Seneca Valley Virus 3Cpro Cleaves PABPC1 to Promote Viral Replication
Abstract
1. Introduction
2. Results
2.1. SVV Infection Cleaved PABPC1 Protein
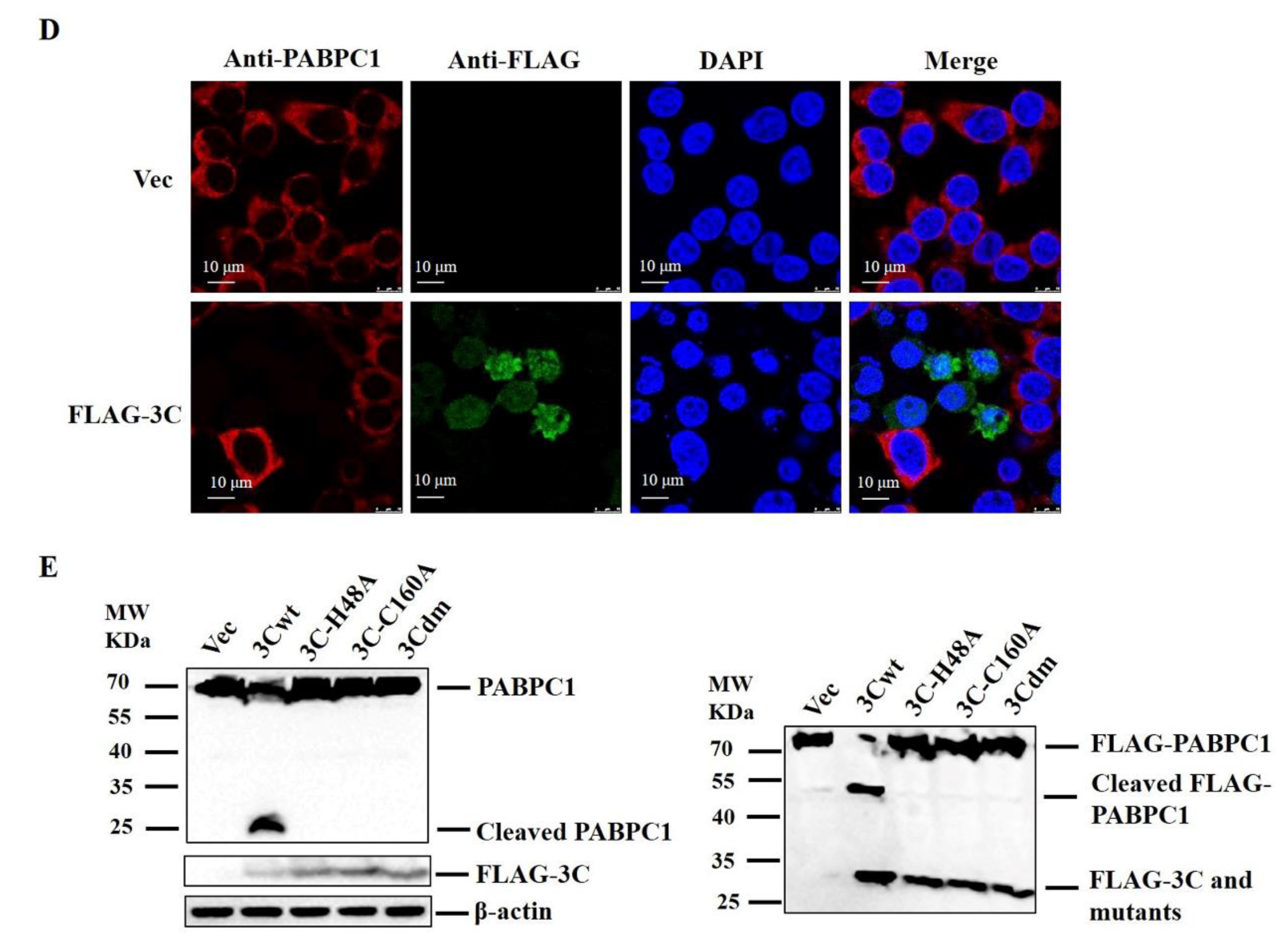
2.2. SVV 3Cpro Cleaved PABPC1, and 3Cpro Protease Activity Was Responsible for this Effect
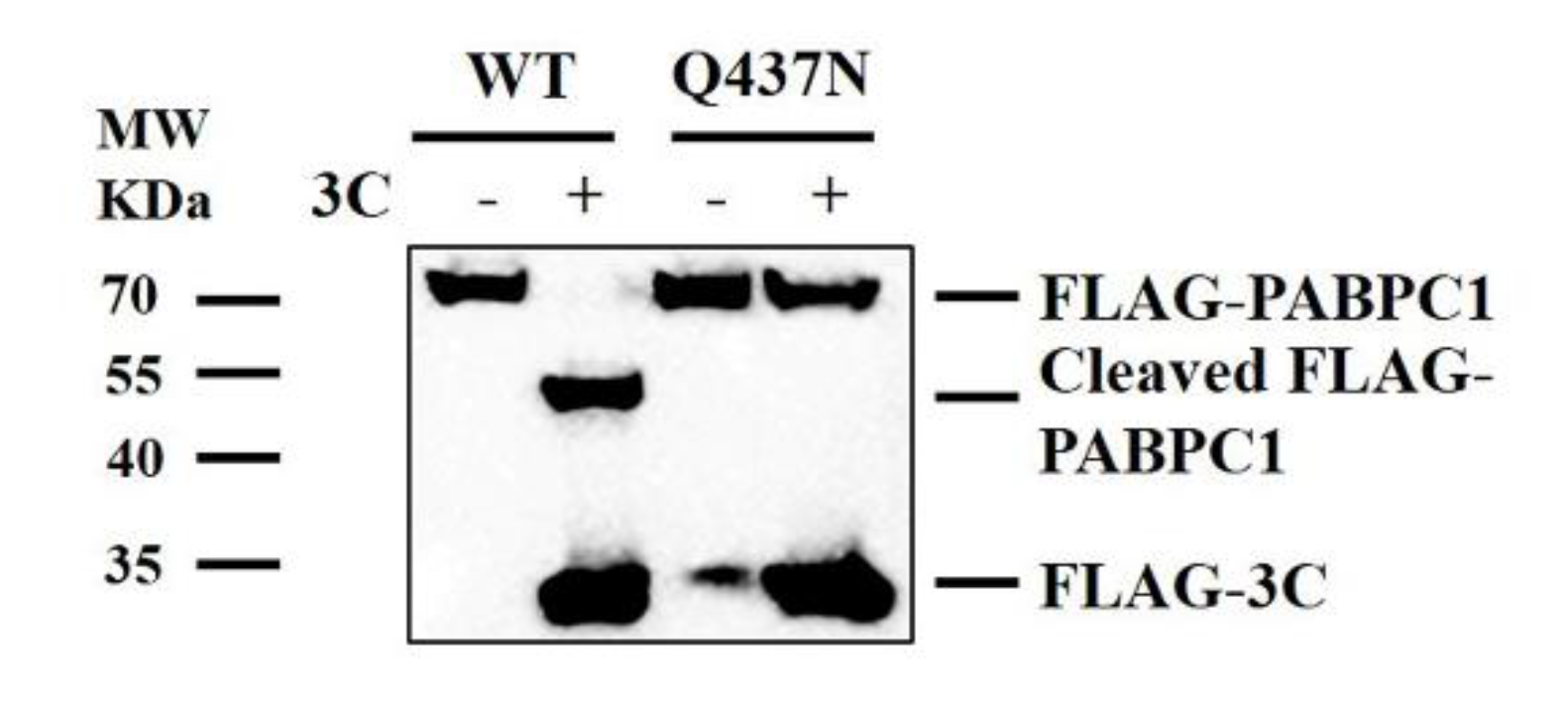
2.3. Cleavage Sites of PABPC1
2.4. SVV 3Cpro Protease Activity Was Essential for Inhibition of the Protein Synthesis Rates
2.5. PABPC1 Inhibited SVV Replication during Viral Infection
3. Discussion
4. Materials and Methods
4.1. Cells, Viruses and Infection
4.2. Plasmids and Antibodies
4.3. Western Blotting
4.4. Indirect Immunofluorescence Microscopy
4.5. Expression and Purification of Recombinant PABPC1 and SVV 3Cpro Proteins
4.6. In Vitro Cleavage Reaction
4.7. Knockdown of PABPC1 Using siRNA
4.8. RNA Extraction and qPCR
4.9. Puromycin Labeling
4.10. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hales, L.M.; Knowles, N.J.; Reddy, P.S.; Xu, L.; Hay, C.; Hallenbeck, P.L. Complete genome sequence analysis of seneca valley virus-001, a novel oncolytic picornavirus. J. Gen. Virol. 2008, 89, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Yang, F.; Chen, P.; Liu, H.; Cao, W.; Zhang, K.; Liu, X.; Zheng, H. Emergence of novel seneca valley virus strains in china, 2017. Transbound. Emerg. Dis. 2017, 64, 1024–1029. [Google Scholar] [CrossRef] [PubMed]
- Leme, R.A.; Alfieri, A.F.; Alfieri, A.A. Update on senecavirus infection in pigs. Viruses 2017, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Leme, R.A.; Zotti, E.; Alcantara, B.K.; Oliveira, M.V.; Freitas, L.A.; Alfieri, A.F.; Alfieri, A.A. Senecavirus a: An emerging vesicular infection in brazilian pig herds. Transbound. Emerg. Dis. 2015, 62, 603–611. [Google Scholar] [CrossRef]
- Tousignant, S.J.P.; Bruner, L.; Schwartz, J.; Vannucci, F.; Rossow, S.; Marthaler, D.G. Longitudinal study of senecavirus a shedding in sows and piglets on a single united states farm during an outbreak of vesicular disease. Bmc Vet. Res. 2017, 13, 277. [Google Scholar] [CrossRef]
- Vannucci, F.A.; Linhares, D.C.; Barcellos, D.E.; Lam, H.C.; Collins, J.; Marthaler, D. Identification and complete genome of seneca valley virus in vesicular fluid and sera of pigs affected with idiopathic vesicular disease, brazil. Transbound. Emerg. Dis. 2015, 62, 589–593. [Google Scholar] [CrossRef]
