrossRef] [PubMed]
- Shubert, C.; Slaughter, J.; Creely, D.; van Belkum, A.; Gayral, J.P.; Dunne, W.M.; Zambardi, G.; Shortridge, D. Population analysis of Escherichia coli isolates with discordant resistance levels by piperacillin-tazobactam broth microdilution and agar dilution testing. Antimicrob. Agents Chemother. 2014, 58, 1779–1781. [Google Scholar] [CrossRef] [PubMed]
- Nichols, K.; Chung, E.K.; Knoderer, C.A.; Buenger, L.E.; Healy, D.P.; Dees, J.; Crumby, A.S.; Kays, M.B. Population pharmacokinetics and pharmacodynamics of extended-infusion piperacillin and tazobactam in critically ill children. Antimicrob. Agents Chemother. 2016, 60, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Rahbar, M.; Van de Velde, S.; Eslami, P.; Mardani, M. Activity of temocillin and comparators against urinary Escherichia coli and Klebsiella pneumoniae from Iran. Eur. J. Clin. Microbiol. Infect. Dis. 2020, in press. [Google Scholar]
- Mullins, B.P.; Kramer, C.J.; Bartel, B.J.; Catlin, J.S.; Gilder, R.E. Comparison of the nephrotoxicity of vancomycin in combination with cefepime, meropenem, or piperacillin/tazobactam: A prospective, multicenter study. Ann. Pharmacother. 2018, 52, 639–644. [Google Scholar] [CrossRef]
- Giacobbe, D.R.; Bassetti, M.; De Rosa, F.G.; Del Bono, V.; Grossi, P.A.; Menichetti, F.; Pea, F.; Rossolini, G.M.; Tumbarello, M.; Viale, P.; et al. ISGRI-SITA (Italian Study Group on Resistant Infections of the Società Italiana Terapia Antinfettiva). Ceftolozane/tazobactam: Place in therapy. Expert Rev. Anti. Infect. Ther. 2018, 16, 307–320. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Wiebe, R.; Dilay, L.; Thomson, K.; Rubinstein, E.; Hoban, D.J.; Noreddin, A.M.; Karlowsky, J.A. Comparative review of the carbapenems. Drugs 2007, 67, 1027–1052. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Lawrence, C.K.; Adam, H.; Schweizer, F.; Zelenitsky, S.; Zhanel, M.; Lagacé-Wiens, P.R.S.; Walkty, A.; Denisuik, A.; Golden, A.; et al. Imipenem-Relebactam and Meropenem-Vaborbactam: Two novel carbapenem-β-lactamase inhibitor combinations. Drugs 2018, 78, 65–98. [Google Scholar] [CrossRef]
- Mashni, O.; Nazer, L.; Le, J. Critical review of double-carbapenem therapy for the treatment of carbapenemase-producing Klebsiella pneumonia. Ann. Pharmacother. 2019, 53, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Abuhussain, S.S.A.; Sutherland, C.A.; Nicolau, D. In vitro potency of antipseudomonal β-lactams against blood and respiratory isolates of P. aeruginosa collected from US hospitals. J. Thorac. Dis. 2019, 11, 1896–1902. [Google Scholar] [CrossRef] [PubMed]
- Breilh, D.; Texier-Maugein, J.; Allaouchiche, B.; Saux, M.C.; Boselli, E. Carbapenems. J. Chemother. 2013, 25, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Papp-Wallace, K.; Endimiani, A.; Magdalena, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, present, and future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef] [PubMed]
- Paczkowska, M.; Garbacki, P.; Zalewski, P.; Talaczyńska, A.; Cielecka-Piontek, J. Meropenem--therapeutic recommendation after twenty years of presence on pharmaceutical market. Postepy Hig. Med. Dosw. 2014, 68, 441–445. [Google Scholar] [CrossRef]
- Wong, G.; Farkas, A.; Sussman, R.; Daroczi, G.; Hope, W.W.; Lipman, J.; Roberts, J.A. Comparison of the accuracy and precision of pharmacokinetic equations to predict free meropenem concentrations in critically ill patients. Antimicrob. Agents Chemother. 2015, 59, 1411–1417. [Google Scholar] [CrossRef]
- Gupta, N.; Limbago, B.M.; Patel, J.B.; Kallen, A.J. Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin. Infect. Dis. 2011, 53, 60–67. [Google Scholar] [CrossRef]
- Wilson, A.P.R. Sparing carbapenem usage. J. Antimicrob. Chemother. 2017, 72, 2410–2417. [Google Scholar] [CrossRef]
- Beigverdi, R.; Sattari-Maraji, A.; Emaneini, M.; Jabalameli, F. Status of carbapenem-resistant Acinetobacter baumannii harboring carbapenemase: First systematic review and meta-analysis from Iran. Infect. Genet. Evol. 2019, 73, 433–443. [Google Scholar] [CrossRef]
- Turton, J.F.; Woodford, N.; Glover, J.; Yarde, S.; Kaufmann, M.E.; Pitt, T.L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like Carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 2006, 44, 2974–2976. [Google Scholar] [CrossRef]
- Turton, J.F.F.; Baddal, B.; Perry, C. Use of the accessory genome for characterization and typing of Acinetobacter baumannii. J. Clin. Microbiol. 2011, 49, 1260–1266. [Google Scholar] [CrossRef] [PubMed]
- Kobs, V.C.; Ferreira, J.A.; Bobrowicz, T.A.; Ferreira, L.E.; Deglmann, R.C.; Westphal, G.A.; França, P.H.C.D. The role of the genetic elements bla oxa and IS Aba 1 in the Acinetobacter calcoaceticus-Acinetobacter baumannii complex in carbapenem resistance in the hospital setting. Rev. Soc. Bras. Med. Trop. 2016, 49, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Maurya, A.P.; Dhar, D.; Basumatary, M.K.; Paul, D.; Ingti, B.; Choudhury, D.; Talukdar, A.D.; Chakravarty, A.; Mishra, S.; Bhattacharjee, A. Expansion of highly stable bla OXA-10 β-lactamase family within diverse host range among nosocomial isolates of Gram-negative bacilli within a tertiary referral hospital of Northeast India. BMC Res. Notes 2017, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Yazdansetad, S.; Najari, E.; Ghaemi, E.A.; Javid, N.; Hashemi, A.; Ardebili, A. Carbapenem-resistant Acinetobacter baumannii isolates carrying blaOXA genes with upstream ISAba1: First report of a novel OXA subclass from Iran. J. Glob. Antimicrob. Resist. 2019, 18, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Noval, M.; Banoub, M.; Claeys, K.C.; Heil, E. The battle is on: New beta-lactams for the treatment of multidrug-resistant gram-negative organisms. Curr. Infect. Dis. Rep. 2020, 22, 1. [Google Scholar] [CrossRef]
- Mugnier, M.D.; Poirel, L.; Naas, T.; Nordmann, P. Worldwide dissemination of the (blaOXA-23) carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 2010, 16, 35–40. [Google Scholar] [CrossRef]
- Effah, C.Y.; Sun, T.; Liu, S.; Wu, Y. Klebsiella pneumoniae: An increasing threat to public health. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1. [Google Scholar] [CrossRef]
- Singh, A.R. Science, Names Giving and Names Calling: Change NDM-1 to PCM. Mens Sana Monogr. 2011, 9, 294–319. [Google Scholar] [CrossRef]
- Pittalis, S.; Ferarro, F.; Puro, V. NDM-1: The superbug? Infez. Med. 2011, 19, 224–234. (In Italian) [Google Scholar]
- Qu, H.; Wang, X.; Ni, Y.; Liu, J.; Tan, R.; Huang, J.; Li, L.; Sun, J. NDM-1-producing Enterobacteriaceae in a teaching hospital in Shanghai, China: IncX3-type plasmids may contribute to the dissemination of blaNDM-1. Int. J. Infect. Dis. 2015, 34, 8–13. [Google Scholar] [CrossRef]
- Greninger, A.L.; Chorny, I.; Knowles, S.; Ng, V.L.; Chaturvedi, V. Draft genome ssequences of four NDM-1-producing Klebsiella pneumoniae strains from a health care nacility in Northern California. Genome Announc. 2015, 3, e00421-15. [Google Scholar] [CrossRef] [PubMed]
- Hamzan, N.; Yean, C.Y.; Rahman, R.A.; Hasan, H.; Rahman, Z.A. Detection of blaIMP4 and blaNDM1 harboring Klebsiella pneumoniae isolates in a university hospital in Malaysia. Emerg. Health Threats J. 2015, 8, 26011. [Google Scholar] [CrossRef] [PubMed]
- Schuelter-Trevisol, F.; Schmitt, G.J.; Araújo, J.M.D.; Souza, L.B.D.; Nazário, J.G.; Januário, R.L.; Mello, R.S.D.; Trevisol, D.J. New Delhi metallo-beta-lactamase-1-producing Acinetobacter spp. infection: Report of a survivor. Rev. Soc. Bras. Med. Trop. 2016, 49, 130–134. [Google Scholar] [CrossRef]
- Kazmierczak, K.M.; Rabine, S.; Hackel, M.; McLaughlin, R.E.; Biedenbach, D.J.; Bouchillon, S.K.; Sahm, D.F.; Bradford, P.A. Multiyear, multinational survey of the incidence and global distribution of metallo-β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Zaman, T.U.; Alrodayyan, M.; Albladi, M.; Aldrees, M.; Siddique, M.I.; Aljohani, S.; Balkhy, H.H. Clonal diversity and genetic profiling of antibiotic resistance among multidrug/carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary care hospital in Saudi Arabia. BMC Infect. Dis. 2018, 18, 205. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.; Sáez, D.; Bautista, V.; Fernández-Romero, S.; Lara, N.; Aracil, B.; Pérez-Vázquez, M.; Campos, J.; Oteo, J.; Aznar, J.E. Spanish Collaborating Group for the Antibiotic Resistance Surveillance Programme: Carbapenemase-producing Escherichia coli is becoming more prevalent in Spain mainly because of the polyclonal dissemination of OXA-48. J. Antimicrob. Chemother. 2016, 71, 2131–2138. [Google Scholar] [CrossRef]
- Çicek, A.Ç.; Düzgün, A.Ö.; Saral, A.; Kayman, T.; Çİzmecİ, Z.; Balcı, P.Ö.; Dal, T.; Fırat, M.; Tosun, İ.; Alıtntop, Y.A.; et al. Detection of class 1 integron in Acinetobacter baumannii isolates collected from nine hospitals in Turkey. Asian Pac. J. Trop. Biomed. 2013, 3, 743–747. [Google Scholar] [CrossRef]
- Iraz, M.; Duzgun, A.O.; Cicek, A.C.; Bonnin, R.A.; Ceylan, A.; Saral, A.; Nordmann, P.; Sandalli, C. Characterization of novel VIM carbapenemase, VIM-38, and first detection of GES-5 carbapenem-hydrolyzing β-lactamases in Pseudomonas aeruginosa in Turkey. Diagn. Microbiol. Infect. Dis. 2014, 78, 292–294. [Google Scholar] [CrossRef]
- Ku, W.W.; Kung, C.H.; Lee, C.H.; Tseng, C.P.; Wu, P.F.; Kuo, S.C.; Chen, T.L.; Lee, Y.T.; Wang, F.D.; Fung, C.P. Evolution of carbapenem resistance in Acinetobacter baumannii: An 18-year longitudinal study from a medical center in northern Taiwan. J. Microbiol. Immunol. Infect. 2015, 48, 57–64. [Google Scholar] [CrossRef][Green Version]
- Temkin, E.; Adler, A.; Lerner, A.; Carmeli, Y. Carbapenem-resistant Enterobacteriaceae: Biology, epidemiology, and management. Ann. N. Y. Acad. Sci. 2015, 1323, 22–42. [Google Scholar] [CrossRef]
- Milan, A.; Furlanis, L.; Cian, F.; Bressan, R.; Luzzati, R.; Lagatolla, C.; Deiana, M.L.; Knezevich, A.; Tonin, E.; Dolzani, L. Epidemic dissemination of a carbapenem-resistant Acinetobacter baumannii clone carrying arma two years after its first isolation in an Italian hospital. Microb. Drug Resist. 2016, 22, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Swathi, C.H.; Chikala, R.; Ratnakar, K.S.; Sritharan, V. A structural, epidemiological & genetic overview of Klebsiella pneumoniae carbapenemases (KPCs). Indian J. Med. Res. 2016, 144, 21–31. [Google Scholar] [PubMed]
- Piazza, A.; Caltagirone, M.; Bitar, I.; Nucleo, E.; Spalla, M.; Fogato, E.; D’Angelo, R.; Pagani, L.; Migliavacca, R. Emergence of Escherichia coli sequence type 131 (ST131) and ST3948 with KPC-2, KPC-3 and KPC-8 carbapenemases from a long-term care and rehabilitation facility (LTCRF) in Northern Italy. Adv. Exp. Med. Biol. 2016, 901, 77–89. [Google Scholar] [PubMed]
- Stansly, P.G.; Shepherd, R.G.; White, J. Polymyxin: A new chemotherapeutic agent. Bull. Johns Hopkins Hosp. 1947, 81, 43–54. [Google Scholar] [PubMed]
- Vaara, M.; Porro, M. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 1996, 40, 1801–1805. [Google Scholar] [CrossRef]
- Falagas, M.E.; Rafailidis, P.I. Attributable mortality of Acinetobacter baumannii: No longer a controversial issue. Crit. Care 2007, 11, 134. [Google Scholar] [CrossRef]
- Falagas, M.E.; Rafailidis, P.I. Nephrotoxicity of colistin: New insight into an old antibiotic. Clin. Infect. Dis. 2009, 48, 1729–1731. [Google Scholar] [CrossRef]
- Kádár, B.; Kocsis, B.; Nagy, K.; Szabó, D. The renaissance of polymyxins. Curr. Med. Chem. 2013, 20, 3759–3773. [Google Scholar] [CrossRef]
- Kelesidis, T.; Falagas, M.E. The safety of polymyxin antibiotics. Expert Opin. Drug Safety 2015, 14, 1687–1701. [Google Scholar] [CrossRef]
- Poulikakos, P.; Tansarli, G.S.; Falagas, M.E. Combination antibiotic treatment versus monotherapy for multidrug-resistant, extensively drug-resistant, and pandrug-resistant Acinetobacter infections: A systematic review. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1675–1685. [Google Scholar] [CrossRef]
- Loho, T.; Dharmayanti, A. Colistin: An antibiotic and its role in multiresistant Gram-negative infections. Acta. Med. Indones. 2015, 47, 157–168. [Google Scholar] [PubMed]
- Cai, Y.; Cha, D.; Wang, R.; Liang, B.; Bai, N. Colistin resistance of Acinetobacter baumannii: Clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 2012, 67, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Lee, W.; Kwa, A.L. Polymyxin B versus colistin: An update. Expert Rev. Anti Infect. Ther. 2015, 13, 1481–1497. [Google Scholar] [CrossRef] [PubMed]
- Vaara, M.; Vaara, T.; Jensen, M.; Helander, I.; Nurminen, M.; Rietschel, E.T.; Mäkelä, P.H. Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett. 1981, 129, 145–149. [Google Scholar] [CrossRef]
- El-Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: an extended review (2000–2019). Emerg Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zeng, X.; Li, X.P.; Liao, X.P.; Liu, Y.H.; Lin, J. Plasmid-mediated colistin resistance in animals: Current status and future directions. Anim. Health Res. Rev. 2017, 18, 136–152. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, H.; Liu, Y.H.; Feng, Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018, 26, 794–808. [Google Scholar] [CrossRef]
