A Snapshot of the Prevalence and Molecular Diversity of Legionella pneumophila in the Water Systems of Israeli Hotels
Abstract
1. Introduction
2. Results
2.1. Legionella Contamination Rates
2.2. Distribution of Serotypes
2.3. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
Author Contributions
Funding
Conflicts of Interest
References
- Fields, B.S.; Benson, R.F.; Besser, R.E. Legionella and Legionnaires’ disease: 25 years of investigation. Clin. Microbiol. Rev. 2002, 15, 506–526. [Google Scholar] [CrossRef] [PubMed]
- Oliva, G.; Sahr, T.; Buchrieser, C. The Life Cycle of L. pneumophila: Cellular Differentiation Is Linked to Virulence and Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- McDade, J.E.; Shepard, C.C.; Fraser, D.W.; Tsai, T.R.; Redus, M.A.; Dowdle, W.R. Legionnaires’ disease: Isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 1977, 297, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Phin, N.; Parry-Ford, F.; Harrison, T.; Stagg, H.R.; Zhang, N.; Kumar, K.; Lortholary, O.; Zumla, A.; Abubakar, I. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect. Dis. 2014, 14, 1011–1021. [Google Scholar] [CrossRef]
- Khodr, A.; Kay, E.; Gomez-Valero, L.; Ginevra, C.; Doublet, P.; Buchrieser, C.; Jarraud, S. Molecular epidemiology, phylogeny and evolution of Legionella. Infect. Genet. Evol. 2016, 43, 108–122. [Google Scholar] [CrossRef]
- Burillo, A.; Pedro-Botet, M.L.; Bouza, E. Microbiology and Epidemiology of Legionnaire’s Disease. Infect. Dis. Clin. N. Am. 2017, 31, 7–27. [Google Scholar] [CrossRef]
- Ratcliff, R.M.; Lanser, J.A.; Manning, P.A.; Heuzenroeder, M.W. Sequence-based classification scheme for the genus Legionella targeting the mip gene. J. Clin. Microbiol. 1998, 36, 1560–1567. [Google Scholar] [CrossRef]
- Yu, V.L.; Plouffe, J.F.; Pastoris, M.C.; Stout, J.E.; Schousboe, M.; Widmer, A.; Summersgill, J.; File, T.; Heath, C.M.; Paterson, D.L.; et al. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: An international collaborative survey. J. Infect. Dis. 2002, 186, 127–128. [Google Scholar] [CrossRef]
- Bartram, J.; Chartier, Y.; Lee, J.V.; Pond, K.; Surman-Lee, S. Legionella and the Prevention of Legionellosis; World Health Organization: Geneva, Switzerland, 2007. [Google Scholar]
- ECDC. Legionnaires’ Disease—Annual Epidemiological Report for 2015. Available online: https://www.ecdc.europa.eu/sites/portal/files/documents/AER_for_2015-legionnaires-disease_0.pdf (accessed on 21 November 2017).
- Smith, S.S.; Ritger, K.; Samala, U.; Black, S.R.; Okodua, M.; Miller, L.; Kozak-Muiznieks, N.A.; Hicks, L.A.; Steinheimer, C.; Ewaidah, S.; et al. Legionellosis Outbreak Associated With a Hotel Fountain. Open Forum Infect. Dis. 2015, 2, ofv164. [Google Scholar] [CrossRef]
- Ahlen, C.; Aas, M.; Krusnell, J.; Iversen, O.J. A single Legionella pneumophila genotype in the freshwater system in a ship experiencing three separate outbreaks of legionellosis in 6 years. Microb. Ecol. Health Dis. 2016, 27, 31148. [Google Scholar]
- Sánchez-Busó, L.; Guiral, S.; Crespi, S.; Moya, V.; Camaró, M.L.; Olmos, M.P.; Adrián, F.; Morera, V.; González-Morán, F.; Vanaclocha, H.; et al. Genomic Investigation of a Legionellosis Outbreak in a Persistently Colonized Hotel. Front. Microbiol. 2016, 6, 1556. [Google Scholar] [CrossRef] [PubMed]
- Chochlakis, D.; Sandalakis, V.; Keramarou, M.; Tselentis, Y.; Psaroulaki, A. Legionellosis: A Walk-through to Identification of the Source of Infection. Cent. Eur. J. Public Health 2017, 25, 235–239. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Borella, P.; Montagna, M.T.; Stampi, S.; Stancanelli, G.; Romano-Spica, V.; Triassi, M.; Marchesi, I.; Bargellini, A.; Tato, D.; Napoli, C.; et al. Legionella Contamination in Hot Water of Italian Hotels. Appl. Environ. Microbiol. 2005, 71, 5805–5813. [Google Scholar] [CrossRef] [PubMed]