- Sun, D.; Vannucci, F.; Knutson, T.P.; Corzo, C.; Marthaler, D.G. Emergence and whole-genome sequence of senecavirus a in colombia. Transbound. Emerg. Dis. 2017, 64, 1346–1349. [Google Scholar] [CrossRef]
- Saeng-Chuto, K.; Rodtian, P.; Temeeyasen, G.; Wegner, M.; Nilubol, D. The first detection of senecavirus a in pigs in thailand, 2016. Transbound. Emerg. Dis. 2018, 65, 285–288. [Google Scholar] [CrossRef]
- Wu, Q.; Zhao, X.; Bai, Y.; Sun, B.; Xie, Q.; Ma, J. The first identification and complete genome of senecavirus an affecting pig with idiopathic vesicular disease in china. Transbound. Emerg. Dis. 2017, 64, 1633–1640. [Google Scholar] [CrossRef]
- Qian, S.; Fan, W.; Qian, P.; Chen, H.; Li, X. Isolation and full-genome sequencing of seneca valley virus in piglets from china, 2016. Virol. J. 2016, 13, 173. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, Q.; Bai, Y.; Chen, G.; Zhou, L.; Wu, Z.; Li, Y.; Zhou, W.; Yang, H.; Ma, J. Phylogenetic and genome analysis of seven senecavirus a isolates in china. Transbound. Emerg. Dis. 2017, 64, 2075–2082. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; Fan, W.; Liu, T.; Wu, M.; Zhang, H.; Cui, X.; Zhou, Y.; Hu, J.; Wei, S.; Chen, H.; et al. Seneca valley virus suppresses host type i interferon production by targeting adaptor proteins mavs, trif, and tank for cleavage. J. Virol. 2017, 91, e00823-17. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C.M.; Poirier, J.T.; Senzer, N.N.; Stephenson, J.; Loesch, D.; Burroughs, K.D.; Reddy, P.S.; Hann, C.L.; Hallenbeck, P.L. Phase i clinical study of seneca valley virus (svv-001), a replication-competent picornavirus, in advanced solid tumors with neuroendocrine features. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 2011, 17, 888–895. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhao, X.; Mao, H.; Baxter, P.A.; Huang, Y.; Yu, L.; Wadhwa, L.; Su, J.M.; Adesina, A.; Perlaky, L.; et al. Intravenous injection of oncolytic picornavirus svv-001 prolongs animal survival in a panel of primary tumor-based orthotopic xenograft mouse models of pediatric glioma. Neuro-Oncology 2013, 15, 1173–1185. [Google Scholar] [CrossRef]
- Reddy, P.S.; Burroughs, K.D.; Hales, L.M.; Ganesh, S.; Jones, B.H.; Idamakanti, N.; Hay, C.; Li, S.S.; Skele, K.L.; Vasko, A.J.; et al. Seneca valley virus, a systemically deliverable oncolytic picornavirus, and the treatment of neuroendocrine cancers. J. Natl. Cancer Inst. 2007, 99, 1623–1633. [Google Scholar] [CrossRef]