- Al-Tawfiq, J.A.; Laxminarayan, R.; Mendelson, M. How should we respond to the emergence of plasmid-mediated colistin resistance in humans and animals? Int. J. Infect. Dis. 2017, 54, 77–84. [Google Scholar] [CrossRef]
- Lima, T.; Domingues, S.; Da Silva, G.J. Plasmid-mediated colistin resistance in Salmonella enterica: A Review. Microorganisms 2019, 7, 55. [Google Scholar] [CrossRef]
- Anyanwu, M.U.; Jaja, I.F.; Nwobi, O.C. Occurrence and characteristics of mobile colistin resistance (mcr) gene-containing isolates from the environment: A Review. Int. J. Environ. Res. Public Health 2020, 17, E1028. [Google Scholar] [CrossRef]
- Karaiskos, I.; Souli, M.; Galani, I.; Giamarellou, H. Colistin: Still a lifesaver for the 21st century? Expert Opin. Drug Metab. Toxicol. 2017, 13, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, G.J.; Domingues, S. Interplay between colistin resistance, virulence and fitness in Acinetobacter baumannii. Antibiotics 2017, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed]
- Bialvaei, A.Z.; Kafil, S.H. Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 2015, 31, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Qin, W.; Lin, J.; Fang, S.; Qiu, J. Antibacterial mechanisms of polymyxin and bacterial resistance. Biomed Res. Int. 2015, 2015, 679109. [Google Scholar] [CrossRef] [PubMed]
- Otter, J.A.; Doumith, M.; Davies, F.; Mookerjee, S.; Dyakova, E.; Gilchrist, M.; Brannigan, E.T.; Bamford, K.; Galletly, T.; Donaldson, H.; et al. Emergence and clonal spread of colistin resistance due to multiple mutational mechanisms in carbapenemase-producing Klebsiella pneumoniae in London. Sci. Rep. 2017, 7, 12711. [Google Scholar] [CrossRef] [PubMed]
- López-Causapé, C.; Cabot, G.; Del Barrio-Tofiño, E.; Oliver, A. The versatile mutational resistome of Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 685. [Google Scholar] [CrossRef] [PubMed]
- Mlynarcik, P.; Kolar, M. Molecular mechanisms of polymyxin resistance and detection of mcr genes. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2019, 163, 28–38. [Google Scholar] [CrossRef]
- Caniaux, I.; van Belkum, A.; Zambardi, G.; Poirel, L.; Gros, M.F. MCR: Modern colistin resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 415–420. [Google Scholar] [CrossRef]
- Feng, Y. Transferability of MCR-1/2 polymyxin resistance: Complex dissemination and genetic mechanism. ACS Infect. Dis. 2018, 4, 291–300. [Google Scholar] [CrossRef]
- Jochumsen, N.; Marvig, R.L.; Damkiær, S.; Jensen, R.L.; Paulander, W.; Molin, S.; Jelsbak, L.; Folkesson, A. The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nature Comm. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, V.B.; Venkataramaiah, M.; Mondal, A.; Rajamohan, G. Functional characterization of AbeD, an RND-type membrane transporter in antimicrobial resistance in Acinetobacter baumannii. PLoS ONE 2015, 10, e0141314. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Bour, M.; Triponney, P.; Muller, J.; Koebel, C.; Bonnin, R.A.; Plésiat, P. Sequential emergence of colistin and rifampicin resistance in an OXA-72- producing outbreak strain of Acinetobacter baumannii. Int. J. Antimicrob. Agents 2019, 53, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Trebosc, V.; Gartenmann, S.; Tötzl, M.; Lucchini, V.; Schellhorn, B.; Pieren, M.; Lociuro, S.; Gitzinger, M.; Tigges, M.; Bumann, D.; et al. Dissecting colistin resistance mechanisms in extensively drug-resistant Acinetobacter baumannii clinical isolates. MBio 2019, 10, e01083-19. [Google Scholar] [CrossRef] [PubMed]
- Romano, K.P.; Warrier, T.; Poulsen, B.E.; Nguyen, P.H.; Loftis, A.R.; Saebi, A.; Pentelute, B.L.; Hung, D.T. Mutations in pmrB confer cross-resistance between the LptD inhibitor POL7080 and colistin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e00511-19. [Google Scholar] [CrossRef]
- Chew, K.L.; La, M.V.; Lin, R.T.P.; Teo, J.W.P. Colistin and polymyxin B susceptibility testing for carbapenem-resistant and mcr-positive Enterobacteriaceae: Comparison of sensititre, microscan, vitek 2, and etest with broth microdilution. J. Clin. Microbiol. 2017, 55, 2609–2616. [Google Scholar] [CrossRef]
- Shankar, C.; Venkatesan, M.; Rajan, R.; Mani, D.; Lal, B.; Prakash, J.A.J.; Anandan, S.; Pragasam, A.K.; Walia, K.; Ohri, V.C.; et al. Molecular characterization of colistin-resistant Klebsiella pneumoniae & its clonal relationship among Indian isolates. Indian J. Med. Res. 2019, 149, 199–207. [Google Scholar]
- Huang, L.; Feng, Y.; Zong, Z. Heterogeneous resistance to colistin in Enterobacter cloacae complex due to a new small transmembrane protein. J. Antimicrob. Chemother. 2019, 74, 2551–2558. [Google Scholar] [CrossRef]
- Yi, L.X.; Liu, Y.Y.; Wu, R.; Liang, Z.S.; Liu, J.H. Research progress on the plasmid-mediated colistin resistance gene mcr-1. Yi Chuan 2017, 39, 110–126. [Google Scholar]
- Feng, S.; Shen, C.; Chen, H.; Zheng, X.; Xia, Y.; Zhong, L.L.; Huang, X.; Wu, X.; Tian, G.B. Co-production of MCR-1 and NDM-5 in Escherichia coli isolated from a colonization case of inpatient. Infect. Drug Resist. 2018, 11, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Idowu, T.; Arthur, G.; Zhanel, G.G.; Schweizer, F. Heterodimeric Rifampicin-Tobramycin conjugates break intrinsic resistance of Pseudomonas aeruginosa to doxycycline and chloramphenicol in vitro and in a Galleria mellonella in vivo model. Eur. J. Med. Chem. 2019, 174, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Idowu, T.; Ammeter, D.; Arthur, G.; Zhanel, G.G.; Schweizer, F. Potentiation of β-lactam antibiotics and β-lactam/β-lactamase inhibitor combinations against MDR and XDR Pseudomonas aeruginosa using non-ribosomal tobramycin-cyclam conjugates. J. Antimicrob. Chemother. 2019, 74, 2640–2648. [Google Scholar] [CrossRef]
- Perez, F.; El Chakhtoura, N.G.; Yasmin, M.; Bonomo, R.A. Polymyxins: To combine or not to combine? Antibiotics 2019, 8, 38. [Google Scholar] [CrossRef]
- Kim, T.H.; Tao, X.; Moya, B.; Jiao, Y.; Basso, K.B.; Zhou, J.; Lang, Y.; Sutaria, D.S.; Zavascki, A.P.; Barth, A.L.; et al. Novel Cassette Assay To Quantify the Outer Membrane Permeability of Five β-Lactams Simultaneously in Carbapenem-Resistant Klebsiella pneumoniae and Enterobacter cloacae. mBio 2020, 11, e03189-19. [Google Scholar] [CrossRef] [PubMed]
- Toala, P.; McDonald, A.; Wilcox, C.; Finland, M. Susceptibility of group D Streptococcus (Enterococcus) to 21 antibiotics in vitro, with special reference to species differences. Am. J. Med. Sci. 1969, 258, 416–430. [Google Scholar] [CrossRef]
- Seligman, S.J. Penicillinase-negative variants of methicillin-resistant Staphylococcus aureus. Nature 1966, 209, 994–996. [Google Scholar] [CrossRef] [PubMed]
- Williamson, R.; Calderwood, S.B.; Moellering, R.C., Jr.; Tomasz, A. Studies on the mechanism of intrinsic resistance to β-lactam antibiotics in group D Streptococci. J. Gen. Microbiol. 1983, 129, 813–822. [Google Scholar] [CrossRef]
- Fuda, C.C.; Fisher, J.F.; Mobashery, S. Beta-lactam resistance in Staphylococcus aureus: The adaptive resistance of a plastic genome. Cell Mol. Life Sci. 2005, 62, 2617–2633. [Google Scholar] [CrossRef]
- Peck, S.M. Problem of the resistant Staphylococcus in infections of the skin. N. Y. State J. Med. 1961, 61, 4168–4184. [Google Scholar]
- Moellering, R.C., Jr. Current treatment options for community-acquired methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 2008, 46, 1032–1037. [Google Scholar] [CrossRef]
- Sakoulas, G.; Moellering, R.C., Jr. Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin. Infect. Dis. 2008, 46, S360–S367. [Google Scholar] [CrossRef]
- Howe, R.A.; Bowker, K.E.; Walsh, T.R.; Feest, T.G.; MacGowan, A.P. Vancomycin-resistant Staphylococcus aureus. Lancet 1998, 351, 602. [Google Scholar] [CrossRef]
- Planet, P.J.; Diaz, L.; Kolokotronis, S.O.; Narechania, A.; Reyes, J.; Xing, G.; Rincon, S.; Smith, H.; Panesso, D.; Ryan, C.; et al. Parallel epidemics of community-associated methicillin-resistant Staphylococcus aureus USA300 infection in North and South America. J. Infect. Dis. 2015, 212, 1874–1882. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef]
- Madhavan, K.; Chieng, L.O.; Armstrong, V.L.; Wang, M.Y. Spondylodiscitis in end-stage renal disease: A systematic review. J. Neurosurg. Spine 2019, 15, 1–9. [Google Scholar] [CrossRef]
- Khan, T.M.; Kok, Y.L.; Bukhsh, A.; Lee, L.H.; Chan, K.G.; Goh, B.H. Incidence of methicillin resistant Staphylococcus aureus (MRSA) in burn intensive care unit: A systematic review. Germs 2018, 8, 113–125. [Google Scholar] [CrossRef]
- Kalligeros, M.; Shehadeh, F.; Karageorgos, S.A.; Zacharioudakis, I.M.; Mylonakis, E. MRSA colonization and acquisition in the burn unit: A systematic review and meta-analysis. Burn 2019, 45, 1528–1536. [Google Scholar] [CrossRef]
- Galar, A.; Weil, A.A.; Dudzinski, D.M.; Muñoz, P.; Siedner, M.J. methicillin-resistant Staphylococcus aureus prosthetic valve endocarditis: Pathophysiology, epidemiology, clinical presentation, diagnosis, and management. Clin. Microbiol. Rev. 2019, 32, e00041-18. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.Z.; Kodiyanplakkal, R.P.L.; Calfee, D.P. Antimicrobial resistance in nephrology. Nat. Rev. Nephrol. 2019, 15, 463–481. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M. The continuing threat of Methicillin-Resistant Staphylococcus aureus. Antibiotics 2019, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Couto, N.; Monchique, C.; Belas, A.; Marques, C.; Gama, L.T.; Pomba, C. Trends and molecular mechanisms of antimicrobial resistance in clinical staphylococci isolated from companion animals over a 16-year period. J. Antimicrob Chemother. 2016, 71, 1479–1487. [Google Scholar] [CrossRef] [PubMed]
- Ruzauskas, M.; Couto, N.; Pavilonis, A.; Klimiene, I.; Siugzdiniene, R.; Virgailis, M.; Vaskeviciute, L.; Anskiene, L.; Pomba, C. Characterization of Staphylococcus pseudointermedius isolated from diseased dogs in Lithuania. Pol. J. Vet. Sci. 2016, 19, 7–14. [Google Scholar] [CrossRef][Green Version]
- Gonzales, P.R.; Pesesky, M.W.; Bouley, R.; Ballard, A.; Biddy, B.A.; Suckow, M.A.; Wolter, W.R.; Schroeder, V.A.; Burnham, C.A.D.; Mobashery, S.; et al. Synergistic, collaterally sensitive β-lactam combinations suppress resistance in MRSA. Nat. Chem. Biol. 2015, 11, 855–861. [Google Scholar] [CrossRef]
- Tenover, F.C.; Sinner, S.W.; Segal, R.E.; Huang, V.; Alexandre, S.S.; McGowan, J.E., Jr.; Weinstein, M.P. Characterization of a Staphylococcus aureus strain with progressive loss of susceptibility to vancomycin and daptomycin during therapy. Int. J. Antimicrob. Agents 2008, 33, 564–568. [Google Scholar] [CrossRef][Green Version]
- Murakami, K.; Tomasz, A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J. Bacteriol. 1989, 171, 874–879. [Google Scholar] [CrossRef]
- Chambers, H.F. Methicillin resistance in staphylococci: Molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 1997, 10, 781–791. [Google Scholar] [CrossRef]
- Tomasz, A.; Drugeon, H.B.; de Lencastre, H.M.; Jabes, D.; McDougall, L.; Bille, J. New mechanism for methicillin resistance in Staphylococcus aureus: Clinical isolates that lack the PBP 2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob. Agents Chemother. 1989, 33, 1869–1874. [Google Scholar] [CrossRef]
- Massidda, O.; Montanari, M.P.; Varaldo, P.E. Evidence for a methicillin-hydrolysing beta-lactamase in Staphylococcus aureus strains with borderline susceptibility to this drug. FEMS Microbiol. Lett. 1992, 71, 223–227. [Google Scholar]
- Tomasz, A. Accelerated evolution: Emergence of multidrug resistant gram-positive bacterial pathogens in the 1990’s. Neth. J. Med. 1998, 52, 219–524. [Google Scholar] [CrossRef]
- Crisóstomo, M.I.; Westh, H.; Tomasz, A.; Chung, M.; Oliveira, D.C.; de Lencastre, H. The evolution of methicillin resistance in Staphylococcus aureus: Similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 2001, 98, 9865–9870. [Google Scholar] [CrossRef]
- Antignac, A.; Tomasz, A. Reconstruction of the phenotypes of methicillin-resistant Staphylococcus aureus by replacement of the staphylococcal cassette chromosome mec with a plasmid-borne copy of Staphylococcus sciuri pbpD gene. Antimicrob. Agents Chemother. 2009, 53, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Hiramatsu, K.; Tomasz, A.; de Lencastre, H.; Perreten, V.; Holden, M.T.; Coleman, D.C.; Goering, R.; Giffard, P.M.; Skov, R.L.; et al. International Working Group on the Classification of Staphylococcal Cassette Chromosome elements (IWGSCC). Guidelines for reporting novel mecA gene homologues. Antimicrob. Agents Chemother. 2012, 56, 4997–4999. [Google Scholar] [CrossRef] [PubMed]
- Rolo, J.; Worning, P.; Nielsen, J.B.; Bowden, R.; Bouchami, O.; Damborg, P.; Guardabassi, L.; Perreten, V.; Tomasz, A.; Westh, H.; et al. Evolutionary origin of the Staphylococcal Cassette Chromosome mec (SCCmec). Antimicrob. Agents Chemother. 2017, 61, e02302-16. [Google Scholar] [CrossRef]
- Tickler, I.A.; Goering, R.V.; Mediavilla, J.R.; Kreiswirth, B.N.; Tenover, F.C. HAI Consortium. Continued expansion of USA300-like methicillin-resistant Staphylococcus aureus (MRSA) among hospitalized patients in the United States. Diagn. Microbiol. Infect. Dis. 2017, 88, 342–347. [Google Scholar] [CrossRef]