- Leoni, E.; De Luca, G.; Legnani, P.P.; Sacchetti, R.; Stampi, S.; Zanetti, F. Legionella waterline colonization: Detection of Legionella species in domestic, hotel and hospital hot water systems. J. Appl. Microbiol. 2005, 98, 373–379. [Google Scholar] [CrossRef]
- Mouchtouri, V.A.; Rudge, J.W. Legionnaires’ Disease in Hotels and Passenger Ships: A Systematic Review of Evidence, Sources, and Contributing Factors. J. Travel Med. 2015, 22, 325–337. [Google Scholar] [CrossRef]
- Mouchtouri, V.; Velonakis, E.; Tsakalof, A.; Kapoula, C.; Goutziana, G.; Vatopoulos, A.; Kremastinou, J.; Hadjichristodoulou, C. Risk Factors for Contamination of Hotel Water Distribution Systems by Legionella Species. Appl. Environ. Microbiol. 2007, 73, 1489–1492. [Google Scholar] [CrossRef]
- Bonetta, S.; Bonetta, S.; Ferretti, E.; Balocco, F.; Carraro, E. Evaluation ofLegionella pneumophila contamination in Italian hotel water systems by quantitative real-time PCR and culture methods. J. Appl. Microbiol. 2010, 108, 1576–1583. [Google Scholar] [CrossRef]
- Fragou, K.; Kokkinos, P.; Gogos, C.; Alamanos, Y.; Vantarakis, A. Prevalence of Legionella spp. in water systems of hospitals and hotels in South Western Greece. Int. J. Environ. Health Res. 2012, 22, 340–354. [Google Scholar] [CrossRef]
- Sepin Özen, N.; Tuğlu Ataman, Ş.; Emek, M. Exploring the Legionella pneumophila positivity rate in hotel water samples from Antalya, Turkey. Environ. Sci. Pollut. Res. 2017, 24, 12238–12242. [Google Scholar] [CrossRef]
- Lee, H.K.; Shim, J.I.; Kim, H.E.; Yu, J.Y.; Kang, Y.H. Distribution of Legionella Species from Environmental Water Sources of Public Facilities and Genetic Diversity of L. pneumophila Serogroup 1 in South Korea. Appl. Environ. Microbiol. 2010, 76, 6547–6554. [Google Scholar] [CrossRef]
- Chochlakis, D.; Sandalakis, V.; Panoulis, C.; Goniotakis, I.; Makridaki, E.; Tselentis, Y.; Psaroulaki, A. Typing ofLegionella strains isolated from environmental samples in Crete, Greece, during the period 2004–2011. J. Water Health 2013, 11, 762. [Google Scholar] [CrossRef] [PubMed]
- Katsiaflaka, A.; Pournaras, S.; Kristo, I.; Mouchtouri, V.A.; Kyritsi, M.; Velonakis, E.; Vatopoulos, A.C.; Hadjichristodoulou, C. Epidemiological Investigation of Legionella pneumophila Serogroup 2 to 14 Isolates from Water Samples by Amplified Fragment Length Polymorphism and Sequence-Based Typing and Detection of Virulence Traits. Appl. Environ. Microbiol. 2016, 82, 6102–6108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moran-Gilad, J.; Mentasti, M.; Lazarovitch, T.; Huberman, Z.; Stocki, T.; Sadik, C.; Shahar, T.; Anis, E.; Valinsky, L.; Harrison, T.G.; et al. Molecular epidemiology of Legionnaires’ disease in Israel. Clin. Microbiol. Infect. 2014, 20, 690–696. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weiss, D.; Boyd, C.; Rakeman, J.L.; Greene, S.K.; Fitzhenry, R.; McProud, T.; Musser, K.; Huang, L.; Kornblum, J.; Nazarian, E.J.; et al. A Large Community Outbreak of Legionnaires’ Disease Associated With a Cooling Tower in New York City, 2015. Public Health Rep. 2017, 132, 241–250. [Google Scholar] [CrossRef]
- Fitzhenry, R.; Weiss, D.; Cimini, D.; Balter, S.; Boyd, C.; Alleyne, L.; Stewart, R.; McIntosh, N.; Econome, A.; Lin, Y.; et al. Legionnaires’ Disease Outbreaks and Cooling Towers, New York City, New York, USA. Emerg. Infect. Dis. 2017, 23, 1769. [Google Scholar] [CrossRef]