- Burke, M.J. Oncolytic seneca valley virus: Past perspectives and future directions. Oncolytic Virotherapy 2016, 5, 81–89. [Google Scholar] [CrossRef]
- Xue, Q.; Liu, H.; Zhu, Z.; Yang, F.; Ma, L.; Cai, X.; Xue, Q.; Zheng, H. Seneca valley virus 3c(pro) abrogates the irf3- and irf7-mediated innate immune response by degrading irf3 and irf7. Virology 2018, 518, 1–7. [Google Scholar] [CrossRef]
- Thompson, S.R.; Sarnow, P. Regulation of host cell translation by viruses and effects on cell function. Curr. Opin. Microbiol. 2000, 3, 366–370. [Google Scholar] [CrossRef]
- Tahiri-Alaoui, A.; Zhao, Y.; Sadigh, Y.; Popplestone, J.; Kgosana, L.; Smith, L.P.; Nair, V. Poly(a) binding protein 1 enhances cap-independent translation initiation of neurovirulence factor from avian herpesvirus. PLoS ONE 2014, 9, e114466. [Google Scholar] [CrossRef][Green Version]
- Warren, K.; Wei, T.; Li, D.; Qin, F.; Warrilow, D.; Lin, M.H.; Sivakumaran, H.; Apolloni, A.; Abbott, C.M.; Jones, A.; et al. Eukaryotic elongation factor 1 complex subunits are critical hiv-1 reverse transcription cofactors. Proc. Natl. Acad. Sci. USA 2012, 109, 9587–9592. [Google Scholar] [CrossRef]
- Smith, R.W.P.; Anderson, R.C.; Larralde, O.; Smith, J.W.S.; Gorgoni, B.; Richardson, W.A.; Malik, P.; Graham, S.V.; Gray, N.K. Viral and cellular mrna-specific activators harness pabp and eif4g to promote translation initiation downstream of cap binding. Proc. Natl. Acad. Sci. USA 2017, 114, 6310–6315. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Ito, K. Genetic analysis of l123 of the trna-mimicking eukaryote release factor erf1, an amino acid residue critical for discrimination of stop codons. Nucleic Acids Res. 2015, 43, 4591–4601. [Google Scholar] [CrossRef] [PubMed]
- Joachims, M.; Van Breugel, P.C.; Lloyd, R.E. Cleavage of poly(a)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 1999, 73, 718–727. [Google Scholar] [CrossRef] [PubMed]
- Kuyumcu-Martinez, N.M.; Joachims, M.; Lloyd, R.E. Efficient cleavage of ribosome-associated poly(a)-binding protein by enterovirus 3c protease. J. Virol. 2002, 76, 2062–2074. [Google Scholar] [CrossRef]
- Suzuki, Y.; Chin, W.X.; Han, Q.; Ichiyama, K.; Lee, C.H.; Eyo, Z.W.; Ebina, H.; Takahashi, H.; Takahashi, C.; Tan, B.H.; et al. Characterization of ryden (c19orf66) as an interferon-stimulated cellular inhibitor against dengue virus replication. PLoS Pathog. 2016, 12, e1005357. [Google Scholar] [CrossRef]