- Harkins, C.P.; Pichon, B.; Doumith, M.; Parkhill, J.; Westh, H.; Tomasz, A.; de Lencastre, H.; Bentley, S.D.; Kearns, A.M.; Holden, M.T. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. 2017, 18, 30. [Google Scholar] [CrossRef]
- Sobral, R.; Tomasz, A. The staphylococcal cell wall. Microbiol. Spectr 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Massidda, O.; Tomasz, A. The cell wall of Streptococcus pneumonia. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Gilmore, M. The Enterococci: Pathogenesis, Molecular Biology and Antimicrobial Resistance; ASM Press: Washington, DC, USA, 2002. [Google Scholar]
- Gordon, K.A.; Beach, M.L.; Biedenbach, D.J.; Jones, R.N.; Rhomberg, P.R.; Mutnick, A.H. Antimicrobial susceptibility patterns of beta-hemolytic and viridans group streptococci: Report from the SENTRY Antimicrobial Surveillance Program (1997–2000). Diagn Microbiol. Infect. Dis. 2002, 43, 157–162. [Google Scholar] [CrossRef]
- Robbins, W.C.; Tompsett, R. Treatment of enterococcal endocarditis and bacteremia; results of combined therapy with penicillin and streptomycin. Am. J. Med. 1951, 10, 278–299. [Google Scholar] [CrossRef]
- Waksman, S.A. Antibiotic substances-contribution of the microbiologist. N. Y. Acad. Sci. 2010, 1213, 107–111. [Google Scholar] [CrossRef]
- Baddour, L.M.; Wilson, W.R.; Bayer, A.S.; Fowler, V.G., Jr.; Bolger, A.F.; Levison, M.E.; Ferrieri, P.; Gerber, M.A.; Tani, L.Y.; Gewitz, M.H.; et al. Infective endocarditis: Diagnosis, antimicrobial therapy, and management of complications: A statement for healthcare professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, and the Councils on Clinical Cardiology, Stroke, and Cardiovascular Surgery and Anesthesia, American Heart Association: Endorsed by the Infectious Diseases Society of America. Circulation 2005, 111, e394. [Google Scholar] [CrossRef]
- Gilmore, M.S.; Lebreton, F.; van Schaik, W. Genomic transition of Enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr. Opin. Microbiol. 2013, 16, 10–16. [Google Scholar] [CrossRef]
- Lebreton, F.; van Schaik, W.; McGuire, A.M.; Godfrey, P.; Griggs, A.; Mazumdar, V.; Corander, J.; Cheng, L.; Saif, S.; Young, S.; et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 2013, 4, e00534-13. [Google Scholar] [CrossRef]
- Lebreton, F.; Willems, R.J.L.; Gilmore, M.S. Enterococcus diversity, origins in nature, and gut colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Lebreton, F.; Valentino, M.D.; Schaufler, K.; Earl, A.M.; Cattoir, V.; Gilmore, M.S. Transferable vancomycin resistance in clade B commensal-type Enterococcus faecium. J Antimicrob Chemother. 2018, 73, 1479–1486. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Cormican, M.; Flamm, R.K.; Mendes, R.E.; Jones, R.N. Temporal and geographic variation in antimicrobial susceptibility and resistance patterns of enterococci: Results from the SENTRY Antimicrobial Surveillance Program, 1997–2016. Open Forum Infect. Dis. 2019, 6, S54–S62. [Google Scholar] [CrossRef]
- Hidron, A.I.; Edwards, J.R.; Patel, J.; Horan, T.C.; Sievert, D.M.; Pollock, D.A.; Fridkin, S.K. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 2008, 29, 996–1011. [Google Scholar] [CrossRef]
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-Resistant Enterococci: A Review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef] [PubMed]
- Coupri, D.; Budin-Verneuil, A.; Hartke, A.; Benachour, A.; Léger, L.; Lequeux, T.; Pfund, E.; Verneuil, N. Genetic and pharmacological inactivation of d-alanylation of teichoic acids sensitizes pathogenic enterococci to β-lactams. J. Antimicrob. Chemother. 2019, 74, 3162–3169. [Google Scholar] [CrossRef]
- Tran, T.T.; Munita, J.M.; Arias, C.A. Mechanisms of drug resistance: Daptomycin resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 32–53. [Google Scholar] [CrossRef]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012, 3, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Brede, D.A.; Diep, D.B.; Nes, I.F.; Lotfipour, F.; Hojabri, Z. Efficient inactivation of multi-antibiotics resistant nosocomial Enterococci by purified hiracin Bacteriocin. Adv. Pharm. Bull. 2015, 5, 393–401. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Faron, M.L.; Ledeboer, N.A.; Buchan, B.W. Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant Enterococcus in the health care setting. J. Clin. Microbiol. 2016, 54, 2436–2447. [Google Scholar] [CrossRef] [PubMed]
- Sauvageot, N.; Mokhtari, A.; Joyet, P.; Budin-Verneuil, A.; Blancato, V.S.; Repizo, G.D.; Henry, C.; Pikis, A.; Thompson, J.; Magni, C.; et al. Enterococcus faecalis uses a phosphotransferase system permease and a host colonization-related ABC transporter for maltodextrin uptake. J. Bacteriol. 2017, 199, e00878-16. [Google Scholar] [CrossRef]
- Macesic, N.; Nelson, B.; Mcconville, T.H.; Giddins, M.J.; Green, D.A.; Stump, S.; Gomez-Simmonds, A.; Annavajhala, M.K.; Uhlemann, A.C. Emergence of Polymyxin Resistance in Clinical Klebsiella pneumoniae Through Diverse Genetic Adaptations: A Genomic, Retrospective Cohort Study. Clin. Infect Dis. 2020, 70, 2084–2091. [Google Scholar] [CrossRef] [PubMed]
- Murray, B.E. Beta-lactamase-producing Enterococci. Antimicrob. Agents Chemother. 1992, 36, 2355–2359. [Google Scholar] [CrossRef]
- Turolla, A.; Sabatino, R.; Fontaneto, D.; Eckert, E.M.; Colinas, N.; Corno, G.; Citterio, B.; Biavasco, F.; Antonelli, M.; Mauro, A.; et al. Defence strategies and antibiotic resistance gene abundance in enterococci under stress by exposure to low doses of peracetic acid. Chemosphere 2017, 185, 480–488. [Google Scholar] [CrossRef]
- Bedenić, B.; Sardelić, S.; Ladavac, M. Multiresistant bacteria. Acta Med. Croatica 2015, 69, 211–216. [Google Scholar]
- Morosini, M.I.; Díez-Aguilar, M.; Cantón, R. Mechanisms of action and antimicrobial activity of ceftobiprole. Rev. Esp. Quimioter 2019, 32, 3–10. [Google Scholar]
- Rajagopal, M.; Walker, S. Envelope structures of gram-positive bacteria. Curr. Top. Microbiol. Immunol. 2017, 404, 1–44. [Google Scholar]
- Kilian, R.; Frasch, H.J.; Kulik, A.; Wohlleben, W.; Stegmann, E. The VanRS homologous two-component system VnlRSAb of the glycopeptide producer Amycolatopsis balhimycina activates transcription of the vanHAXSc genes in Streptomyces coelicolor, but not in A. balhimycina. Microb. Drug Resist. 2016, 22, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.D.; Foster, E.E.; Wallace, A.G.; Kim, S.J. Peptidoglycan O-acetylation increases in response to vancomycin treatment in vancomycin-resistant Enterococcus faecalis. Sci. Rep. 2017, 7, 46500. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.D.; Foster, E.E.; Yang, H.; Kim, S.J. Quantification of the d-Ala-d-Lac-terminated peptidoglycan structure in vancomycin-resistant Enterococcus faecalis using a combined solid-state nuclear magnetic resonance and mass spectrometry analysis. Biochemistry 2017, 56, 612–622. [Google Scholar] [CrossRef]
- Chang, J.D.; Foster, E.E.; Thadani, A.N.; Ramirez, A.J.; Kim, S.J. Inhibition of Staphylococcus aureus cell wall biosynthesis by desleucyl-oritavancin: A quantitative peptidoglycan composition analysis by mass spectrometry. J. Bacteriol. 2017, 199, e00278-17. [Google Scholar] [CrossRef]
- Ladjouzi, R.; Bizzini, A.; Lebreton, F.; Sauvageot, N.; Rincé, A.; Benachour, A.; Hartke, A. Analysis of the tolerance of pathogenic enterococci and Staphylococcus aureus to cell wall active antibiotics. J. Antimicrob. Chemother. 2013, 68, 2083–2091. [Google Scholar] [CrossRef]
- Master, R.N.; Deane, J.; Opiela, C.; Sahm, D.F. Recent trends in resistance to cell envelope-active antibacterial agents among key bacterial pathogens. Ann. N. Y. Acad. Sci. 2013, 1277, 1–7. [Google Scholar] [CrossRef]
- Dulberger, C.L.; Rubin, E.J.; Boutte, C.C. The mycobacterial cell envelope - a moving target. Nat. Rev. Microbiol. 2020, 18, 47–59. [Google Scholar] [CrossRef]
- Hung, W.W.; Chen, Y.H.; Tseng, S.P.; Jao, Y.T.; Teng, L.J.; Hung, W.C. Using groEL as the target for identification of Enterococcus faecium clades and 7 clinically relevant Enterococcus species. J. Microbiol. Immunol. Infect. 2019, 52, 255–264. [Google Scholar] [CrossRef]
- Paganelli, F.L.; de Been, M.; Braat, J.C.; Hoogenboezem, T.; Vink, C.; Bayjanov, J.; Rogers, M.R.C.; Huebner, J.; Bonten, M.J.M.; Willems, R.J.L.; et al. Distinct SagA from hospital-associated clade A1 Enterococcus faecium strains contributes to biofilm formation. Appl. Environ. Microbiol. 2015, 81, 6873–6882. [Google Scholar] [CrossRef]
- Montealegre, M.C.; Roh, J.H.; Rae, M.; Davlieva, M.G.; Singh, K.V.; Shamoo, Y.; Murray, B.E. Differential penicillin-binding protein 5 (PBP5) Levels in the Enterococcus faecium clades with different levels of ampicillin resistance. Antimicrob. Agents Chemother. 2016, 61, e02034-16. [Google Scholar] [CrossRef]
- Freitas, A.R.; Tedim, A.P.; Novais, C.; Coque, T.M.; Peixe, L. Distribution of putative virulence markers in Enterococcus faecium: Towards a safety profile review. J. Antimicrob. Chemother. 2018, 73, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Huo, W.; Adams, H.M.; Trejo, C.; Badia, R.; Palmer, K.L. A Type I restriction-modification system associated with Enterococcus faecium subspecies separation. Appl. Environ. Microbiol. 2019, 85, e02174-18. [Google Scholar] [CrossRef]
- Palmer, K.L.; Godfrey, P.; Griggs, A.; Kos, V.N.; Zucker, J.; Desjardins, C.; Cerqueira, G.; Gevers, D.; Walker, S.; Wortman, J.; et al. Comparative genomics of enterococci: Variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. MBio 2012, 3, e00318-11. [Google Scholar] [CrossRef]
- Qin, X.; Galloway-Peña, J.R.; Sillanpaa, J.; Roh, J.H.; Nallapareddy, S.R.; Chowdhury, S.; Bourgogne, A.; Choudhury, T.; Muzny, D.M.; Buhay, C.J.; et al. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 2012, 12, 135. [Google Scholar] [CrossRef] [PubMed]
- Uttley, A.H.; Woodford, N.; Johnson, A.P.; Cookson, B.; George, R.C. Vancomycin-resistant Enterococci. Lancet 1993, 342, 615. [Google Scholar] [CrossRef]
- Top, J.; Willems, R.; Bonten, M. Emergence of CC17 Enterococcus faecium: From commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 2008, 52, 297–308. [Google Scholar] [CrossRef]
- Kim, E.B.; Marco, M.L. Nonclinical and clinical Enterococcus faecium strains, but not Enterococcus faecalis strains, have distinct structural and functional genomic features. Appl. Environ. Microbiol. 2014, 80, 154–165. [Google Scholar] [CrossRef]
- Douglas, A.P.; Marshall, C.; Baines, S.L.; Ritchie, D.; Szer, J.; Madigan, V.; Chan, H.T.; Ballard, S.A.; Howden, B.P.; Buising, K.; et al. Utilizing genomic analyses to investigate the first outbreak of vanA vancomycin-resistant Enterococcus in Australia with emergence of daptomycin non-susceptibility. J. Med. Microbiol. 2019, 68, 303–308. [Google Scholar] [CrossRef]
- Zhang, X.; Paganelli, F.L.; Bierschenk, D.; Kuipers, A.; Bonten, M.J.; Willems, R.J.; Van Schaik, W. Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium. PLoS Genet. 2012, 8, e1002804. [Google Scholar] [CrossRef]
- Gilmore, M.S.; Rauch, M.; Ramsey, M.M.; Himes, P.R.; Varahan, S.; Manson, J.M.; Lebreton, F.; Hancock, L.E. Pheromone killing of multidrug-resistant Enterococcus faecalis V583 by native commensal strains. Proc. Natl. Acad. Sci. USA 2015, 112, 7273–7278. [Google Scholar] [CrossRef]
- Nilsson, O. Vancomycin resistant Enterococci in farm animals’ occurrence and importance. Infect. Ecol. Epidemiol. 2012, 2, 16959. [Google Scholar] [CrossRef] [PubMed]
- Mutters, N.T.; Mersch-Sundermann, V.; Mutters, R.; Brandt, C.; Schneider-Brachert, W.; Frank, U. Control of the spread of vancomycin-resistant Enterococci in hospitals: Epidemiology and clinical relevance. Dtsch. Arztebl. Int. 2013, 110, 725–731. [Google Scholar] [PubMed]
- Levitus, M.; Perera, T.B. Vancomycin-Resistant Enterococci (VRE); StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, E.S.; Paras, M.L.; Noubary, F.; Walensky, R.P.; Hooper, D.C. Natural history of colonization with methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE): A systematic review. BMC Infect. Dis. 2014, 14, 177. [Google Scholar] [CrossRef]
- Remschmidt, C.; Behnke, M.; Kola, A.; Diaz, L.A.P.; Rohde, A.M.; Gastmeier, P.; Schwab, F. The effect of antibiotic use on prevalence of nosocomial vancomycin-resistant Enterococci an ecologic study. Antimicrob. Resist. Infect. Control. 2017, 6, 95. [Google Scholar] [CrossRef][Green Version]
- Lewis, J.D.; Barros, A.J.; Sifri, C.D. Comparison of risk factors and outcomes of daptomycin-susceptible and -nonsusceptible vancomycin-resistant Enterococcus faecium infections in liver transplant recipients. Transpl. Infect. Dis. 2018, 20, e12856. [Google Scholar] [CrossRef]
- Chen, C.H.; Xu, X.G. Genetic characteristics of vancomycin resistance gene cluster in Enterococcus spp. Yi Chuan 2015, 37, 452–457. [Google Scholar]
- Bayjanov, J.R.; Baan, J.; Rogers, M.R.C.; Troelstra, A.; Willems, R.J.L.; van Schaik, W. Enterococcus faecium genome dynamics during long-term asymptomatic patient gut colonization. Microbial Genom. 2019, 7, e000277. [Google Scholar] [CrossRef]