- Petzold, M.; Ehricht, R.; Slickers, P.; Pleischl, S.; Brockmann, A.; Exner, M.; Monecke, S.; Luck, C. Rapid genotyping of Legionella pneumophila serogroup 1 strains by a novel DNA microarray-based assay during the outbreak investigation in Warstein, Germany 2013. Int. J. Hyg. Environ. Health 2017, 220, 673–678. [Google Scholar] [CrossRef]
- Bassett, M.T.; Balter, S. Regulating Cooling Towers to Prevent Outbreaks of Legionnaires’ Disease. Public Health Rep. 2017, 132, 133–135. [Google Scholar] [CrossRef]
- Llewellyn, A.C.; Lin, B.; Lucas, C.E.; Roberts, S.E.; Brown, E.W.; Nayak, B.S.; Raphael, B.H.; Winchell, J.M. Distribution of Legionella and bacterial community composition among regionally diverse US cooling towers. PLoS ONE 2017, 12, e0189937. [Google Scholar] [CrossRef]
- Wuthrich, D.; Gautsch, S.; Spieler-Denz, R.; Dubuis, O.; Gaia, V.; Moran-Gilad, J.; Hinic, V.; Seth-Smith, H.M.; Nickel, C.H.; Tschudin-Sutter, S.; et al. Air-conditioner cooling towers as complex reservoirs and continuous source of Legionella pneumophila infection evidenced by a genomic analysis study in 2017, Switzerland. Eur. Commun. Dis. Bull. 2019, 24, 1800192. [Google Scholar] [CrossRef]
- Amemura-Maekawa, J.; Kikukawa, K.; Helbig, J.H.; Kaneko, S.; Suzuki-Hashimoto, A.; Furuhata, K.; Chang, B.; Murai, M.; Ichinose, M.; Ohnishi, M.; et al. Distribution of monoclonal antibody subgroups and sequence-based types among Legionella pneumophila serogroup 1 isolates derived from cooling tower water, bathwater, and soil in Japan. Appl. Environ. Microbiol. 2012, 78, 4263–4270. [Google Scholar] [CrossRef]
- Lawrence, A.; Eglezos, S.; Huston, W. Environmental Legionella spp. collected in urban test sites of South East Queensland, Australia, are virulent to human macrophages in vitro. Res. Microbiol. 2016, 167, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Napoli, C.; Fasano, F.; Iatta, R.; Barbuti, G.; Cuna, T.; Montagna, M.T. Legionella spp. and legionellosis in southeastern Italy: Disease epidemiology and environmental surveillance in community and health care facilities. BMC Public Health 2010, 10, 660. [Google Scholar] [CrossRef] [PubMed]
- Perola, O.; Kauppinen, J.; Kusnetsov, J.; Karkkainen, U.M.; Luck, P.C.; Katila, M.L. Persistent Legionella pneumophila colonization of a hospital water supply: Efficacy of control methods and a molecular epidemiological analysis. APMIS 2005, 113, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Cooper, I.R.; White, J.; Mahenthiralingam, E.; Hanlon, G.W. Long-term persistence of a single Legionella pneumophila strain possessing the mip gene in a municipal shower despite repeated cycles of chlorination. J. Hosp. Infect. 2008, 70, 154–159. [Google Scholar] [CrossRef]
- Vekens, E.; Soetens, O.; De Mendonca, R.; Echahidi, F.; Roisin, S.; Deplano, A.; Eeckhout, L.; Achtergael, W.; Pierard, D.; Denis, O.; et al. Sequence-based typing of Legionella pneumophila serogroup 1 clinical isolates from Belgium between 2000 and 2010. Eurosurveillance 2012, 17, 20302. [Google Scholar]
- David, S.; Rusniok, C.; Mentasti, M.; Gomez-Valero, L.; Harris, S.R.; Lechat, P.; Lees, J.; Ginevra, C.; Glaser, P.; Ma, L.; et al. Multiple major disease-associated clones of Legionella pneumophila have emerged recently and independently. Genome Res. 2016, 26, 1555–1564. [Google Scholar] [CrossRef]
- Qin, T.; Zhou, H.; Ren, H.; Guan, H.; Li, M.; Zhu, B.; Shao, Z. Distribution of sequence-based types of legionella pneumophila serogroup 1 strains isolated from cooling towers, hot springs, and potable water systems in China. Appl. Environ. Microbiol. 2014, 80, 2150–2157. [Google Scholar] [CrossRef]