- Belsham, G.J.; McInerney, G.M.; Ross-Smith, N. Foot-and-mouth disease virus 3c protease induces cleavage of translation initiation factors eif4a and eif4g within infected cells. J. Virol. 2000, 74, 272–280. [Google Scholar] [CrossRef]
- Kuyumcu-Martinez, N.M.; Van Eden, M.E.; Younan, P.; Lloyd, R.E. Cleavage of poly(a)-binding protein by poliovirus 3c protease inhibits host cell translation: A novel mechanism for host translation shutoff. Mol. Cell. Biol. 2004, 24, 1779–1790. [Google Scholar] [CrossRef]
- Sun, D.; Wang, M.; Wen, X.; Cheng, A.; Jia, R.; Sun, K.; Yang, Q.; Wu, Y.; Zhu, D.; Chen, S.; et al. Cleavage of poly(a)-binding protein by duck hepatitis a virus 3c protease. Sci. Rep. 2017, 7, 16261. [Google Scholar] [CrossRef]
- Bonderoff, J.M.; Larey, J.L.; Lloyd, R.E. Cleavage of poly(a)-binding protein by poliovirus 3c proteinase inhibits viral internal ribosome entry site-mediated translation. J. Virol. 2008, 82, 9389–9399. [Google Scholar] [CrossRef]
- Kobayashi, M.; Arias, C.; Garabedian, A.; Palmenberg, A.C.; Mohr, I. Site-specific cleavage of the host poly(a) binding protein by the encephalomyocarditis virus 3c proteinase stimulates viral replication. J. Virol. 2012, 86, 10686–10694. [Google Scholar] [CrossRef][Green Version]
- Schmidt, E.K.; Clavarino, G.; Ceppi, M.; Pierre, P. Sunset, a nonradioactive method to monitor protein synthesis. Nat. Methods 2009, 6, 275–277. [Google Scholar] [CrossRef] [PubMed]
- Avanzino, B.C.; Fuchs, G.; Fraser, C.S. Cellular cap-binding protein, eif4e, promotes picornavirus genome restructuring and translation. Proc. Natl. Acad. Sci. USA 2017, 114, 9611–9616. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wei, T.; Jin, H.; Rose, A.; Wang, R.; Lin, M.H.; Spann, K.; Harrich, D. Binding of the eukaryotic translation elongation factor 1a with the 5’utr of hiv-1 genomic rna is important for reverse transcription. Virol. J. 2015, 12, 118. [Google Scholar] [CrossRef]
- Wang, D.; Fang, L.; Li, P.; Sun, L.; Fan, J.; Zhang, Q.; Luo, R.; Liu, X.; Li, K.; Chen, H.; et al. The leader proteinase of foot-and-mouth disease virus negatively regulates the type i interferon pathway by acting as a viral deubiquitinase. J. Virol. 2011, 85, 3758–3766. [Google Scholar] [CrossRef] [PubMed]
- Piron, M.; Vende, P.; Cohen, J.; Poncet, D. Rotavirus rna-binding protein nsp3 interacts with eif4gi and evicts the poly(a) binding protein from eif4f. Embo J. 1998, 17, 5811–5821. [Google Scholar] [CrossRef]