- Gorrie, C.; Higgs, C.; Carter, G.; Stinear, T.P.; Howden, B. Genomics of vancomycin-resistant Enterococcus faecium. Microb. Genom. 2019, 5, e000283. [Google Scholar] [CrossRef]
- Melegh, S.; Nyül, A.; Kovács, K.; Kovács, T.; Ghidán, Á.; Dombrádi, Z.; Szabó, J.; Berta, B.; Lesinszki, V.; Pászti, J.; et al. Dissemination of VanA-Type Enterococcus faecium isolates in Hungary. Microb. Drug Resist. 2018, 24, 1376–1390. [Google Scholar] [CrossRef]
- Lebreton, F.; Depardieu, F.; Bourdon, N.; Fines-Guyon, M.; Berger, P.; Camiade, S.; Leclercq, R.; Courvalin, P.; Cattoir, V. D-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 2011, 55, 4606–4612. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Tanimoto, K.; Shibayama, K.; Arakawa, Y.; Fujimoto, S.; Ike, Y.; Tomita, H. Identification of VanN-type vancomycin resistance in an Enterococcus faecium isolate from chicken meat in Japan. Antimicrob Agents Chemother. 2012, 56, 6389–6392. [Google Scholar] [CrossRef]
- Boyd, D.A.; Lévesque, S.; Picard, A.C.; Golding, G.R. Vancomycin-resistant Enterococcus faecium harbouring vanN in Canada: A case and complete sequence of pEfm12493 harbouring the vanN operon. J. Antimicrob. Chemother. 2015, 70, 2163–2165. [Google Scholar] [CrossRef]
- Devriese, L.A.; Cauwerts, K.; Hermans, K.; Wood, A.M. Enterococcus cecorum septicaemia as a cause of bone and joint lesions resulting in lameness in broiler chickens. Vlaams Diergeneeskundig Tijdschrift 2002, 71, 219–221. [Google Scholar]
- Wood, A.M.; MacKenzie, G.; McGillveray, N.C.; Brown, L.; Devriese, L.A.; Baele, M. Isolation of Enterococcus cecorum from bone lesions in broiler chickens. Vet. Record. 2002, 150, 27. [Google Scholar]
- Chadfield, M.S.; Christensen, J.P.; Christensen, H.; Bisgaard, M. Characterization of Streptococci and Enterococci associated with septicaemia in broiler parents with a high prevalence of endocarditis. Avian Pathol. 2004, 33, 610–617. [Google Scholar] [CrossRef]
- Debnam, A.L.; Jackson, C.R.; Avellaneda, G.E.; Barrett, J.B.; Hofacre, C.L. Effect of growth promotant usage on Enterococci species on a poultry farm. Avian Dis. 2005, 49, 361–365. [Google Scholar] [CrossRef]
- Thayer, S.G.; Waltman, W.D.; Wages, D.P. Streptococcus and Enterococcus. In Diseases of Poultry, 12th ed.; Saif, Y.M., Glisson, J.R., McDougald, L.R., Nolan, L.K., Swayne, D.E., Eds.; Blackwell Publishing Professional: Ames, IA, USA, 2008; pp. 900–908. [Google Scholar]
- Aziz, T.; Barnes, H.J. Is spondylitis an emerging disease in broilers? World Poultry 2007, 23, 44–45. [Google Scholar]
- Aziz, T.; Barnes, H.J. Spondylitis is emerging in broilers. World Poultry 2009, 25, 19. [Google Scholar]
- DeHerdt, P.; Defoort, P.; Steelant, J.V.; Swam, H.; Tanghe, L.; Goethem, S.V.; Vanrobaeys, M. Enterococcus cecorum osteomyelitis and arthritis in broiler chickens. Vlaams Diergeneeskundig Tijdschrift 2008, 78, 44–48. [Google Scholar]
- Stalker, M.J.; Brash, M.L.; Weisz, A.; Ouckama, R.M.; Slavic, D. Arthritis and osteomyelitis associated with Enterococcus cecorum infection in broiler and broiler breeder chickens in Ontario, Canada. J. Vet. Diag. Invest. 2010, 22, 643–645. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.T.; Martin, M.P.; Barnes, H.J. Experimental reproduction of Enterococcus spondylitis in male broiler breeder chickens. Avian Dis. 2011, 55, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Boerlin, P.; Nicholson, V.; Brash, M.; Slavic, D.; Boyen, F.; Sanei, B.; Butaye, P. Diversity of Enterococcus cecorum from chickens. Vet. Microbiol. 2012, 157, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Jung, A.; Chen, L.R.; Suyemoto, M.M.; Barnes, H.J.; Borst, L.B. A review of Enterococcus cecorum infection in poultry. Avian Dis. 2018, 62, 261–271. [Google Scholar] [CrossRef]
- Griffith, R.S. Introduction to vancomycin. Rev. Infect. Dis. 1981, 3, S200–S204. [Google Scholar] [CrossRef]
- Xu, L.; Huang, H.; Wei, W.; Zhong, Y.; Tang, B.; Yuan, H.; Zhu, L.; Huang, W.; Ge, M.; Yang, S.; et al. Complete genome sequence and comparative genomic analyses of the vancomycin-producing Amycolatopsis orientalis. BMC Genom. 2014, 15, 363. [Google Scholar] [CrossRef]
- Chen, X.Y.; Xu, R.X.; Zhou, X.; Liu, Y.; Hu, C.Y.; Xie, X.F. Acute kidney injury associated with concomitant vancomycin and piperacillin/tazobactam administration: A systematic review and meta-analysis. Int. Urol. Nephrol. 2018, 50, 2019–2026. [Google Scholar] [CrossRef]
- Grabel, Z.J.; Boden, A.; Segal, D.N.; Boden, S.; Milby, A.H.; Heller, J.G. The impact of prophylactic intraoperative vancomycin powder on microbial profile, antibiotic regimen, length of stay, and reoperation rate in elective spine surgery. Spine J. 2019, 19, 261–266. [Google Scholar] [CrossRef]
- Mühlberg, E.; Umstätter, F.; Kleist, C.; Domhan, C.; Mier, W.; Uhl, P. Renaissance of vancomycin: Approaches for breaking antibiotic resistance in multidrug-resistant bacteria. Can. J. Microbiol. 2020, 66, 11–16. [Google Scholar] [CrossRef]
- Haenni, M.; Saras, E.; Châtre, P.; Meunier, D.; Martin, S.; Lepage, G.; Ménard, M.F.; Lebreton, P.; Rambaud, T.; Madec, J.Y. vanA in Enterococcus faecium, Enterococcus faecalis, and Enterococcus casseliflavus detected in French cattle. Foodborne Pathog. Dis. 2009, 6, 1107–1111. [Google Scholar] [CrossRef]
- Gardete, S.; Tomasz, A. Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Investig. 2014, 124, 2836–2840. [Google Scholar] [CrossRef] [PubMed]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 2017, 90, 269–281. [Google Scholar]
- Manges, A.R.; Steiner, T.S.; Wright, A.J. Fecal microbiota transplantation for the intestinal decolonization of extensively antimicrobial-resistant opportunistic pathogens: A review. Infect. Dis. 2016, 48, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Satlin, M.J.; Walsh, T.J. Multidrug-resistant Enterobacteriaceae, Pseudomonas aeruginosa, and vancomycin-resistant Enterococcus: Three major threats to hematopoietic stem cell transplant recipients. Transpl. Infect. Dis. 2017, 19, e12762. [Google Scholar] [CrossRef] [PubMed]
- Vehreschild, M.J.G.T.; Haverkamp, M.; Biehl, L.M.; Lemmen, S.; Fätkenheuer, G. Vancomycin-resistant enterococci (VRE): A reason to isolate? Infection 2019, 47, 7–11. [Google Scholar] [CrossRef]
- Stogios, P.J.; Savchenko, A. Molecular mechanisms of vancomycin resistance. Protein Sci. 2020, 29, 654–669. [Google Scholar] [CrossRef]
- Thally, F.P.; DeBruin, M.F. Development of daptomycin for Gram-positive infections. J. Antimicrob. Chemother. 2000, 46, 523–526. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Contreras, G.A.; Munita, J.M.; Arias, C.A. Novel strategies for the management of vancomycin-resistant enterococcal infections. Curr. Infect. Dis. Rep. 2019, 21, 22. [Google Scholar] [CrossRef]
- Micklefield, J. Daptomycin structure and mechanism of action revealed. Chem. Biol. 2004, 11, 887–888. [Google Scholar] [CrossRef]
- Eisenstein, B.I.; Oleson, F.B., Jr.; Baltz, R.H. Daptomycin: From the mountain to the clinic, with essential help from Francis Tally, MD. Clin Infect Dis. 2010, 50, S10–S15. [Google Scholar] [CrossRef] [PubMed]
- Fowler, V.G., Jr.; Boucher, H.W.; Corey, G.R.; Abrutyn, E.; Karchmer, A.W.; Rupp, M.E.; Levine, D.P.; Chambers, H.F.; Tally, F.P.; Vigliani, G.A.; et al. S. aureus Endocarditis and Bacteremia Study Group. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 2006, 355, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, P.; Raja, A.; LaBonte, J.; Lebbos, J. Daptomycin. Nat. Rev. Drug Discov. 2003, 2, 943–944. [Google Scholar] [PubMed]
- Arbeit, R.D.; Maki, D.; Tally, F.P.; Campanaro, E.; Eisenstein, B.I. The safety and efficacy of daptomycin for the treatment of complicated skin and skin-structure infections. Clin. Infect. Dis. 2004, 38, 1673–1681. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Lye, D.C.; Yahav, D.; Sud, A.; Robinson, J.O.; Nelson, J.; Archuleta, S.; Roberts, M.A.; Cass, A.; Paterson, D.L.; et al. Effect of Vancomycin or Daptomycin With vs Without an Antistaphylococcal β-Lactam on Mortality, Bacteremia, Relapse, or Treatment Failure in Patients With MRSA Bacteremia: A Randomized Clinical Trial Australasian Society for Infectious Diseases Clinical Research Network. JAMA 2020, 323, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Heidary, M.; Khosravi, A.D.; Khoshnood, S.; Nasiri, M.J.; Soleimani, S.; Goudarzi, M. Daptomycin. J. Antimicrob. Chemother. 2018, 73, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Saw, S. Daptomycin; StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- McHenney, M.A.; Baltz, R.H. Gene transfer and transposition mutagenesis in Streptomyces roseosporus: Mapping of insertions that influence daptomycin or pigment production. Microbiology 1996, 142, 2363–2373. [Google Scholar] [CrossRef]
- Mchenney, M.A.; Hosted, T.J.; Dehoff, B.S.; Rosteck, P.R.; Baltz, R.H. Molecular cloning and physical mapping of the daptomycin gene cluster from Streptomyces roseosporus. J Bacteriol. 1998, 180, 143–151. [Google Scholar] [CrossRef]
- Taylor, S.D.; Palmer, M. The action mechanism of daptomycin. Bioorg. Med. Chem. 2016, 24, 6253–6268. [Google Scholar] [CrossRef]
- Cantón, R.; Ruiz-Garbajosa, P.; Chaves, R.L.; Johnson, A.P. A potential role for daptomycin in enterococcal infections: What is the evidence? J. Antimicrob. Chemother. 2010, 65, 1126–1136. [Google Scholar] [CrossRef]
- Arias, C.A.; Panesso, D.; McGrath, D.M.; Qin, X.; Mojica, M.F.; Miller, C.; Diaz, L.; Tran, T.T.; Rincon, S.; Barbu, E.M.; et al. Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 2011, 365, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Hachmann, A.B.; Sevim, E.; Gaballa, A.; Popham, D.L.; Antelmann, H.; Helmann, J.D. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 2011, 55, 4326–4337. [Google Scholar] [CrossRef] [PubMed]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal infection—Treatment and antibiotic resistance. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 87–134. [Google Scholar]
- Mercuro, N.J.; Davis, S.L.; Zervos, M.J.; Herc, E.S. Combatting resistant enterococcal infections: A pharmacotherapy review. Expert Opin. Pharmacother. 2018, 19, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.; Panesso, D.; Mishra, N.N.; Mileykovskaya, E.; Guan, Z.; Munita, J.M.; Reyes, J.; Diaz, L.; Weinstock, G.M.; Murray, B.E.; et al. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio 2013, 4, e00281-13. [Google Scholar] [CrossRef]
- Cattoir, V.; Giard, J.C. Antibiotic resistance in Enterococcus faecium clinical isolates. Expert Rev. Anti Infect. Ther. 2014, 12, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.Y.; Kim, H.M.; Chung, D.R.; Kim, S.H.; Huh, H.J.; Kang, C.I.; Peck, K.R.; Lee, N.Y.; Song, J.H. Resistance mechanisms and clinical characteristics of linezolid-resistant Enterococcus faecium isolates: A single-centre study in South Korea. J. Glob. Antimicrob. Resist. 2018, 12, 44–47. [Google Scholar] [CrossRef]
- Yim, J.; Smith, J.R.; Rybak, M.J. Role of combination antimicrobial therapy for vancomycin-resistant Enterococcus faecium infections: Review of the current evidence. Pharmacotherapy 2017, 37, 579–592. [Google Scholar] [CrossRef]
- Mwangi, M.M.; Wu, S.W.; Zhou, Y.; Sieradzki, K.; de Lencastre, H.; Richardson, P.; Bruce, D.; Rubin, E.; Myers, E.; Siggia, E.D.; et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 2007, 104, 9451–9456. [Google Scholar] [CrossRef]
- Casanova, G.N.; Ruiz, S.M.; Bellido, M.J.L. Mechanisms of resistance to daptomycin in Staphylococcus aureus. Rev. Esp. Quimioter 2017, 30, 391–396. [Google Scholar]
- Jiang, J.H.; Dexter, C.; Cameron, D.R.; Monk, I.R.; Baines, S.L.; Abbott, I.J.; Spelman, D.W.; Kostoulias, X.; Nethercott, C.; Howden, B.P.; et al. Evolution of daptomycin resistance in coagulase-negative staphylococci involves mutations of the essential two-component regulator WalKR. Antimicrob. Agents Chemother. 2019, 63, e01926-18. [Google Scholar] [CrossRef]
- Taglialegna, A.; Varela, M.C.; Rosato, R.R.; Rosato, A.E. VraSR and virulence trait modulation during daptomycin resistance in methicillin-resistant Staphylococcus aureus infection. mSphere 2019, 4, e00557-18. [Google Scholar] [CrossRef]
- Matono, T.; Hayakawa, K.; Hirai, R.; Tanimura, A.; Yamamoto, K.; Fujiya, Y.; Mawatari, M.; Kutsuna, S.; Takeshita, N.; Mezaki, K.; et al. Emergence of a daptomycin-non-susceptible Enterococcus faecium strain that encodes mutations in DNA repair genes after high-dose daptomycin therapy. BMC Res. Notes 2016, 9, 197. [Google Scholar] [CrossRef][Green Version]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogen without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Iyer, A.; Madder, A.; Singh, I. Teixobactins: A new class of 21st century antibiotics to combat multidrug-resistant bacterial pathogens. Future Microbiol. 2019, 14, 457–460. [Google Scholar] [CrossRef]
- Li, Y.X.; Zhong, Z.; Hou, P.; Zhang, W.P.; Qian, P.Y. Resistance to nonribosomal peptide antibiotics mediated by D-stereospecific peptidases. Nat. Chem. Biol. 2018, 14, 381–387. [Google Scholar] [CrossRef]