- Lévesque, S.; Lalancette, C.; Bernard, K.; Pacheco, A.L.; Dion, R.; Longtin, J.; Tremblay, C. Molecular Typing of Legionella pneumophila Isolates in the Province of Quebec from 2005 to 2015. PLoS ONE 2016, 11, e0163818. [Google Scholar] [CrossRef]
- Kozak-Muiznieks, N.A.; Lucas, C.E.; Brown, E.; Pondo, T.; Taylor, T.H., Jr.; Frace, M.; Miskowski, D.; Winchell, J.M. Prevalence of sequence types among clinical and environmental isolates of Legionella pneumophila serogroup 1 in the United States from 1982 to 2012. J. Clin. Microbiol. 2014, 52, 201–211. [Google Scholar] [CrossRef]
- Sanchez-Buso, L.; Coscolla, M.; Palero, F.; Camaro, M.L.; Gimeno, A.; Moreno, P.; Escribano, I.; Lopez Perezagua, M.M.; Colomina, J.; Vanaclocha, H.; et al. Geographical and Temporal Structures of Legionella pneumophila Sequence Types in Comunitat Valenciana (Spain), 1998 to 2013. Appl. Environ. Microbiol. 2015, 81, 7106–7113. [Google Scholar] [CrossRef]
- Quero, S.; Parraga-Nino, N.; Barrabeig, I.; Sala, M.R.; Pedro-Botet, M.L.; Monso, E.; Jane, M.; Sabria, M.; Garcia-Nunez, M. Population structure of Environmental and Clinical Legionella pneumophila isolates in Catalonia. Sci. Rep. 2018, 8, 6241. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Health Central Laboratories Annual Report, pp. 66–67. Available online: https://www.health.gov.il/PublicationsFiles/LAB_JER2017.pdf (accessed on 25 May 2020).
- Harrison, T.G.; Moran-Gilad, J.; Mentasti, M.; David, S.; Afshar, B.; Andersen, P.S.; Stegger, M.; Valinsky, L.; Yakunin, E.; Uldum, S.A. Use of whole genome sequence (WGS) data to investigate anomalies in the ESGLI Legionella pneumophila DNA-sequence based typing (SBT) method. In Proceedings of the 2nd ESCMID Study Group for Legionella Infections Meeting (ESGLI), Barcelona, Spain, 17–19 September 2014. [Google Scholar]
- Qadreyah Al-Matawah, S.A.-Z. Søren Uldum Sequence-Based Typing for Legionella Pneumophila Isolated from Water Systems of Residential Facilities in Kuwait. J. Environ. Sci. 2016, 2, 016. [Google Scholar]
- Yakunin, E.; Ohayon, S.; Schnaidman, B.; Marva, E.; Agmon, V.; Eizenkraft, A.; Wagnert, L.; Grotto, I.; Valinsky, L.; Moran-Gilad, J. Prevalence and Diversity of Legionella pneumophila in the Defence Setting. In Proceedings of the Israeli Society for Microbiology Annual Meeting, Beit Dagan, Israel, 13–14 September 2017. [Google Scholar]
- Leoni, E.; Legnani, P.P. Comparison of selective procedures for isolation and enumeration of Legionella species from hot water systems. J. Appl. Microbiol. 2001, 90, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, A.K.T. Determination of viable legionellae in engineered water systems: Do we find what we are looking for? Water Res. 2016, 93, 276–288. [Google Scholar] [CrossRef]
- International Organization for Standardization. Water Quality—Enumeration of Legionella; ISO 11731:2017; ISO: Geneva, Switzerland, 2017. [Google Scholar]
- Guidelines on Preventing the Proliferation of the Legionella Bacteria in Water Systems. Available online: https://www.health.gov.il/Subjects/Environmental_Health/drinking_water/Pages/Legionella.aspx (accessed on 25 May 2020).
- International Organization for Standardization. Water Quality-Detection and Enumeration of Legionella-Part 2: Direct Membrane Filtration method for Waters with Low Bacterial Counts; ISO 11731-2:2004; ISO: Geneva, Switzerland, 2004. [Google Scholar]
- Public Health England. mip Sequencing Database. Available online: https://www.gov.uk/guidance/phe-data-and-analysis-tools (accessed on 19 November 2018).