- Kuyumcu-Martinez, M.; Belliot, G.; Sosnovtsev, S.V.; Chang, K.O.; Green, K.Y.; Lloyd, R.E. Calicivirus 3c-like proteinase inhibits cellular translation by cleavage of poly(a)-binding protein. J. Virol. 2004, 78, 8172–8182. [Google Scholar] [CrossRef]
- Zhang, B.; Morace, G.; Gauss-Muller, V.; Kusov, Y. Poly(a) binding protein, c-terminally truncated by the hepatitis a virus proteinase 3c, inhibits viral translation. Nucleic Acids Res. 2007, 35, 5975–5984. [Google Scholar] [CrossRef][Green Version]
- Rodriguez Pulido, M.; Serrano, P.; Saiz, M.; Martinez-Salas, E. Foot-and-mouth disease virus infection induces proteolytic cleavage of ptb, eif3a,b, and pabp rna-binding proteins. Virology 2007, 364, 466–474. [Google Scholar] [CrossRef]
- Lloyd, R.E. Translational control by viral proteinases. Virus Res. 2006, 119, 76–88. [Google Scholar] [CrossRef]
- Li, S.; Feng, S.; Wang, J.H.; He, W.R.; Qin, H.Y.; Dong, H.; Li, L.F.; Yu, S.X.; Li, Y.; Qiu, H.J. Eef1a interacts with the ns5a protein and inhibits the growth of classical swine fever virus. Viruses 2015, 7, 4563–4581. [Google Scholar] [CrossRef]
- McKinney, C.; Yu, D.; Mohr, I. A new role for the cellular pabp repressor paip2 as an innate restriction factor capable of limiting productive cytomegalovirus replication. Genes Dev. 2013, 27, 1809–1820. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.K.; Krausslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A segment of the 5’ nontranslated region of encephalomyocarditis virus rna directs internal entry of ribosomes during in vitro translation. J. Virol. 1988, 62, 2636–2643. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mrna directed by a sequence derived from poliovirus rna. Nature 1988, 334, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Miles, L.A.; Burga, L.N.; Gardner, E.E.; Bostina, M.; Poirier, J.T.; Rudin, C.M. Anthrax toxin receptor 1 is the cellular receptor for Seneca Valley virus. J. Clin. Investig. 2017, 127, 2957–2967. [Google Scholar] [CrossRef]
- Zhu, Z.; Shi, Z.; Yan, W.; Wei, J.; Shao, D.; Deng, X.; Wang, S.; Li, B.; Tong, G.; Ma, Z. Nonstructural protein 1 of influenza a virus interacts with human guanylate-binding protein 1 to antagonize antiviral activity. PLoS ONE 2013, 8, e55920. [Google Scholar] [CrossRef]