- Karas, J.A.; Chen, F.; Schneider-Futschik, E.K.; Kang, Z.; Hussein, M.; Swarbrick, J.; Hoyer, D.; Giltrap, A.M.; Payne, R.J.; Li, J.; et al. Synthesis and structure-activity relationships of teixobactin. Ann. N. Y. Acad. Sci. 2020, 1459, 86–105. [Google Scholar] [CrossRef]
- Yang, H.; Pishenko, A.V.; Li, X.; Nowick, J.S. Design, synthesis, and study of lactam and ring-expanded analogues of teixobactin. J. Org. Chem. 2020, 85, 1331–1339. [Google Scholar] [CrossRef]
- He, Y.; Fan, A.; Han, M.; Zhang, Y.; Tong, Y.; Zheng, G.; Zhu, S. New perspectives on the treatment of mycobacterial infections using antibiotics. Appl. Microbiol. Biotechnol. 2020, 104, 4197–4209. [Google Scholar] [CrossRef]
- Ötvös, L., Jr.; Wade, J.D. Current challenges in peptide-based drug discovery. Front. Chem. 2014, 2, 62. [Google Scholar] [CrossRef]
- Lázár, V.; Martins, A.; Spohn, R.; Daruka, L.; Grézal, G.; Pál, C. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 2018, 3, 718–731. [Google Scholar] [CrossRef]
- Ötvös, L., Jr. Antibacterial peptides isolated from insects. J. Pept. Sci. 2000, 6, 497–511. [Google Scholar]
- Ötvös, L., Jr.; Wade, J.D.; Lin, F.; Condie, B.A.; Hanrieder, J.; Hoffmann, R. Designer antibacterial peptides kill fluoroquinolone-resistant clinical isolates. J. Med. Chem. 2005, 48, 5349–5359. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef]
- Jenssen, H.; Fjell, C.D.; Cherkasov, A.; Hancock, R.E. QSAR modeling and computer-aided design of antimicrobial peptides. J. Pept. Sci. 2008, 14, 110–114. [Google Scholar] [CrossRef]
- Schoppet, M.; Tailhades, J.; Kulkarni, K.; Cryle, M.J. Precursor manipulation in glycopeptide antibiotic biosynthesis: Are β-amino acids compatible with the oxidative cyclization cascade? J. Org. Chem. 2018, 83, 7206–7214. [Google Scholar] [CrossRef]
- Spänig, S.; Heider, D. Encodings and models for antimicrobial peptide classification for multi-resistant pathogens. BioData Min 2019, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Knappe, D.; Wend, E.; Ostorházi, E.; Hoffmann, R. In vivo Efficacy and Pharmacokinetics of Optimized Apidaecin Analogs. Front. Chem. 2017, 5, 15. [Google Scholar] [CrossRef]
- Hicks, R.P.; Abercrombie, J.J.; Wong, R.K.; Leung, K.P. Antimicrobial peptides containing unnatural amino acid exhibit potent bactericidal activity against ESKAPE pathogens. Bioorg. Med. Chem. 2012, 21, 205–214. [Google Scholar] [CrossRef]
- Ötvös, L., Jr. The short proline-rich antibacterial peptide family. Cell Mol. Life Sci. 2002, 59, 1138–1150. [Google Scholar] [CrossRef]
- Taniguchi, M.; Ochiai, A.; Kondo, H.; Fukuda, S.; Ishiyama, Y.; Saitoh, E.; Kato, T.; Tanaka, T. Pyrrhocoricin, a proline-rich antimicrobial peptide derived from insect, inhibits the translation process in the cell-free Escherichia coli protein synthesis system. J. Biosci. Bioeng. 2016, 121, 591–598. [Google Scholar] [CrossRef]
- Berrthold, N.; Hoffmann, R. Cellular uptake of apidaecin 1b and related analogs in Gram-negative bacteria reveals novel antibacterial mechanism for proline-rich antimicrobial peptides. Protein Pept. Lett. 2014, 21, 391–398. [Google Scholar] [CrossRef]
- Wu, S.; Wang, J.; Zhu, L.; Ren, H.; Yang, X. A novel apidaecin Api-PR19 synergizes with the gut microbial community to maintain intestinal health and promote growth performance of broilers. J. Anim. Sci. Biotechnol. [CrossRef] [PubMed]
- Knappe, D.; Ruden, S.; Langanke, S.; Tikkoo, T.; Ritzer, J.; Mikut, R.; Martin, L.L.; Hoffmann, R.; Hilpert, K. Optimization of oncocin for antibacterial activity using a SPOT synthesis approach: Extending the pathogen spectrum to Staphylococcus aureus. Amino Acids. 2016, 48, 269–280. [Google Scholar] [CrossRef]
- Ötvös, L., Jr.; Ostorházi, E.; Szabó, D.; Zumbrun, S.D.; Miller, L.L.; Halasohoris, S.A.; Desai, P.D.; Int Veldt, S.M.; Kraus, C.N. Synergy Between Proline-Rich Antimicrobial Peptides and Small Molecule Antibiotics Against Selected Gram-Negative Pathogens in vitro and in vivo. Front Chem. 2108, 6, 309. [Google Scholar] [CrossRef] [PubMed]
- Ostorházi, E.; Hoffmann, R.; Herth, N.; Wade, J.D.; Kraus, C.N.; Ötvös, L., Jr. Advantage of a Narrow Spectrum Host Defense (Antimicrobial) Peptide Over a Broad Spectrum Analog in Preclinical Drug Development. Front Chem. 2018, 6, 359. [Google Scholar] [CrossRef] [PubMed]
- Karlowsky, J.A.; Steenbergen, J.; Zhanel, G.G. Microbiology and preclinical review of omadacycline. Clin. Infect. Dis. 2019, 69, S6–S15. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Esquivel, J.; Zelenitsky, S.; Lawrence, C.K.; Adam, H.J.; Golden, A.; Hink, R.; Berry, L.; Schweizer, F.; Zhanel, M.A.; et al. Omadacycline: A novel oral and intravenous aminomethylcycline antibiotic agent. Drugs 2020, 80, 285–313. [Google Scholar] [CrossRef]
- Lengyel, K.; Lang, E.; Fodor, A.; Szállás, E.; Schumann, P.; Stackebrandt, E. Description of four novel species of Xenorhabdus, family Enterobacteriaceae: Xenorhabdus budapestensis sp. nov., Xenorhabdus ehlersii sp. nov., Xenorhabdus innexi sp. nov., and Xenorhabdus szentirmaii sp. nov. Syst. Appl. Microbiol. 2005, 28, 115–122. [Google Scholar] [CrossRef]
- Furgani, G.; Böszörményi, E.; Fodor, A.; Máthé Fodor, A.; Forst, S.; Hogan, J.S.; Katona, Z.; Klein, M.G.; Stackebrandt, E.; Szentirmai, A.; et al. Xenorhabdus antibiotics: A comparative analysis and potential utility for controlling mastitis caused by bacteria. J. Appl. Microbiol. 2008, 104, 745–758. [Google Scholar] [CrossRef]
- Böszörményi, E.; Érsek, T.; Fodor, A.; Fodor, A.M.; Földes, L.S.; Hevesi, M.; Hogan, J.S.; Katona, Z.; Klein, M.G.; Kormány, A.; et al. Isolation and activity of Xenorhabdus antimicrobial compounds against the plant pathogens Erwinia amylovora and Phytophthora nicotianae. J. Appl. Microbiol. 2009, 107, 746–759. [Google Scholar] [CrossRef]
- Gualtieri, M.; Ogier, J.C.; Pagès, S.; Givaudan, A.; Gaudriault, S. Draft genome sequence and annotation of the entomopathogenic\bacterium Xenorhabdus szentirmaii Strain DSM16338. Genome Announ. 2014, 2, e00190-14. [Google Scholar] [CrossRef]
- Vozik, D.; Bélafi-Bakó, K.; Hevesi, M.; Böszörményi, E.; Fodor, A. Effectiveness of a peptide-rich fraction from xenorhabdus budapestensis culture against fire blight disease on apple blossoms. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 2015, 43, 547–553. [Google Scholar] [CrossRef]
- Fuchs, S.W.; Sachs, C.C.; Kegler, C.; Nollmann, F.I.; Karas, M.; Bode, H.B. Neutral loss fragmentation pattern, based screening for arginine-rich natural products in Xenorhabdus and Photorhabdus. Anal. Chem. 2012, 84, 6948–6955. [Google Scholar] [CrossRef]
- Fuchs, S.W.; Grundmann, F.; Kurz, M.; Kaiser, M.; Bode, H.B. Fabclavines: Bioactive peptide–polyketide-polyamino hybrids from Xenorhabdus. Chem. BioChem. 2014, 15, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Wenski, S.L.; Kolbert, D.; Grammbitter, G.L.C.; Bode, H.B. Fabclavine biosynthesis in X. szentirmaii: Shortened derivatives and characterization of the thioester reductase FclG and the condensation domain-like protein FclL. J. Ind. Microbiol. Biotechnol. 2019, 46, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Donmez Ozkan, H.; Cimen, H.; Ulug, D.; Wenski, S.; Yigit Ozer, S.; Telli, M.; Aydin, N.; Bode, H.B.; Hazir, S. Nematode-associated bacteria: Production of antimicrobial agent as a presumptive nominee for curing endodontic infections caused by Enterococcus faecalis. Front. Microbiol. 2019, 10, 2672. [Google Scholar] [CrossRef] [PubMed]
- Kajla, M.K.; Barrett-Wilt, G.A.; Paskewitz, S.M. Bacteria: A novel source for potent mosquito feeding-deterrents. Sci. Adv. 2019, 5, eaau6141. [Google Scholar] [CrossRef]
- Ciezki, K.; Wesener, S.; Jaber, D.; Mirza, S.; Forst, S. ngrA-dependent natural products are required for interspecies competition and virulence in the insect pathogenic bacterium Xenorhabdus szentirmaii. Microbiology 2019, 165, 538–553. [Google Scholar] [CrossRef]
- Gajdács, M. The concept of an ideal antibiotic: Implications for drug design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef]
- Pál, C.; Papp, B.; Lercher, M.J. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 2005, 37, 1372–1375. [Google Scholar] [CrossRef]
- Waclaw, B. Evolution of drug resistance in bacteria. Adv. Exp. Med. Biol. 2016, 915, 49–67. [Google Scholar]
- Liu, J.; Gefen, O.; Ronin, I.; Bar-Meir, M.; Balaban, N.Q. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 2020, 10, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef]
- Fridman, O.; Goldberg, A.; Ronin, I.; Shoresh, N.; Balaban, N.Q. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 2014, 513, 418–421. [Google Scholar] [CrossRef]
- Kim, J.S.; Wood, T.K. Persistent persister misperceptions. Front. Microbiol. 2016, 7, 2134. [Google Scholar] [CrossRef]
- Girgis, H.S.; Harris, K.; Tavazoie, S. Large mutational target size for rapid emergence of bacterial persistence. Proc. Natl Acad. Sci. USA 2012, 109, 12740–12745. [Google Scholar] [CrossRef]
- Moyed, H.S.; Bertrand, K.P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983, 155, 768–775. [Google Scholar] [CrossRef]
- Slattery, A.; Victorsen, A.H.; Brown, A.; Hillman, K.; Phillips, G.J. Isolation of highly persistent mutants of Salmonella enterica serovar typhimurium reveals a new toxin-antitoxin module. J. Bacteriol. 2013, 195, 647–657. [Google Scholar] [CrossRef]
- Shan, Y.; Lazinski, D.; Rowe, S.; Camilli, A.; Lewis, K. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. MBio 2015, 6, e00078-15. [Google Scholar] [CrossRef]
- McDermott, W. Microbial persistence. Yale J. Biol. Med. 1958, 30, 257–291. [Google Scholar]
- Johnson, P.J.; Levin, B.R. Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS Genet. 2013, 9, e1003123. [Google Scholar] [CrossRef]
- Chowdhury, N.; Kwan, B.W.; Wood, T.K. Persistence increases in the absence of the alarmone guanosine tetraphosphate by reducing cell growth. Sci. Rep. 2016, 6, 20519. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Wang, X.; O’Connor, H.F.; Benedik, M.J.; Wood, T.K. Bacterial persistence increases as environmental fitness decreases. Microb. Biotechnol. 2012, 5, 509–522. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Kaldalu, N.; Hauryliuk, V.; Tenson, T. Persisters–as elusive as ever. Appl. Microbiol. Biotechnol. 2016, 100, 6545–6553. [Google Scholar] [CrossRef]
- Shan, Y.; Gandt, A.B.; Rowe, S.E.; Deisinger, J.P.; Conlon, B.P.; Lewis, K. ATP-dependent persister formation in Escherichia coli. MBio 2017, 8, e02267-16. [Google Scholar] [CrossRef]
- Hobby, G.L.; Meyer, K.; Chaffee, E. Observations on the mechanism of action of penicillin. Exp. Biol. Med. 1942, 50, 281–285. [Google Scholar] [CrossRef]
- Bigger, J.W. Treatment of staphylococcal infections with penicillin by intermittent sterilization. Lancet 1944, 244, 497–500. [Google Scholar] [CrossRef]
- Van den Bergh, B.; Michiels, J.E.; Wenseleers, T.; Windels, E.M.; Boer, P.V.; Kestemont, D.; De Meester, L.; Verstrepen, K.J.; Verstraeten, N.; Fauvart, M.; et al. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat. Microbiol. 2016, 1, 16020. [Google Scholar] [CrossRef]
- Cohen, N.R.; Lobritz, M.A.; Collins, J.J. Microbial persistence and the road to drug resistance. Cell Host Microb. 2013, 13, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Levin-Reisman, I.; Ronin, I.; Gefen, O.; Braniss, I.; Shoresh, N.; Balaban, N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science 2017, 355, 826–830. [Google Scholar] [CrossRef]
- Shah, D.; Zhang, Z.; Khodursky, A.; Kaldalu, N.; Kurg, K.; Lewis, K. Persisters: A distinct physiological state of E. coli. BMC Microbiol. 2006, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.; Shan, Y. Why tolerance invites resistance. Science 2017, 355, 796. [Google Scholar] [CrossRef] [PubMed]
- Wiuff, C.; Andersson, D.I. Antibiotic treatment in vitro of phenotypically tolerant bacterial populations. J. Antimicrob. Chemother. 2007, 59, 254–263. [Google Scholar] [CrossRef]
- Kwan, B.W.; Valenta, J.A.; Benedik, M.J.; Wood, T.K. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 2013, 57, 1468–1473. [Google Scholar] [CrossRef]
- Kwan, B.W.; Osbourne, D.O.; Hu, Y.; Benedik, M.J.; Wood, T.K. Phosphodiesterase DosP increases persistence by reducing cAMPwhich reduces the signal indole. Biotechnol. Bioeng. 2015, 112, 588–600. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007, 5, 48–56. [Google Scholar] [CrossRef]
- Lewis, K. Multidrug tolerance of biofilms and persister cells. In Bacterial Biofilms; Romeo, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 322, pp. 107–131. [Google Scholar]
- Vahdani, M.; Azimi, L.; Asghari, B.; Bazmi, F.; Lari, R.A. Phenotypic screening of extended-spectrum ß-lactamase and metallo-ß-lactamase in multidrug-resistant Pseudomonas aeruginosa from infected burns. Annal. Burns Fire Disast. 2012, 25, 78–81. [Google Scholar]
- Defraine, V.; Fauvart, M.; Michiels, J. Fighting bacterial persistence: Current and emerging anti-persister strategies and therapeutics. Drug Resist. Update 2018, 38, 12–26. [Google Scholar] [CrossRef]
- Cabral, D.J.; Wurster, J.I.; Belenky, P. Antibiotic persistence as a metabolic adaptation: Stress, metabolism, the host, and new directions. Pharmaceuticals 2018, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-how P. aeruginosa can escape antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef]