- Gaia, V.; Fry, N.K.; Afshar, B.; Luck, P.C.; Meugnier, H.; Etienne, J.; Peduzzi, R.; Harrison, T.G. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 2005, 43, 2047–2052. [Google Scholar] [CrossRef]
- Ratzow, S.; Gaia, V.; Helbig, J.H.; Fry, N.K.; Luck, P.C. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 2007, 45, 1965–1968. [Google Scholar] [CrossRef]
- Public Health England. L. pneumophila Sequenced-Based Typing. Available online: https://webarchive.nationalarchives.gov.uk/20190501130700/http://bioinformatics.phe.org.uk/legionella/legionella_sbt/php/sbt_homepage.php (accessed on 20 November 2019).
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
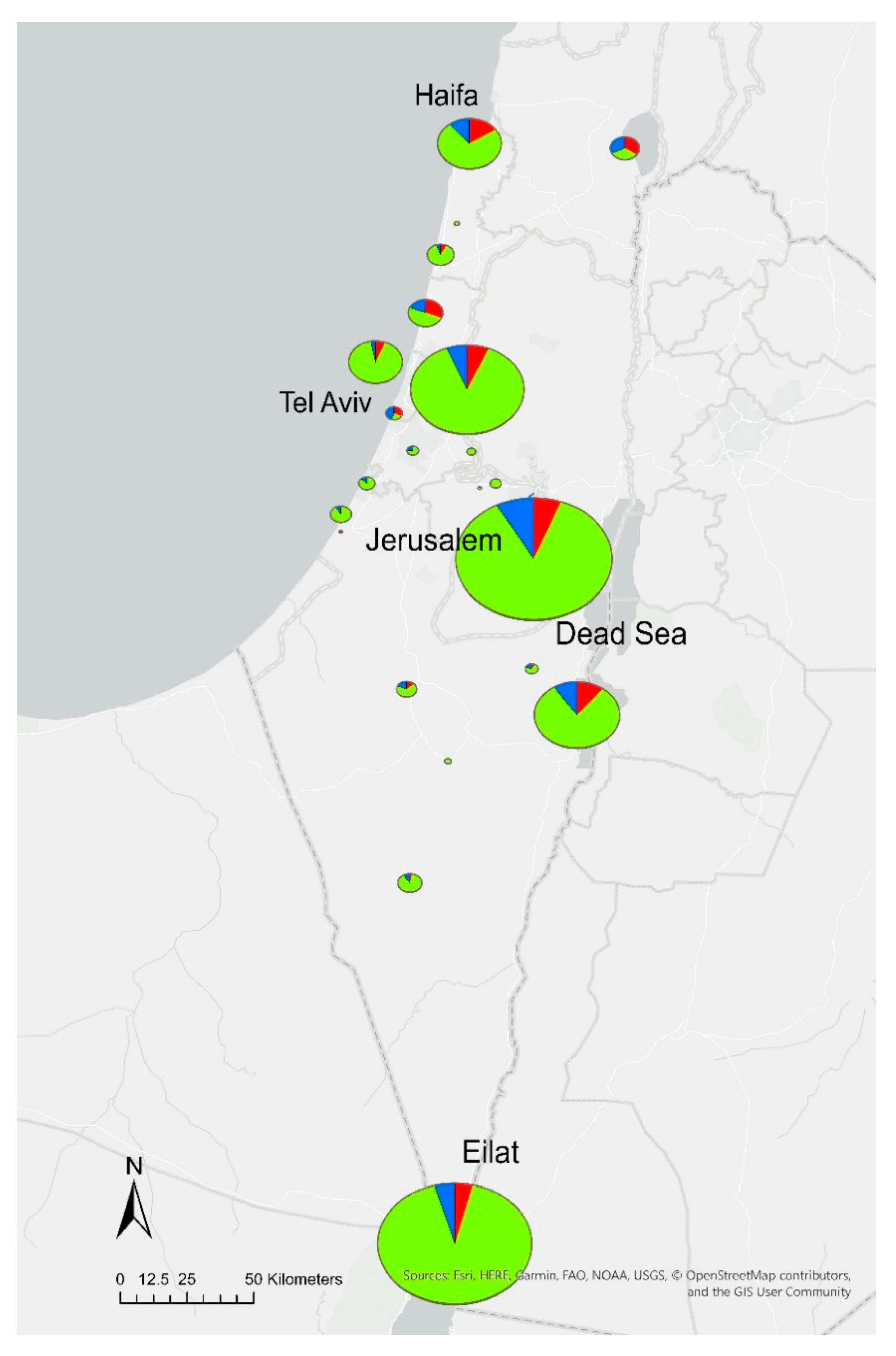
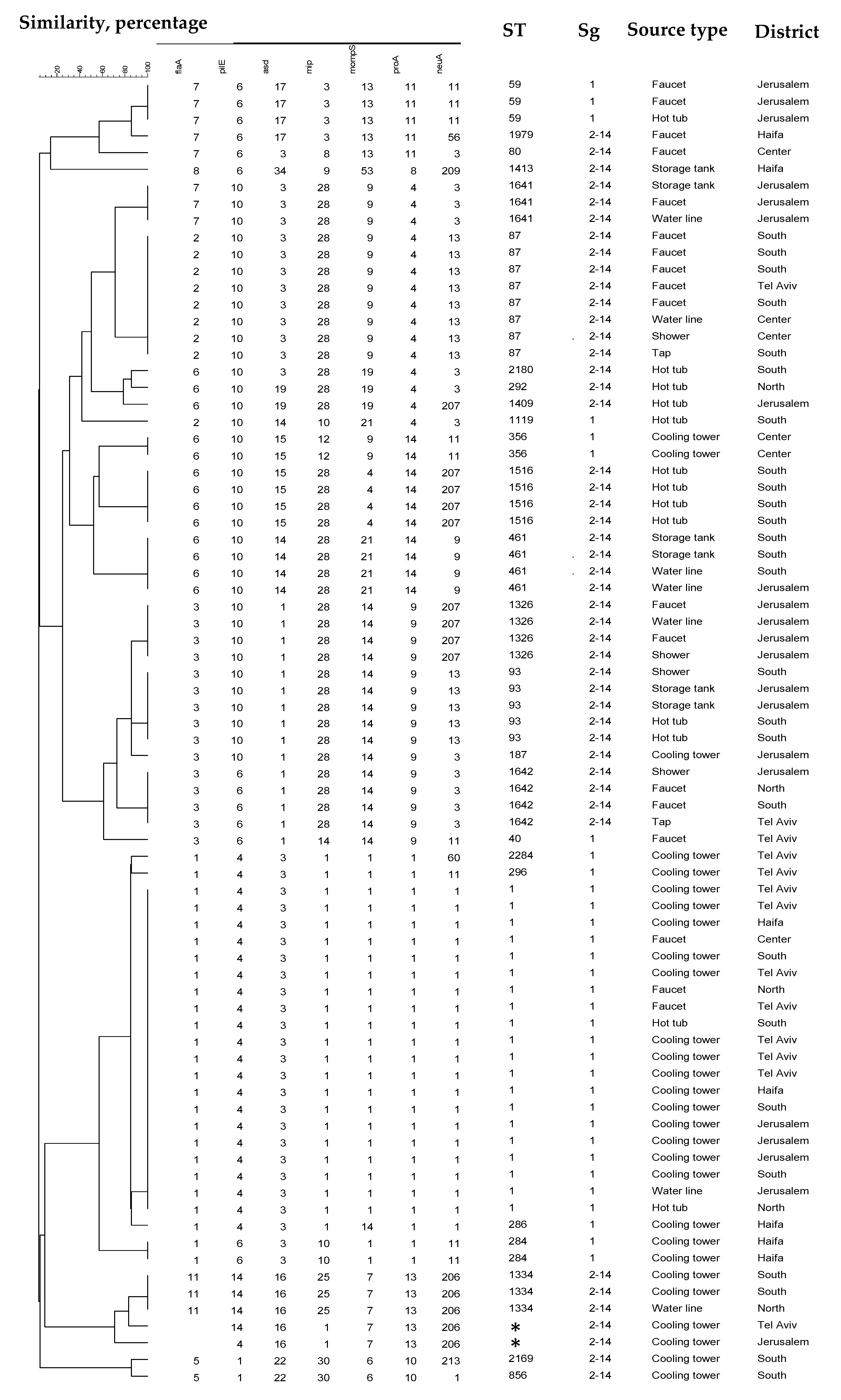
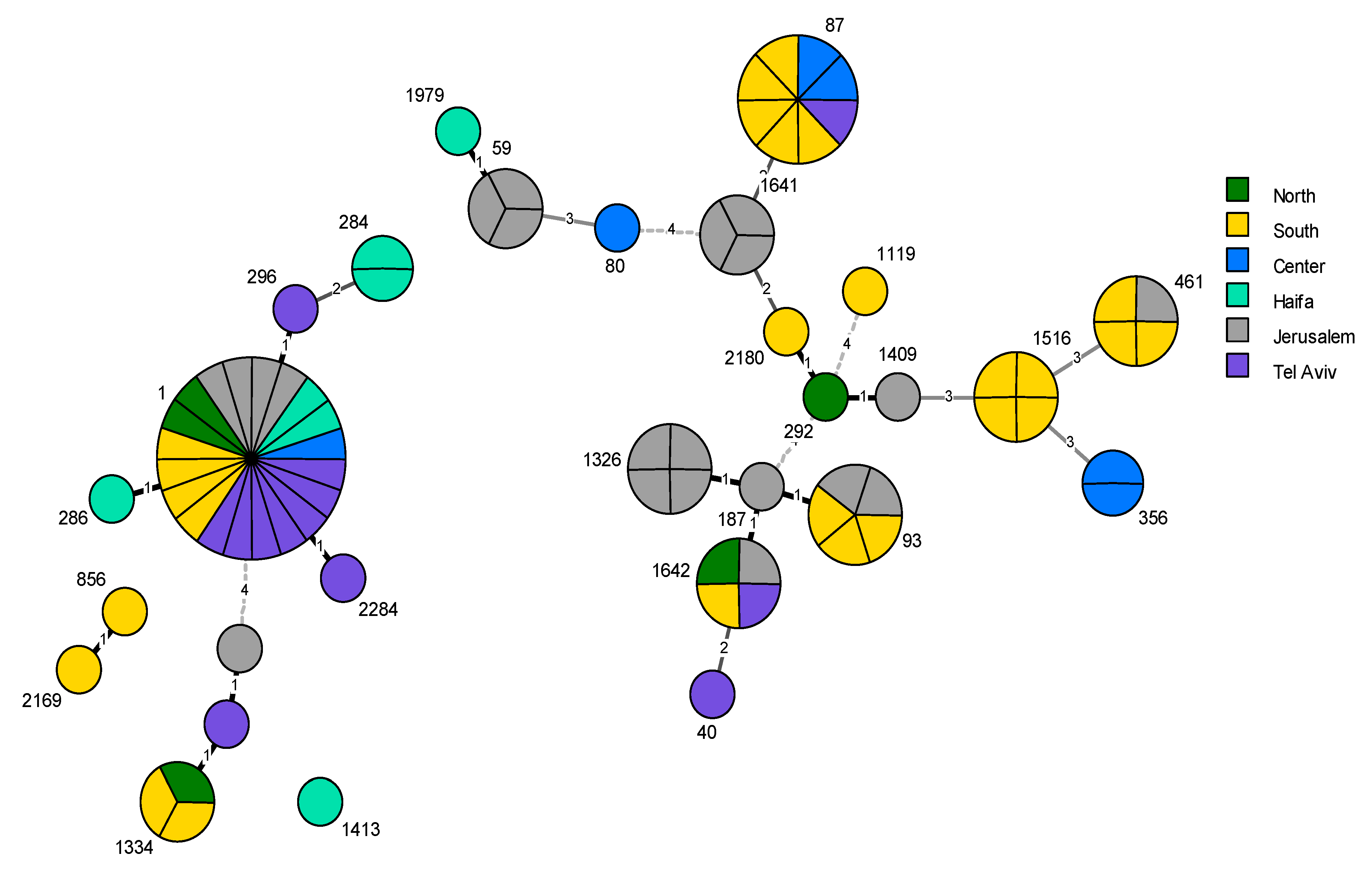
| District | Total Tested | Positive Samples | Exceeding Samples 1 | |||
|---|---|---|---|---|---|---|
| No. of Hotels | No. of Samples | No. of Hotels | No. of Samples (% Per District) | No. of Hotels | No. of Samples (% Per District) | |
| North | 3 | 42 | 2 | 27 (64) | 1 | 14 (33) |
| Center | 9 | 207 | 6 | 54 (26) | 4 | 30 (14) |
| South | 78 | 1139 | 44 | 151 (13) | 28 | 72 (6) |
| Haifa | 9 | 201 | 7 | 52 (26) | 4 | 30 (15) |
| Tel Aviv | 20 | 447 | 12 | 63 (14) | 8 | 32 (7) |
| Jerusalem | 49 | 794 | 30 | 123 (15) | 17 | 52 (7) |
| Total | 168 a | 2830 | 101 b | 470 (17) | 62 c | 230 (8) |