- Sui, C.; Jiang, D.; Wu, X. Crispr-cas9 mediated rnase l knockout regulates cellular function of pk-15 cells and increases prv replication. Biomed Res. Int. 2019, 2019, 7398208. [Google Scholar] [CrossRef]
- Shang, L.; Zhang, S.; Yang, X.; Sun, J.; Li, L.; Cui, Z.; He, Q.; Guo, Y.; Sun, Y.; Yin, Z. Biochemical characterization of recombinant enterovirus 71 3c protease with fluorogenic model peptide substrates and development of a biochemical assay. Antimicrob. Agents Chemother. 2015, 59, 1827–1836. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, G.; Yang, F.; Cao, W.; Mao, R.; Du, X.; Zhang, X.; Li, C.; Li, D.; Zhang, K.; et al. Foot-and-mouth disease virus viroporin 2b antagonizes rig-i-mediated antiviral effects by inhibition of its protein expression. J. Virol. 2016, 90, 11106–11121. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time pcr data by the comparative c(t) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Liu, H.; Xue, Q.; Cao, W.; Yang, F.; Ma, L.; Liu, W.; Zhang, K.; Liu, X.; Zhu, Z.; Zheng, H. Foot-and-mouth disease virus nonstructural protein 2b interacts with cyclophilin a, modulating virus replication. Faseb J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018, 32, 6706–6723. [Google Scholar] [CrossRef]




| Primers | Sequences (5′-3′) | Target Gene |
|---|---|---|
| SVV-F | AGAATTTGGAAGCCATGCTCT | SVV gene |
| SVV-R | GAGCCAACATAGARACAGATTGC | |
| PABPC1-F | GGTTATGATGGAGGGTGGTCGC | human PABPC1 gene |
| PABPC1-R | GGGGTTGATTACAGGGTTGGGA | |
| GAPDH-F | CGGGAAGCTTGTGATCAATGG | human GAPDH gene |
| GAPDH-R | GGCAGTGATGGCATGGACTG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, Q.; Liu, H.; Zhu, Z.; Xue, Z.; Liu, X.; Zheng, H. Seneca Valley Virus 3Cpro Cleaves PABPC1 to Promote Viral Replication. Pathogens 2020, 9, 443. https://doi.org/10.3390/pathogens9060443
Xue Q, Liu H, Zhu Z, Xue Z, Liu X, Zheng H. Seneca Valley Virus 3Cpro Cleaves PABPC1 to Promote Viral Replication. Pathogens. 2020; 9(6):443. https://doi.org/10.3390/pathogens9060443
Chicago/Turabian StyleXue, Qiao, Huisheng Liu, Zixiang Zhu, Zhaoning Xue, Xiangtao Liu, and Haixue Zheng. 2020. "Seneca Valley Virus 3Cpro Cleaves PABPC1 to Promote Viral Replication" Pathogens 9, no. 6: 443. https://doi.org/10.3390/pathogens9060443
APA StyleXue, Q., Liu, H., Zhu, Z., Xue, Z., Liu, X., & Zheng, H. (2020). Seneca Valley Virus 3Cpro Cleaves PABPC1 to Promote Viral Replication. Pathogens, 9(6), 443. https://doi.org/10.3390/pathogens9060443