- Yan, J.; Bassler, B.L. Surviving as a community: Antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe. 2019, 26, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Pál, C.; Papp, B.; Lercher, M.J.; Gillings, M.R.; Paulsen, I.T.; Tetu, S.G. Genomics and the evolution of antibiotic resistance. Ann. N. Y. Acad. Sci. 2017, 1388, 92–107. [Google Scholar]
- Fam, N.; Leflon-Guibout, V.; Fouad, S.; Aboul-Fadl, L.; Marcon, E.; Desouky, D.; El-Defrawy, I.; Abou-Aitta, A.; Klena, J.; Nicolas-Chanoine, M.H. CTX-M-15-producing clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microb. Drug Resist. 2011, 17, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lan, F.; Lu, Y.; Li, B. n Characterization of Integrons and Antimicrobial Resistance in Escherichia coli Sequence Type 131 Isolates. Can. J. Infect Dis. Med. Microbiol. 2020, 2020, 3826186. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Miller, O.K. A bacterium’s enemy isn’t your friend. Nature 2018, 563, 637–638. [Google Scholar] [CrossRef]
- Kallonen, T.; Brodrick, H.J.; Harris, S.R.; Corander, J.; Brown, N.M.; Martin, V.; Peacock, S.J.; Parkhill, J. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 2017, 27, 1437–1449. [Google Scholar] [CrossRef]
- McNally, A.; Kallonen, T.; Connor, C.; Abudahab, K.; Aanensen, D.M.; Horner, C.; Peacock, S.J.; Parkhill, J.; Croucher, N.J.; Corander, J. Diversification of colonization factors in a multidrug-resistant Escherichia coli lineage evolving under negative frequency-dependent selection. MBio 2019, 10, e00644-19. [Google Scholar] [CrossRef]
- Mendes, R.E.; Bell, J.M.; Turnidge, J.D.; Castanheira, M.; Jones, R.N. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: Report from the SENTRY Surveillance Program. J. Antimicrob. Chemother. 2009, 63, 55–59. [Google Scholar] [CrossRef]
- Fridman, O.; Goldberg, A.; Balaban, N.Q. Whack an E. coli with the morbidostat. Genome Biol. 2012, 13, 140. [Google Scholar] [CrossRef] [PubMed]
- Toprak, E.; Veres, A.; Yildiz, S.; Pedraza, J.M.; Chait, R.; Paulsson, J.; Kishony, R. Building a morbidostat: An automated continuous-culture device for studying bacterial drug resistance under dynamically sustained drug inhibition. Nat. Protoc. 2013, 8, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.C.; Lee, Y.T.; Wang, C.Y.; Yang, Y.T. Design and use of a low cost, automated morbidostat for adaptive evolution of bacteria under antibiotic drug selection. J. Vis. Exp. 2016, 115, e54426. [Google Scholar] [CrossRef] [PubMed]
- Nyerges, Á.; Csörgő, B.; Draskovits, G.; Kintses, B.; Szili, P.; Ferenc, G.; Révész, T.; Ari, E.; Nagy, I.; Bálint, B.; et al. Directed evolution of multiple genomic loci allows the prediction of antibiotic resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 5726–5735. [Google Scholar] [CrossRef] [PubMed]
- Knöppel, A.; Knopp, M.; Albrecht, L.M.; Lundin, E.; Lustig, U.; Näsvall, J.; Andersson, D.I. Genetic adaptation to growth under laboratory conditions in Escherichia coli and Salmonella enteri. Front. Microbiol. 2018, 9, 756. [Google Scholar] [CrossRef] [PubMed]
- Knöppel, A.; Näsvall, J.; Andersson, D.I. Evolution of antibiotic resistance without antibiotic exposure. Antimicrob. Agents. Chemother. 2017, 61, e01495-17. [Google Scholar] [CrossRef]
- Toprak, E.; Veres, A.; Michel, J.B.; Chait, R.; Hartl, D.L.; Kishony, R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 2011, 44, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Dößelmann, B.; Willmann, M.; Steglich, M.; Bunk, B.; Nübel, U.; Peter, S.; Neher, R.A. Rapid and consistent evolution of colistin resistance in extensively drug-resistant Pseudomonas aeruginosa during morbidostat culture. Antimicrob. Agents Chemother. 2017, 61, e00043-17. [Google Scholar] [CrossRef]
- Tan, K.; Nguyen, J.; Nguyen, K.; Huse, H.K.; Nieberg, P.H.; Wong-Beringer, A. Prevalence of the carbapenem-heteroresistant phenotype among ESBL-producing Escherichia coli and Klebsiella pneumoniae clinical isolates. J. Antimicrob. Chemother. 2020, 75, 1506–1512. [Google Scholar] [CrossRef] [PubMed]
- Barlow, M.; Hall, B.G. Experimental prediction of the evolution of cefepime resistance from the CMY-2 AmpC beta-lactamase. Genetics 2003, 164, 23–29. [Google Scholar]
- Barlow, M.; Hall, B.G. Experimental prediction of the natural evolution of antibiotic resistance. Genetics 2003, 163, 1237–1241. [Google Scholar]
- Courpon-Claudinon, A.; Lefort, A.; Panhard, X.; Clermont, O.; Dornic, Q.; Fantin, B.; Mentré, F.; Wolff, M.; Denamur, E.; Branger, C. Bacteraemia caused by third-generation cephalosporin-resistant in France: Prevalence, molecular epidemiology and clinical features. Clin. Microbiol. Infect. 2011, 17, 557–565. [Google Scholar] [CrossRef] [PubMed][Green Version]
- MacLean, R.C.; Buckling, A. The distribution of fitness effects of beneficial mutations in Pseudomonas aeruginosa. PLoS Genet. 2009, 5, e1000406. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.; Magalhães, S.; Gordo, I. Cost of antibiotic resistance and the geometry of adaptation. Mol. Biol. Evol. 2012, 29, 1417–1428. [Google Scholar] [CrossRef]
- Storz, J.F. Compensatory mutations and epistasis for protein function. Curr. Opin. Struct. Biol. 2018, 50, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Chevereau, G.; Dravecka, M.; Batur, T.; Guvenek, A.; Ayhan, D.H.; Toprak, E.; Bollenbach, T. Quantifying the determinants of evolutionary dynamics leading to drug resistance. PLoS Biol. 2015, 13, e1002299. [Google Scholar] [CrossRef]
- Lázár, V.; Singh, G.P.; Spohn, R.; Nagy, I.; Horváth, B.; Hrtyan, M.; Busa-Fekete, R.; Bogos, B.; Méhi, O.; Csörgő, B.; et al. Bacterial evolution of antibiotic hypersensitivity. Mol. Syst. Biol. 2013, 9, 700. [Google Scholar] [CrossRef]
- Lázár, V.; Nagy, I.; Spohn, R.; Csörgő, B.; Györkei, Á.; Nyerges, Á.; Horváth, B.; Vörös, A.; Busa-Fekete, R.; Hrtyan, M.; et al. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat. Commun. 2014, 5, 4352. [Google Scholar] [CrossRef]
- Pál, C.; Papp, B.; Lázár, V. Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 2015, 23, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Kintses, B.; Méhi, O.; Ari, E.; Számel, M.; Györkei, Á.; Jangir, P.K.; Nagy, I.; Pál, F.; Fekete, G.; Tengölics, R.; et al. Phylogenetic barriers to horizontal transfer of antimicrobial peptide resistance genes in the human gut microbiota. Nat. Microbiol. 2019, 4, 447–458. [Google Scholar] [CrossRef]
- Dunai, A.; Spohn, R.; Farkas, Z.; Lázár, V.; Györkei, Á.; Apjok, G.; Boross, G.; Szappanos, B.; Grézal, G.; Faragó, A.; et al. Rapid decline of bacterial drug-resistance in an antibiotic-free environment through phenotypic reversion. Elife 2019, 8, e47088. [Google Scholar] [CrossRef]
- Charretier, Y.; Diene, S.M.; Baud, D.; Chatellier, S.; Santiago-Allexant, E.; van Belkum, A.; Guigon, G.; Schrenzel, J. Colistin heteroresistance and involvement of the PmrAB regulatory system in Acinetobacter baumannii. Antimicrob Agents Chemother. 2018, 62, e00788-18. [Google Scholar] [CrossRef] [PubMed]
- Apjok, G.; Boross, G.; Nyerges, Á.; Fekete, G.; Lázár, V.; Papp, B.; Pál, C.; Csörgő, B. Limited evolutionary conservation of the phenotypic effects of antibiotic resistance mutations. Mol. Biol. Evol. 2019, 36, 1601–1611. [Google Scholar] [CrossRef]
- Kernéis, S.; Valade, S.; Woerther , P.L. Back into the wild: How resistant pathogens become susceptible again? Intensive Care Med. 2020, 46, 361–363. [Google Scholar] [CrossRef] [PubMed]
- Horváth, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Faure, E.; Kwong, K.; Nguyen, D. Pseudomonas aeruginosa in chronic lung infections: How to adapt within the host? Front. Immunol. 2018, 9, 2416. [Google Scholar] [CrossRef] [PubMed]
- Mensa, J.; Barberán, J.; Soriano, A.; Llinares, P.; Marco, F.; Cantón, R.; Bou, G.; del Castillo, J.G.; Maseda, E.; Azanza, J.R.; et al. Antibiotic selection in the treatment of acute invasive infections by Pseudomonas aeruginosa: Guidelines by the Spanish Society of Chemotherapy. Rev. Esp. Quimioter 2018, 31, 78–100. [Google Scholar]
- Falcone, G.; Garzelli, C.; Serra, M.C. Aspects of Pseudomonas aeruginosa opportunism. Giornale di batteriol. virol. ed immunol. 1979, 72, 62–71. [Google Scholar]
- Koontz, C.S.; Chang, M.C.; Meredith, J.W. Effects of empiric antibiotic administration for suspected pneumonia on subsequent opportunistic pulmonary infections/ discussion. Am. Surg. 2000, 66, 1110. [Google Scholar]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.L.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.R.; Ducret, V.; Leoni, S.; Perron, K. Pseudomonas aeruginosa zinc homeostasis: Key issues for an opportunistic pathogen. Biochim. Biophys. Acta. Gene Regul. Mech. 2019, 1862, 722–733. [Google Scholar] [CrossRef]
- Azam, M.W.; Khan, A.U. Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov. 2019, 24, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Baltrus, D.A.; Dougherty, K.; Diaz, B.; Murillo, R. Evolutionary Plasticity of AmrZ Regulation in Pseudomonas. mSphere 2018, 3, e00132-18. [Google Scholar] [CrossRef]
- Brüggemann, H.; Migliorini, L.B.; Sales, R.O.D.; Koga, P.C.M.; Souza, A.V.D.; Jensen, A.; Poehlein, A.; Brzuszkiewicz, E.; Doi, A.M.; Pasternak, J.; et al. Comparative genomics of nonoutbreak Pseudomonas aeruginosa strains underlines genome plasticity and geographic relatedness of the global clone ST235. Genome Biol. Evol. 2018, 10, 1852–1857. [Google Scholar] [CrossRef] [PubMed]
- Freschi, L.; Bertelli, C.; Jeukens, J.; Moore, M.P.; Kukavica-Ibrulj, I.; Emond-Rheault, J.G.; Hamel, J.; Fothergill, J.L.; Tucker, N.P.; McClean, S.; et al. Genomic characterization of an international Pseudomonas aeruginosa reference panel indicates that the two major groups draw upon distinct mobile gene pools. FEMS Microbiol. Lett. 2018, 365, 120. [Google Scholar] [CrossRef] [PubMed]
- Watkins, S.C.; Sible, E.; Putonti, C. Pseudomonas PB1-Like Phages: Whole genomes from metagenomes offer insight into an abundant group of bacteriophages. Viruses 2018, 10, E331. [Google Scholar] [CrossRef] [PubMed]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019, 6, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Lupo, A.; Haenni, M.; Madec, J.Y. Antimicrobial resistance in Acinetobacter spp. and Pseudomonas spp. Microbiol. Spectr. Microbiol Spectr. 2018, 6, ARBA-0007-2017. [Google Scholar] [CrossRef]
- Subedi, D.; Vijay, A.K.; Willcox, M. Overview of mechanisms of antibiotic resistance in Pseudomonas aeruginosa: An ocular perspective. Clin. Exp. Optom. 2018, 101, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Gaviard, C.; Jouenne, T.; Hardouin, J. Proteomics of Pseudomonas aeruginosa: The increasing role of post-translational modifications. Expert Rev. Proteom. 2018, 15, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Guitor, A.K.; Wright, G.D. Antimicrobial resistance and respiratory infections. Chest 2018, 154, 1202–1212. [Google Scholar] [CrossRef]
- Housseini, B.; Issa, K.; Phan, G.; Broutin, I. Functional mechanism of the efflux pumps transcription regulators from Pseudomonas aeruginosa based on 3D Structures. Front. Mol. Biosci. 2018, 5, 57. [Google Scholar] [CrossRef]
- Jabalameli, F.; Taki, E.; Emaneini, M.; Beigverdi, R. Prevalence of metallo-β-lactamase-encoding genes among carbapenem-resistant Pseudomonas aeruginosa strains isolated from burn patients in Iran. Rev. Soc. Bras. Med. Trop. 2018, 51, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Nehme, D.; Li, X.Z.; Elliot, R.; Poole, K. Assembly of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: Identification and characterization of mutations in mexA compromising MexA multimerization and interaction with MexB. J. Bacteriol. 2004, 186, 2973–2983. [Google Scholar] [CrossRef]
- Wang, Y.; Venter, H.; Ma, S. Efflux pump inhibitors: A novel approach to combat efflux-mediated drug resistance in bacteria. Curr. Drug Targets 2016, 17, 702–719. [Google Scholar] [CrossRef]
- Al-Wrafy, F.; Brzozowska, E.; Górska, S.; Gamian, A. Pathogenic factors of Pseudomonas aeruginosa—The role of biofilm in pathogenicity and as a target for phage therapy. Postepy High Med. Dosw. 2016, 71, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.R.; Isabella, V.M.; Lewis, K. Pseudomonas aeruginosa biofilms in disease. Microb. Ecol. 2014, 68, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef]
- Grosso-Becerra, M.V.; Santos-Medellín, C.; González-Valdez, A.; Méndez, J.L.; Delgado, G.; Morales-Espinosa, R.; Servín-González, L.; Alcaraz, L.D.; Soberón-Chávez, G. Pseudomonas aeruginosa clinical and environmental isolates constitute a single population with high phenotypic diversity. BMC Genom. 2014, 15, 318. [Google Scholar] [CrossRef] [PubMed]
- Jeukens, J.; Kukavica-Ibrulj, I.; Emond-Rheault, J.G.; Freschi, L.; Levesque, R.C. Comparative genomics of a drug-resistant Pseudomonas aeruginosa panel and the challenges of antimicrobial resistance prediction from genomes. FEMS Microbiol. Lett. 2017, 364, fnx161. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Ronco, E.; Naas, T.; Duport, C.; Michel-Briand, Y.; Labia, R. Characterization of a novel extended-spectrum beta-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1993, 37, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.; Kumari, H.; Balasubramanian, D.; Mathee, K. Cell-wall recycling and synthesis in Escherichia coli and Pseudomonas aeruginosa—Their role in the development of resistance. J. Med. Microbiol. 2018, 67, 1–21. [Google Scholar] [CrossRef]