| Source Type | No. of Samples | Positive Samples | Exceeding Samples 1 | |||||
|---|---|---|---|---|---|---|---|---|
| Cold Water | Hot Water | Mixed Water 2 | Total | No. of Samples | Per Category (%) | No. of Samples | Per Category (%) | |
| Outlet | 649 | 1084 | 1733 | 277 | 16% | 84 | 5% | |
| Main water line | 96 | 383 | 479 | 68 | 14% | 35 | 7% | |
| Cooling tower | 232 | 232 | 87 | 38% | 74 | 32% | ||
| Hot tub | 5 | 9 | 204 | 218 | 36 | 17% | 36 | 17% |
| Fountain | 24 | 4 | 28 | 1 | 4% | 0 | 0% | |
| Pool | 11 | 11 | 0 | 0% | 0 | 0% | ||
| Air conditioning | 4 | 4 | 1 | 25% | 1 | 25% | ||
| Not available 3 | 38 | 87 | 125 | 0 | 0% | 0 | 0% | |
| Total: | 1059 | 1567 | 204 | 2830 | 470 | 17% | 230 | 8% |
| Author | Country of Study | Year of Publication | Geography | Sample Selection | Sample Size | Time of Study | Laboratory Methods |
|---|---|---|---|---|---|---|---|
| Borella et al. | Italy | 2005 | Five representative cities, northern, central, and southern Italian regions | The hotels were selected based on the water distribution systems in the cities, the characteristics of the buildings, and hotel cooperation. | 119 water samples from 40 hotels (3–5 samples from each hotel) | September 2003–July 2004 | Legionella isolation, enumeration and serotyping; PFGE analysis; physical and chemical water analyses |