- Aeschlimann, J.R. The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other gram-negative bacteria: Insights from the Society of Infectious Diseases Pharmacists. Pharmacother. J. Human Pharmac. Drug Ther. 2003, 23, 916–924. [Google Scholar] [CrossRef]
- Vila, J.; Martínez, J.L. Clinical impact of the over-expression of efflux pump in nonfermentative Gram-negative bacilli, development of efflux pump inhibitors. Curr. Drug Targets 2008, 9, 797–807. [Google Scholar] [CrossRef]
- Fischer, N.; Raunest, M.; Schmidt, T.H.; Koch, D.C.; Kandt, C. Efflux pump-mediated antibiotics resistance: Insights from computational structural biology. Interdiscip. Sci. 2014, 6, 1–12. [Google Scholar] [CrossRef]
- Puzari, M.; Chetia, P. RND efflux pump mediated antibiotic resistance in Gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa: A major issue worldwide. World J. Microbiol. Biotechnol. 2017, 33, 24. [Google Scholar] [CrossRef]
- Vahaboglu, H.; Coskunkan, F.; Tansel, O.; Ozturk, R.; Sahin, N.; Koksal, I.; Kocazeybek, B.; Tatman-Otkun, M.; Leblebicioglu, H.; Ozinel, M.A.; et al. Clinical importance of extended-spectrum beta-lactamase (PER-1-type)-producing Acinetobacter spp. and Pseudomonas aeruginosa strains. J. Med. Microbiol. 2001, 50, 642–645. [Google Scholar] [CrossRef]
- Kidd, J.M.; Kuti, J.L.; Nicolau, D.P. Novel pharmacotherapy for the treatment of hospital-acquired and ventilator-associated pneumonia caused by resistant gram-negative bacteria. Expert Opin. Pharmacother. 2018, 19, 397–408. [Google Scholar] [CrossRef]
- Szabó, D.; Szentandrássy, J.; Juhász, Z.S.; Katona, K.; Nagy, K.; Rókusz, L. Imported PER-1 producing Pseudomonas aeruginosa, PER-1 producing Acinetobacter baumannii and VIM-2-producing Pseudomonas aeruginosa strains in Hungary. Ann. Clin. Microbiol. Antimicrob. 2008, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Lagacé-Wiens, P.R.S.; Adam, H.J.; Poutanen, S.; Baxter, M.R.; Denisuik, A.J.; Golden, A.R.; Nichol, K.A.; Walkty, A.; Karlowsky, J.A.; Mulvey, M.R.; et al. Trends in antimicrobial resistance over 10 years among key bacterial pathogens from Canadian hospitals: Results of the CANWARD study 2007-16. J. Antimicrob. Chemother. 2019, 74, iv22–iv31. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Golden, A.R.; Zelenitsky, S.; Wiebe, K.; Lawrence, C.K.; Adam, H.J.; Idowu, T.; Domalaon, R.; Schweizer, F.; Zhanel, M.A.; et al. Cefiderocol: A siderophore cephalosporin with activity against carbapenem-resistant and multidrug-resistant gram-negative bacilli. Drugs 2019, 79, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Wozniak, T.M.; Barnsbee, L.; Lee, X.J.; Pacella, R.E. Using the best available data to estimate the cost of antimicrobial resistance: A systematic review. Antimicrob. Resist. Infect. Control 2019, 8, 26. [Google Scholar] [CrossRef]
- Hirakata, Y.; Srikumar, R.; Poole, K.; Gotoh, N.; Suematsu, T.; Kohno, S.; Kamihira, S.; Hancock, R.E.; Speert, D.P. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 2002, 196, 109–118. [Google Scholar] [CrossRef]
- Nehme, D.; Poole, K. Interaction of the MexA and MexB components of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: Identification of MexA extragenic suppressors of a T578I mutation in MexB. Antimicrob. Agents Chemother. 2005, 49, 4375–4378. [Google Scholar] [CrossRef][Green Version]
- Jeukens, J.; Boyle, B.; Kukavica-Ibrulj, I.; Ouellet, M.M.; Aaron, S.D.; Charette, S.J.; Fothergill, J.L.; Tucker, N.P.; Winstanley, C.; Levesque, R.C. Comparative genomics of isolates of a Pseudomonas aeruginosa epidemic strain associated with chronic lung infections of cystic fibrosis patients. PLoS ONE 2014, 9, e87611. [Google Scholar] [CrossRef]
- Martis, N.; Leroy, S.; Blanc, V. Colistin in multi-drug resistant Pseudomonas aeruginosa blood-stream infections: A narrative review for the clinician. J. Infect. 2014, 69, 1–12. [Google Scholar] [CrossRef]
- Catry, B.; Cavaleri, M.; Baptiste, K.; Grave, K.; Grein, K.; Holm, A.; Jukes, H.; Liebana, E.; Navas, A.L.; Mackay, D.; et al. Use of colistin-containing products within the European Union and European Economic Area (EU/EEA): Development of resistance in animals and possible impact on human and animal health. J. Antimicrob. Agents. 2015, 46, 297–306. [Google Scholar] [CrossRef]
- Hadadi-Fishani, M.; Khaledi, A.; Fatemi-Nasab, Z.S. Corelation between biofilm formation and antibiotic resistance in Pseudomonas aeruginosa: A meta-analysis. Infez. Med. 2020, 28, 47–54. [Google Scholar]
- Valli, R.X.E.; Lyng, M.; Kirkpatrick, C.L. There Is No Hiding if You Seq: Recent breakthroughs in Pseudomonas aeruginosa Research Revealed by Genomic and Transcriptomic Next-Generation Sequencing. J. Med. Microbiol. 2020, 69, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Lai, Y.; Bougouffa, S.; Zeling, X.Z.; Yan, A. Comparative genome and transcriptome analysis reveal distinctive surface characteristics and unique physiological potentials of Pseudomonas aeruginosa ATCC 27853. BMC Genom. 2017, 18, 459. [Google Scholar] [CrossRef] [PubMed]
- Bianconi, I.; Jeukens, J.; Freschi, L.; Alcalá-Franco, B.; Facchini, M.; Boyle, B.; Molinaro, A.; Kukavica-Ibrulj, I.; Tümmler, B.; Levesque, R.C.; et al. Comparative genomics and biological characterization of sequential Pseudomonas aeruginosa isolates from persistent airways infection. BMC Genom. 2015, 16, 1105. [Google Scholar] [CrossRef] [PubMed]
- Freschi, L.; Jeukens, J.; Kukavica-Ibrulj, I.; Boyle, B.; Dupont, M.J.; Laroche, J.; Larose, S.; Maaroufi, H.; Fothergill, J.L.; Moore, M.; et al. Clinical utilization of genomics data produced by the international Pseudomonas aeruginosa consortium. Front. Microbiol. 2015, 6, 1036. [Google Scholar] [CrossRef]
- Cornforth, D.M.; Dees, J.L.; Ibberson, C.B.; Huse, H.K.; Mathiesen, I.H.; Kirketerp-Møller, K.; Wolcott, R.D.; Rumbaugh, K.P.; Bjarnsholt, T.; Whiteley, M. Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl. Acad. Sci. USA 2018, 115, E5125–E5134. [Google Scholar] [CrossRef]
- Kempf, M.; Rolain, J.M. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: Clinical impact and therapeutic options. Int. J. Antimicrob. Agents 2012, 39, 105–114. [Google Scholar] [CrossRef]
- Dubern, J.F.; Cigana, C.; De Simone, M.; Lazenby, J.; Juhas, M.; Schwager, S.; Bianconi, I.; Döring, G.; Eberl, L.; Williams, P.; et al. Integrated whole-genome screening for Pseudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ. Microbiol. 2015, 17, 4379–4393. [Google Scholar] [CrossRef]
- Beijerinck, M. Pigmenten als oxydatieproducten gevormd door bacterien. Versl. Koninklijke Akad. Wetensch. Amsterdam 1911, 19, 1092–1103. [Google Scholar]
- Lessel, E.F. Subcommittee on nomenclature of Moraxella and allied bacteria. Int. J. Syst. Bacteriol. 1971, 21, 213–214. [Google Scholar] [CrossRef][Green Version]
- Lautrop, H. Bergey’s Manual of Determinative Bacteriology; Williams & Wilkins Co.: Baltimore, MD, USA, 1974. [Google Scholar]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Bouvet, P.J.M.; Grimont, P.A.D. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Evol. Microbiol. 1986, 36, 228–240. [Google Scholar]
- Robenshtok, E.; Paul, M.; Leibovici, L.; Fraser, A.; Pitlik, S.; Ostfeld, I.; Samra, Z.; Perez, S.; Lev, B.; Weinberger, M. The significance of Acinetobacter baumannii bacteraemia compared with Klebsiella pneumoniae bacteraemia: Risk factors and outcomes. J. Hosp. Infect. 2006, 64, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef] [PubMed]
- MarÝ-Almirall, M.; Cosgaya, C.; Higgins, P.G.; Van Assche, A.; Telli, M.; Huys, G.; Lievens, B.; Seifert, H.; Dijkshoorn, L.; Roca, I.; et al. MALDI-TOF/MS identification of species from the Acinetobacter baumannii (Ab) group revisited: Inclusion of the novel A. seifertii and A. dijkshoorniae species. Clin. Microbiol. Infect. 2017, 23, 210.e1–210.e9. [Google Scholar] [CrossRef] [PubMed]
- Wareth, G.; Neubauer, H.; Sprague, L.D. Acinetobacter baumannii—A neglected pathogen in veterinary and environmental health in Germany. Vet. Res. Commun. 2019, 43, 1–6. [Google Scholar] [CrossRef]
- Van der Kolk, J.H.; Endimiani, A.; Graubner, C.; Gerber, V.; Perreten, V. Acinetobacter in veterinary medicine, with an emphasis on Acinetobacter baumannii. J. Glob. Antimicrob. Resist. 2019, 16, 59–71. [Google Scholar] [CrossRef]
- Peleg, A.Y.; de Breij, A.; Adams, M.D.; Cerqueira, G.M.; Mocali, S.; Galardini, M.; Nibbering, P.H.; Earl, A.M.; Ward, D.V.; Paterson, D.L.; et al. The success of Acinetobacter species; genetic, metabolic and virulence attributes. PLoS ONE 2012, 7, e46984. [Google Scholar] [CrossRef]
- Zarrilli, R.; Pournaras, S.; Giannouli, M.; Tsakris, A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. J. Antimicrob. Agents 2013, 41, 11–19. [Google Scholar] [CrossRef]
- Lin, M.F.; Lan, C.Y. Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J. Clin. Cases 2014, 2, 787–814. [Google Scholar] [CrossRef]
- Gales, A.C.; Seifert, H.; Gur, D.; Castanheira, M.; Jones, R.N.; Sader, H.S. Antimicrobial susceptibility of Acinetobacter calcoaceticus-Acinetobacter baumannii complex and Stenotrophomonas maltophilia clinical isolates: Results from the SENTRY antimicrobial surveillance program (1997–2016). Open Forum Infect. Dis. 2019, 6, S34–S46. [Google Scholar] [CrossRef] [PubMed]
- Flamm, R.K.; Shortridge, D.; Castanheira, M.; Sader, H.S.; Pfaller, M.A. In Vitro Activity of Minocycline against U.S. Isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus species complex, Stenotrophomonas maltophilia, and Burkholderia cepacia complex: Results from the SENTRY Antimicrobial Surveillance Program (2014–2018). Antimicrob Agents Chemother. 2019, 63, e01154-19. [Google Scholar] [PubMed]
- Eichenberger, E.M.; Thaden, J.T. Epidemiology and mechanisms of resistance of extensively drug resistant gram-negative bacteria. Antibiotics 2019, 8, E37. [Google Scholar] [CrossRef] [PubMed]
- Kochar, M.; Crosatti, M.; Harrison, E.M.; Rieck, B.; Chan, J.; Constantinidou, C.; Pallen, M.; Ou, H.Y.; Rajakumar, K. Deletion of TnAbaR23 results in both expected and unexpected antibiogram changes in a multidrug-resistant Acinetobacter baumannii strain. Antimicrob Agents Chemother. 2012, 56, 1845–1853. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, C.H.; Nastro, M.; Famiglietti, A. Carbapenemases in Acinetobacter baumannii. Review of their dissemination in Latin America. Rev. Argent. Microbiol. 2018, 50, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Gniadek, T.J.; Carroll, K.C.; Simner, P.J. Carbapenem-resistant non-glucose-fermenting gram-negative Bacilli: The missing piece to the puzzle. J. Clin. Microbiol. 2016, 54, 1700–1710. [Google Scholar] [CrossRef]
- Clark, N.M.; Zhanel, G.G.; Lynch, J.P. Emergence of antimicrobial resistance among Acinetobacter species: A global threat. Curr. Opin. Crit. Care 2016, 22, 491–499. [Google Scholar] [CrossRef]
- Seiffert, S.N.; Perreten, H.M.V.; Endimiani, A. Extended spectrum cephalosporin resistant Gram-negative organisms in livestock: An emerging problem for human health. Drug Resist. Update 2013, 16, 22–45. [Google Scholar] [CrossRef]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef]
- Chan, J.Z.; Halachev, M.R.; Loman, N.J.; Constantinidou, C.; Pallen, M.J. Defining bacterial species in the genomic era: Insights from the genus Acinetobacter. BMC Microbiol. 2012, 12, 302. [Google Scholar] [CrossRef]
- Sahl, J.W.; Gillece, J.D.; Schupp, J.M.; Waddell, V.G.; Driebe, E.M.; Engelthaler, D.M.; Keim, P. Evolution of a pathogen: A comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS ONE 2013, 8, e54287. [Google Scholar] [CrossRef]
- Vilacoba, E.; Almuzara, M.; Gulone, L.; Traglia, G.M.; Figueroa, S.A.; Sly, G.; Fernández, A.; Centrón, D.; Ramírez, M.S. Emergence and spread of plasmid-borne tet(B):ISCR2 in minocycline-resistant Acinetobacter baumannii isolates. Antimicrob. Agents Chemother. 2013, 57, 651–654. [Google Scholar] [CrossRef]
- Vilacoba, E.; Déraspe, M.; Traglia, G.M.; Roy, P.H.; Ramírez, M.S.; Centrón, D. Draft genome sequence of an international clonal lineage 1 Acinetobacter baumannii strain from Argentina. Genome Announc. 2014, 2, e01190-14. [Google Scholar] [CrossRef]
- Aron, A.T.; Heffern, M.C.; Lonergan, Z.R.; Vander Wal, M.N.; Blank, B.R.; Spangler, B.; Zhang, Y.; Park, H.M.; Stahl, A.; Renslo, A.R.; et al. In vivo bioluminescence imaging of labile iron accumulation in a murine model of Acinetobacter baumannii infection. Proc. Natl. Acad. Sci. USA 2017, 114, 12669–12674. [Google Scholar] [CrossRef]
- Post, V.; Hall, R.M. AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009, 53, 2667–2671. [Google Scholar] [CrossRef]
- Farrugia, D.N.; Elbourne, L.D.; Hassan, K.A.; Eijkelkamp, B.A.; Tetu, S.G.; Brown, M.H.; Shah, B.S.; Peleg, A.Y.; Mabbutt, B.C.; Paulsen, I.T. The complete genome and phenome of a community-acquired Acinetobacter baumannii. PLoS ONE 2013, 8, e58628. [Google Scholar] [CrossRef]
- Gallagher, L.A.; Lee, S.A.; Manoil, C. Importance of core genome functions for an extreme antibiotic resistance trait. MBio. 2017, 8, e01655-17. [Google Scholar] [CrossRef]
- Karah, N.; Sundsfjord, A.; Towner, K.; Samuelsen, O. Insights into the global molecular epidemiology of carbapenem non susceptible clones of Acinetobacter baumannii. Drug Resist. Update 2012, 15, 237–247. [Google Scholar] [CrossRef]
- Dhabaan, G.N.; AbuBakar, S.; Cerqueira, G.M.; Al-Haroni, M.; Pang, S.P.; Hassan, H. Imipenem treatment induces expression of important genes and phenotypes in a resistant Acinetobacter baumannii isolate. Antimicrob. Agents Chemother. 2015, 60, 1370–1376. [Google Scholar] [CrossRef]
- Adams, M.D.; Goglin, K.; Molyneaux, N.; Hujer, K.M.; Lavender, H.; Jamison, J.J.; MacDonald, I.J.; Martin, K.M.; Russo, T.; Campagnari, A.A.; et al. Comparative genome sequence analysis of multidrug resistant Acinetobacter baumannii. J. Bacteriol. 2008, 190, 8053–8064. [Google Scholar] [CrossRef]
- Hasani, A.; Sheikhalizadeh, V.; Ahangarzadeh Rezaee, M.; Rahmati-Yamchi, M.; Hasani, A.; Ghotaslou, R.; Goli, H.R. Frequency of aminoglycoside-modifying enzymes and ArmA among different sequence groups of Acinetobacter baumannii in Iran. Microb. Drug Resist. 2016, 22, 347–353. [Google Scholar] [CrossRef]
- Caputo, A.; Fournier, P.E.; Raoult, D. Genome and pan-genome analysis to classify emerging bacteria. Biol. Direct 2019, 14, 5. [Google Scholar] [CrossRef]
- Diancourt, L.; Passe, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The population structure of Acinetobacter baumannii: Expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 2010, 5, e10034. [Google Scholar] [CrossRef]
- Zordan, S.; Prenger-Berninghoff, E.; Weiss, R.; van der Reijden, T.; van den Broek, P.; Baljer, G.; Dijkshoorn, L. Multidrug resistant Acinetobacter baumannii in veterinary clinics, Germany. Emerg. Infect. Dis. 2011, 17, 1751–1754. [Google Scholar] [CrossRef]
- Joshi, S.G.; Litake, G.M. Acinetobacter baumannii: An emerging pathogenic threat to public health. World J. Clin. Infect. Dis. 2013, 3, 25–36. [Google Scholar] [CrossRef]
- Repizo, G.D.; Viale, A.M.; Borges, V.; Cameranesi, M.M.; Taib, N.; Espariz, M.; Brochier-Armanet, C.; Gomes, J.P.; Salcedo, S.P. The environmental Acinetobacter baumannii isolate DSM30011 reveals clues into the preantibiotic era genome diversity, virulence potential, and niche range of a predominant nosocomial pathogen. Genome Biol. Evol. 2017, 9, 2292–2307. [Google Scholar] [CrossRef]
- Vila, J.; Martı, S.; Sánchez-Céspedes, J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2007, 59, 1210–1215. [Google Scholar] [CrossRef]
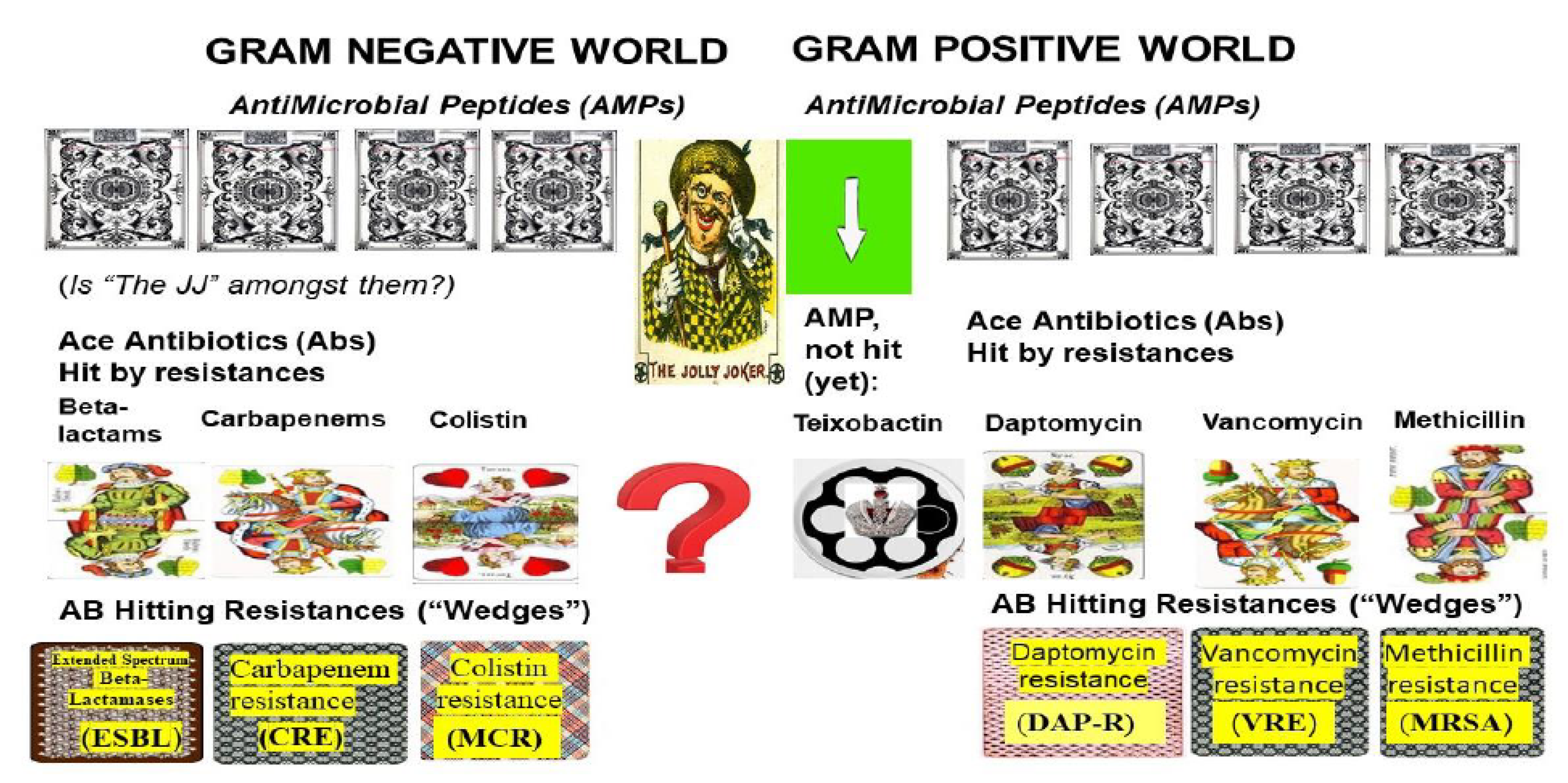

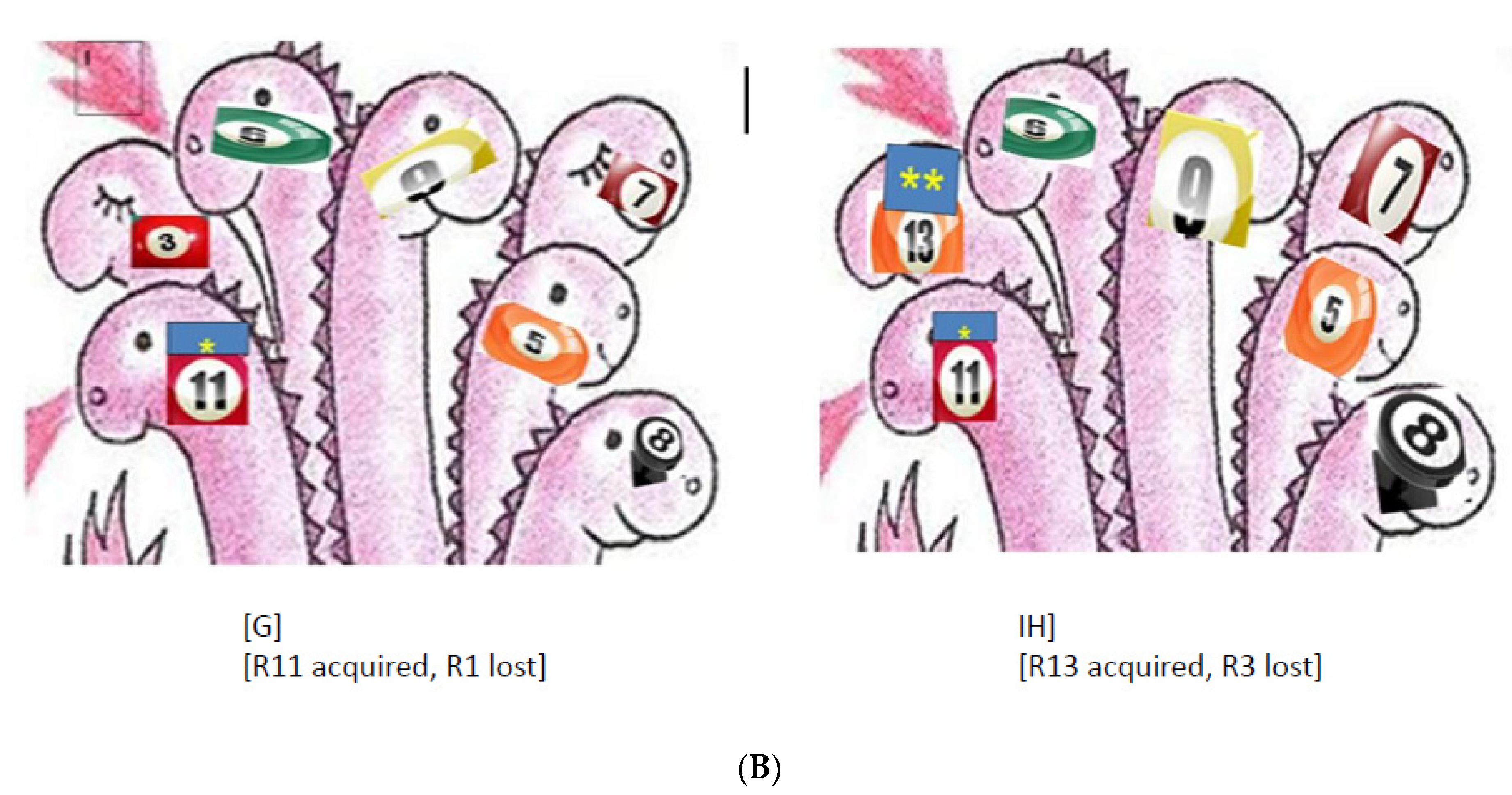
| Questions to PubMed (Entries) | Answers (Items): Articles: A, Reviews: R) | |||||
|---|---|---|---|---|---|---|
| ALL | 2019 | 2018 | 2009 | 2001 | ||
| A | R | R | R | R | R | |
| Beta-lactams | 130,408 | 9814 | 121 | 325 | 345 | 332 |
| Extended Spectrum Beta-Lactam Resistance (ESBL) | 5906 | 510 | 46 | 64 | 20 | 11 |
| ESBL Klebsiella | 2763 | 192 | 11 | 13 | 10 | 8 |
| ESBL Escherichia coli | 3952 | 191 | 5 | 20 | 12 | 6 |
| ESBL Acinetobacter | 505 | 64 | 5 | 4 | 6 | 1 |
| ESBL Pseudomonas | 853 | 98 | 7 | 8 | 6 | 0 |
| Carbapenems | 17,031 | 1992 | 135 | 193 | 114 | 34 |
| Carbapenem Resistance (CRE) | 11,981 | 1461 | 118 | 164 | 95 | 20 |
| CRE Enterobacteriaceae | 2085 | 344 | 50 | 64 | 7 | 0 |
| CRE Klebsiella | 3067 | 292 | 22 | 42 | 19 | 4 |
| CRE Acinetobacter | 2959 | 334 | 36 | 32 | 23 | 3 |
| CRE Pseudomonas | 3142 | 377 | 28 | 36 | 22 | 5 |
| Colistin | 7027 | 653 | 72 | 78 | 21 | 4 |
| Colistin resistance | 4640 | 509 | 62 | 63 | 18 | 3 |
| mcr colistin resistance | 625 | 39 | 7 | 13 | 0 | 0 |
| Colistin resistance, Enterobacter | 1712 | 150 | 20 | 29 | 2 | 0 |
| Colistin resistance, Klebsiella | 914 | 111 | 12 | 13 | 3 | 0 |
| Colistin resistance, Acinetobacter | 1363 | 185 | 22 | 17 | 9 | 1 |
| Colistin resistance, Pseudomonas | 1123 | 154 | 10 | 14 | 11 | 2 |
| Questions to PubMed (Entries) | Answers (Items): Articles: A, Reviews: R) | |||||
|---|---|---|---|---|---|---|
| ALL | 2019 | 2018 | 2009 | 2001 | ||
| A | R | R | R | R | R | |
| Acinetobacter MDR | 1261 | 174 | 25 | 25 | 10 | 0 |
| Acinetobacter MDR/XDR | 18 | 2 | 0 | 0 | 0 | 0 |
| Acinetobacter Pandrug-resistant | 96 | 23 | 2 | 4 | 2 | 0 |
| Pseudomonas MDR | 1310 | 165 | 19 | 27 | 9 | 2 |
| Pseudomonas MDR/XDR | 25 | 4 | 0 | 1 | 0 | 0 |
| Pseudomonas Pandrug-resistant | 52 | 1 | 1 | 2 | 1 | 0 |
| Klebsiella MDR | 1056 | 100 | 18 | 14 | 4 | 1 |
| Klebsiella MDR/XDR | 9 | 2 | 1 | 1 | 0 | 0 |
| Klebsiella Pandrug-resistant | 48 | 11 | 1 | 1 | 0 | 0 |
| Enterobacter MDR | 272 | 14 | 5 | 1 | 1 | 0 |
| Enterobacter MDR/XDR | 8 | 2 | 1 | 0 | 0 | 0 |
| Enterobacter Pandrug-resistant | 1 | 0 | 0 | 0 | 0 | 0 |
| cephalosporin resistant Enterobacteriaceae MDR | 121 | 12 | 2 | 3 | 0 | 0 |
| cephalosporin resistant Enterobacteriaceae MDR/XDR | 1 | 0 | 0 | 0 | 0 | 0 |
| cephalosporin-resistant Enterobacteriaceae Pandrug-resistant | 0 | 0 | 0 | 0 | 0 | 0 |
| Salmonella typhi MDR | 177 | 23 | 2 | 3 | 1 | 0 |
| Salmonella typhi MDR/XDR | 8 | 1 | 0 | 0 | 1 | 0 |
| Salmonella typhi Pandrug-resistant | 0 | 0 | 0 | 0 | 0 | 0 |
| Escherichia coli MDR | 1721 | 107 | 12 | 12 | 6 | 3 |
| Escherichia coli MDR/XDR | 8 | 1 | 0 | 0 | 0 | 0 |
| Escherichia coli Pandrug-resistant | 24 | 3 | 0 | 0 | 0 | 0 |
| Questions to PubMed (Entries) | Evoked Items: Articles: A, Reviews: R) | |||||
|---|---|---|---|---|---|---|
| ALL | 2019 | 2018 | 2009 | 2001 | ||
| A | R | R | R | R | R | |
| Helicobacter pylori MDR | 54 | 2 | 0 | 1 | 0 | 0 |
| Helicobacter pylori MDR/XDR | 0 | 0 | 0 | 0 | 0 | 0 |
| Helicobacter pylori Pandrug-resistant | 1 | 1 | 0 | 0 | 0 | 0 |
| clarithromycin-resistant Helicobacter pylori | 316 | 33 | 3 | 5 | 0 | 1 |
| clarithromycin-resistant Helicobacter pylori MDR | 4 | 0 | 0 | 0 | 0 | 0 |
| clarithromycin-resistant Helicobacter pylori MDR/XDR | 0 | 0 | 0 | 0 | 0 | 0 |
| clarithromycin-resistant Helicobacter pylori Pandrug-resistant | 0 | 0 | 0 | 0 | 0 | 0 |
| Campylobacter MDR | 72 | 5 | 0 | 1 | 1 | 0 |
| Campylobacter MDR/XDR | 0 | 0 | 0 | 0 | 0 | 0 |
| Campylobacter Pandrug-resistant | 0 | 0 | 0 | 0 | 0 | 0 |
| fluoroquinolone-resistant Campylobacter | 69 | 10 | 4 | 3 | 2 | 3 |
| fluoroquinolone-resistant Campylobacter MDR | 4 | 0 | 0 | 0 | 0 | 0 |
| fluoroquinolone-resistant Campylobacter MDR/XDR | 0 | 0 | 0 | 0 | 0 | 0 |
| fluoroquinolone-resistant Campylobacter Pandrug-resistant | 0 | 0 | 0 | 0 | 0 | 0 |
| Neisseria gonorrhoeae MDR | 44 | 3 | 0 | 0 | 0 | 0 |
| Neisseria gonorrhoeae MDR/XDR | 0 | 0 | 0 | 0 | 0 | 0 |
| Neisseria gonorrhoeae Pandrug-resistant | 0 | 0 | 0 | 0 | 0 | 0 |
| Questions to PubMed (Entries) | Evoked Items: Articles: A, Reviews: R) | |||||
|---|---|---|---|---|---|---|
| ALL | 2019 | 2018 | 2009 | 2001 | ||
| A | R | R | R | R | R | |
| A. Temporal Distribution of PubMed Cited Reviews on MDR-related issues with MRSA Staphylococcus aureus | ||||||
| S. aureus MRSA | 31,827 | 2098 | 141 | 249 | 384 | 161 |
| S. aureus MRSA Antibiotic Resistance | 15,822 | 2085 | 35 | 83 | 148 | 109 |
| S. aureus Vancomycin Resistance | 6448 | 1119 | 22 | 36 | 73 | 60 |
| S. aureus MRSA VAN-R | 4591 | 885 | 13 | 28 | 50 | 42 |
| S. aureus Daptomycin resistance | 1462 | 319 | 11 | 17 | 26 | 1 |
| S. aureus MRSA DAP-R | 1169 | 264 | 10 | 13 | 18 | 2 |
| S. aureus Carbapenem Resistance | 1189 | 187 | 16 | 19 | 9 | 3 |
| Teixobactin | 69 | 13 | 1 | 4 | 0 | 0 |
| Teixobactin, MRSA | 15 | 3 | 0 | 0 | 0 | 0 |
| S. aureus MDR | 1122 | 119 | 9 | 17 | 9 | 2 |
| MDR/XDR Gram-positive | 21 | 5 | 1 | 2 | 0 | 0 |
| MDR/XDR S. aureus | 3 | 2 | 1 | 1 | 0 | 0 |
| B. Temporal Distribution of PubMed Cited Reviews on MDR-related Issues in the genus Enterococcus | ||||||
| Enterococcus Antibiotic Resistance | 9167 | 1060 | 26 | 50 | 51 | 51 |
| Enterococcus MDR | 418 | 44 | 6 | 5 | 0 | 2 |
| Enterococcus MDR/XDR | 2 | 2 | 1 | 1 | 0 | 0 |
| Enterococcus methicillin-resistant | 2253 | 453 | 18 | 22 | 23 | 19 |
| Enterococcus Vancomycin Resistance | 5355 | 713 | 55 | 77 | 53 | 58 |
| Enterococcus VRE | 2247 | 274 | 10 | 20 | 6 | 14 |
| E. faecium Vancomycin Resistance | 2635 | 226 | 10 | 12 | 8 | 14 |
| E. faecium VRE | 985 | 83 | 5 | 6 | 2 | 7 |
| E. faecalis Vancomycin Resistance | 1824 | 241 | 6 | 11 | 6 | 4 |
| E. faecalis VRE | 564 | 45 | 1 | 4 | 1 | 3 |
| E. caecum Vancomycin Resistance | 13 | 0 | 0 | 0 | 0 | 0 |
| E. gallinarum Vancomycin Resistance | 278 | 10 | 11 | 2 | 6 | 7 |
| Enterococcus Daptomycin resistance | 584 | 120 | 30 | 42 | 35 | 7 |
| E. faecium Daptomycin resistance | 347 | 47 | 3 | 2 | 4 | 4 |
| E. faecalis Daptomycin resistance | 295 | 33 | 1 | 2 | 2 | 0 |
| E. caecum Daptomycin Resistance | 2 | 0 | 0 | 0 | 0 | 0 |
| E. gallinarum Daptomycin Resistance | 11 | 1 | 0 | 0 | 0 | 0 |
| Enterococcus carbapenem resistance | 524 | 72 | 7 | 5 | 2 | 2 |
| C. Antibiotic Multiresistance (MDR), Extreme Large Spectrum (XDR) Resistance, and Pan-resistance in Gram-Positive ESKAPE Pathogens | ||||||
| Gram-positive bacteria MDR | 4292 | 542 | 11 | 42 | 34 | 12 |
| Gram-positive bacteria MDR/XDR | 21 | 5 | 1 | 2 | 0 | 0 |
| Gram-positive bacteria PDR | 17 | 6 | 0 | 1 | 0 | 0 |
| S. aureus MDR | 1122 | 119 | 9 | 17 | 9 | 2 |
| S. aureus MDR/XDR | 3 | 2 | 1 | 1 | 0 | 0 |
| S. aureus Pandrug-resistant | 19 | 9 | 0 | 2 | 0 | 0 |
| S. aureus MRSA MDR | 715 | 84 | 6 | 14 | 8 | 1 |
| S. aureus MRSA MDR/XDR | 2 | 2 | 1 | 3 | 0 | 0 |
| S. aureus MRSA Pandrug-resistant | 12 | 5 | 0 | 1 | 0 | 0 |
| Enterococcus MDR | 418 | 44 | 6 | 4 | 0 | 2 |
| Enterococcus MDR/XDR Enterococcus | 3 | 1 | 1 | 0 | 0 | 0 |
| Enterococcus Pandrug-resistant | 9 | 4 | 0 | 1 | 0 | 0 |
| Enterococcus VRE MDR | 92 | 12 | 3 | 4 | 0 | 0 |
| Enterococcus VRE MDR/XDR | 1 | 1 | 1 | 0 | 0 | 0 |
| Enterococcus VRE Pandrug-resistant | 3 | 0 | 0 | 0 | 0 | 0 |
| Questions to PubMed (Entries) | Evoked Items: Articles: A, Reviews: R | |||||
|---|---|---|---|---|---|---|
| ALL | 2019 | 2018 | 2009 | 2001 | ||
| A | R | R | R | R | R | |
| Acinetobacter | ||||||
| Intrinsic resistance | 243 | 46 | 2 | 3 | 2 | 0 |
| Acquired Resistance | 1208 | 243 | 9 | 14 | 16 | 6 |
| Resistance genes | 1808 | 103 | 11 | 10 | 3 | 1 |
| Plasmids, Genomic Islands | 27 | 4 | 0 | 1 | 0 | 0 |
| Antibiotic Resistance mechanisms | 932 | 206 | 14 | 13 | 11 | 2 |
| Antibiotic Resistance, Genome plasticity | 24 | 4 | 0 | 0 | 0 | 0 |
| Horizontal Gene Transfer | 210 | 16 | 0 | 1 | 1 | 0 |
| Pangenome pangenomic | 9 | 1 | 0 | 0 | 0 | 0 |
| Clonal Global Distribution | 16 | 1 | 0 | 0 | 0 | 0 |
| Diseases | 4237 | 530 | 45 | 50 | 36 | 18 |
| Host range | 73 | 9 | 1 | 0 | 0 | 0 |
| Collateral Sensitivity | 5 | 1 | 0 | 0 | 0 | 0 |
| Pseudomonas | ||||||
| Intrinsic resistance | 577 | 117 | 8 | 11 | 3 | 4 |
| Acquired Resistance | 2249 | 437 | 25 | 32 | 27 | 12 |
| Resistance genes | 4647 | 233 | 12 | 17 | 4 | 6 |
| Plasmids, Genomic Islands | 39 | 2 | 0 | 0 | 0 | 0 |
| Antibiotic Resistance mechanisms | 2075 | 411 | 16 | 35 | 16 | 10 |
| Resistance, Genome plasticity | 36 | 4 | 1 | 0 | 0 | 0 |
| Horizontal Gene transfer | 512 | 44 | 2 | 3 | 2 | 1 |
| Pangenome | 50 | 4 | 0 | 0 | 0 | 0 |
| Clonal Global Distribution | 6 | 0 | 0 | 0 | 0 | 0 |
| Diseases | 22.950 | 2628 | 92 | 157 | 129 | 04 |
| Host range | 404 | 48 | 2 | 8 | 4 | 1 |
| Collateral Sensitivity | 10 | 1 | 0 | 0 | 0 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fodor, A.; Abate, B.A.; Deák, P.; Fodor, L.; Gyenge, E.; Klein, M.G.; Koncz, Z.; Muvevi, J.; Ötvös, L.; Székely, G.; et al. Multidrug Resistance (MDR) and Collateral Sensitivity in Bacteria, with Special Attention to Genetic and Evolutionary Aspects and to the Perspectives of Antimicrobial Peptides—A Review. Pathogens 2020, 9, 522. https://doi.org/10.3390/pathogens9070522
Fodor A, Abate BA, Deák P, Fodor L, Gyenge E, Klein MG, Koncz Z, Muvevi J, Ötvös L, Székely G, et al. Multidrug Resistance (MDR) and Collateral Sensitivity in Bacteria, with Special Attention to Genetic and Evolutionary Aspects and to the Perspectives of Antimicrobial Peptides—A Review. Pathogens. 2020; 9(7):522. https://doi.org/10.3390/pathogens9070522
Chicago/Turabian StyleFodor, András, Birhan Addisie Abate, Péter Deák, László Fodor, Ervin Gyenge, Michael G. Klein, Zsuzsanna Koncz, Josephat Muvevi, László Ötvös, Gyöngyi Székely, and et al. 2020. "Multidrug Resistance (MDR) and Collateral Sensitivity in Bacteria, with Special Attention to Genetic and Evolutionary Aspects and to the Perspectives of Antimicrobial Peptides—A Review" Pathogens 9, no. 7: 522. https://doi.org/10.3390/pathogens9070522
APA StyleFodor, A., Abate, B. A., Deák, P., Fodor, L., Gyenge, E., Klein, M. G., Koncz, Z., Muvevi, J., Ötvös, L., Székely, G., Vozik, D., & Makrai, L. (2020). Multidrug Resistance (MDR) and Collateral Sensitivity in Bacteria, with Special Attention to Genetic and Evolutionary Aspects and to the Perspectives of Antimicrobial Peptides—A Review. Pathogens, 9(7), 522. https://doi.org/10.3390/pathogens9070522