| Lee et al. | South Korea | 2010 | Seven geographic regions throughout South Korea | The number of samples and isolates depended on the number of facilities located in each region | 4938 water samples from water systems of different settings, including hotels. | June–September 2008 | Legionella isolation, enumeration and serotyping; molecular identification of L. spp (16S rRNA, mip, or rpoB); SBT |
| Napoli et al. | Italy | 2010 | Southeastern Italy | Representative samples from different building types and water systems. Re-inspection samples excluded. | 13,286 water samples, including 5009 samples from 305 hotels | January 2000–December 2009 | Legionella isolation, enumeration, and serotyping |
| Bonetta et al. | Italy | 2010 | Northern, central, and southern Italy | Samples representative of 18 towns and types of water systems. | 76 water samples from 19 hotels | October 2006–February 2007 | Legionella isolation, enumeration, and serotyping; real-time PCR; physical and chemical analyses |
| Chochlakis et al. | Greece | 2013 | Four regions of Crete island | Eight to 15 representative samples from each hotel, depending on hotel size and water system type. | 1494 water samples from 124 hotels | 2004–2011 | Legionella isolation, enumeration, and serotyping; molecular identification of Legionella spp (16S rRNA, mip); MALDI-TOF mass spectrometry; SBT; |
| Sepin Özen et al. | Turkey | 2017 | Antalya region | Samples from different water systems | 1403 water samples from 54 hotels | January–December 2010 | Legionella isolation, enumeration, and serotyping |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yakunin, E.; Kostyal, E.; Agmon, V.; Grotto, I.; Valinsky, L.; Moran-Gilad, J. A Snapshot of the Prevalence and Molecular Diversity of Legionella pneumophila in the Water Systems of Israeli Hotels. Pathogens 2020, 9, 414. https://doi.org/10.3390/pathogens9060414
Yakunin E, Kostyal E, Agmon V, Grotto I, Valinsky L, Moran-Gilad J. A Snapshot of the Prevalence and Molecular Diversity of Legionella pneumophila in the Water Systems of Israeli Hotels. Pathogens. 2020; 9(6):414. https://doi.org/10.3390/pathogens9060414
Chicago/Turabian StyleYakunin, Eugenia, Eszter Kostyal, Vered Agmon, Itamar Grotto, Lea Valinsky, and Jacob Moran-Gilad. 2020. "A Snapshot of the Prevalence and Molecular Diversity of Legionella pneumophila in the Water Systems of Israeli Hotels" Pathogens 9, no. 6: 414. https://doi.org/10.3390/pathogens9060414
APA StyleYakunin, E., Kostyal, E., Agmon, V., Grotto, I., Valinsky, L., & Moran-Gilad, J. (2020). A Snapshot of the Prevalence and Molecular Diversity of Legionella pneumophila in the Water Systems of Israeli Hotels. Pathogens, 9(6), 414. https://doi.org/10.3390/pathogens9060414




