Genome Subtraction and Comparison for the Identification of Novel Drug Targets against Mycobacterium avium subsp. hominissuis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Removal of Duplicate Sequences after Proteome Retrieval
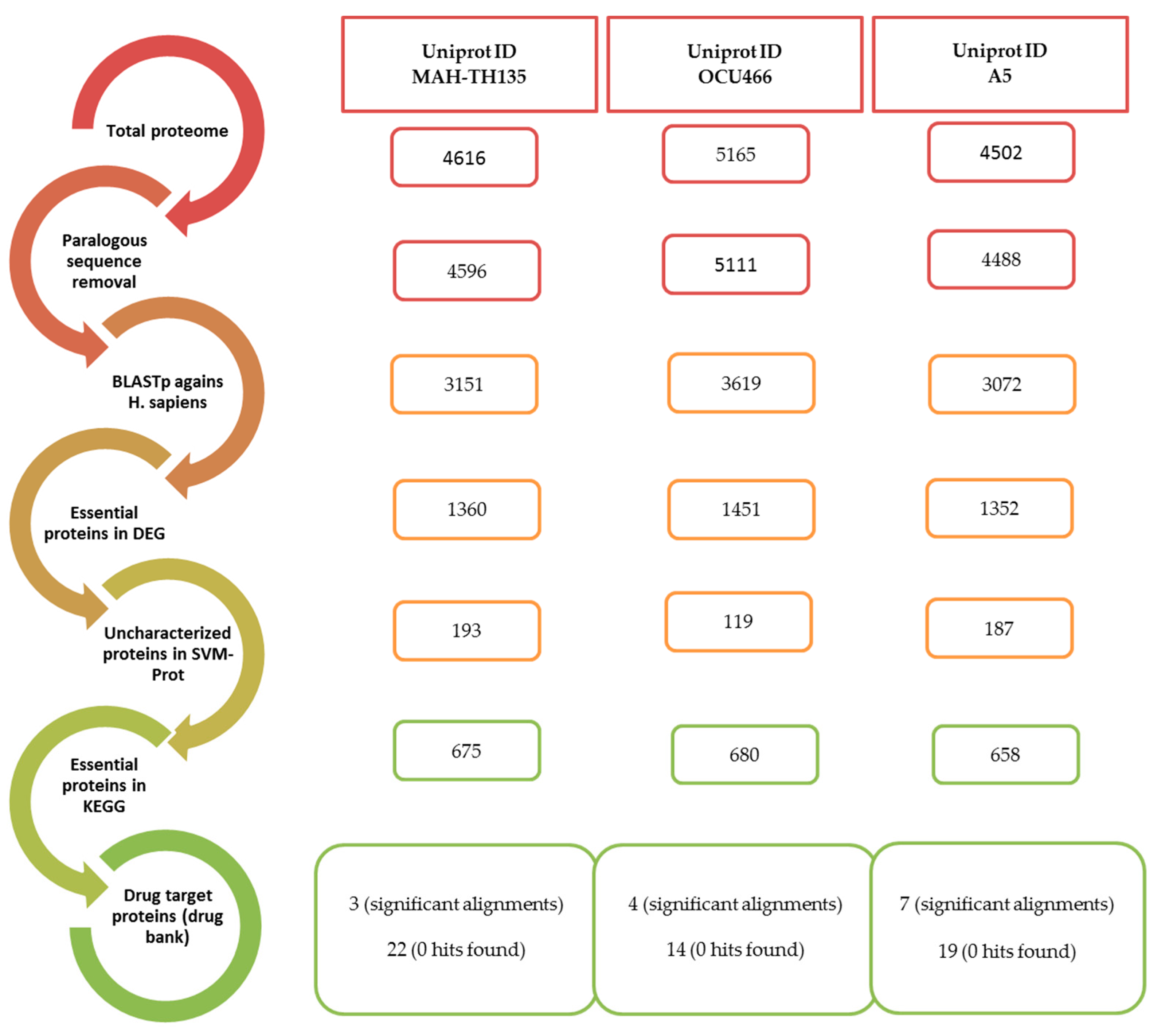
2.2. Searching of Essential, Non-Homologous and Druggable Proteins
2.3. Characterization of Essential Non-Homologous Proteins
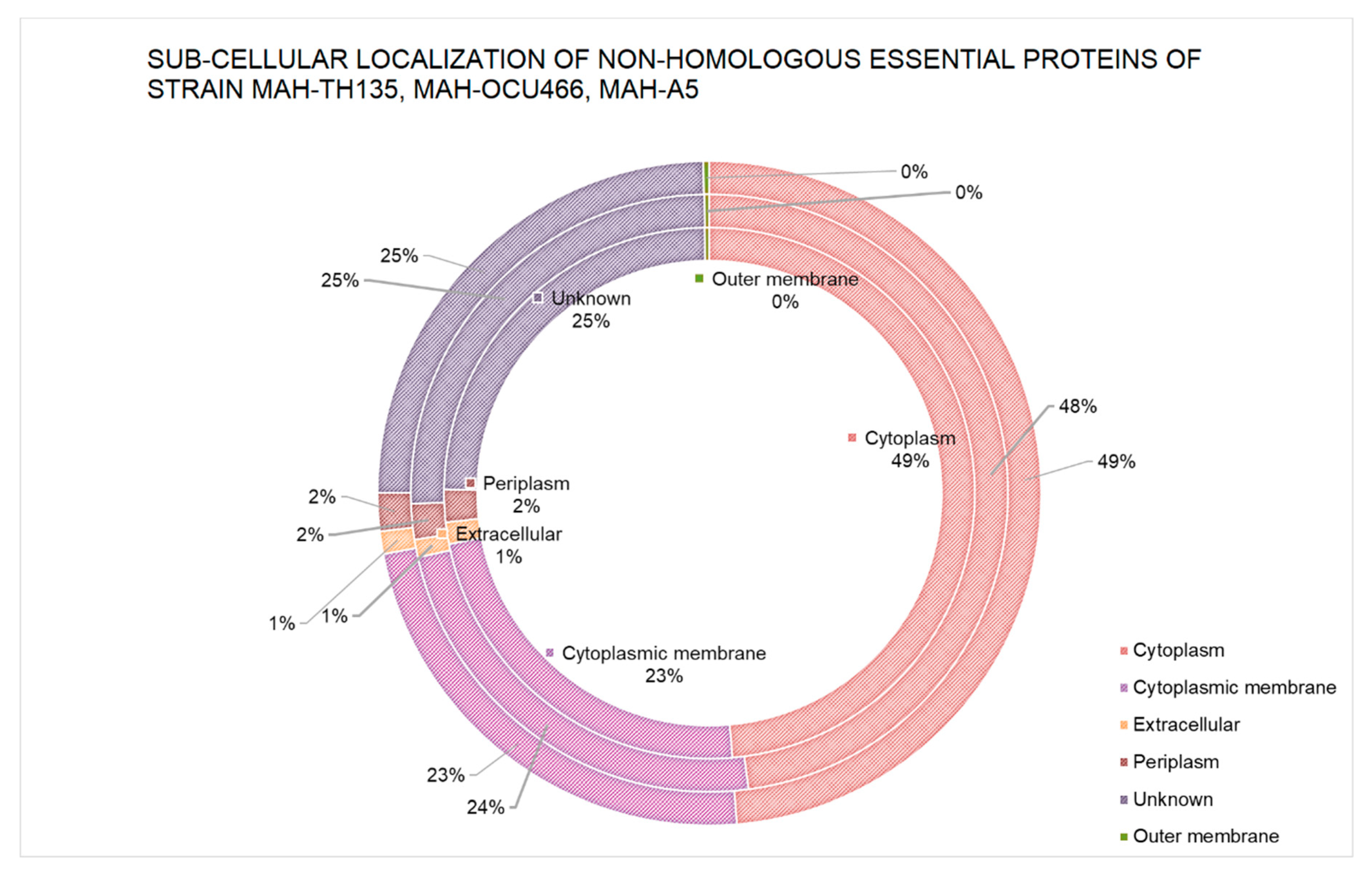
2.3.1. Subcellular Localization
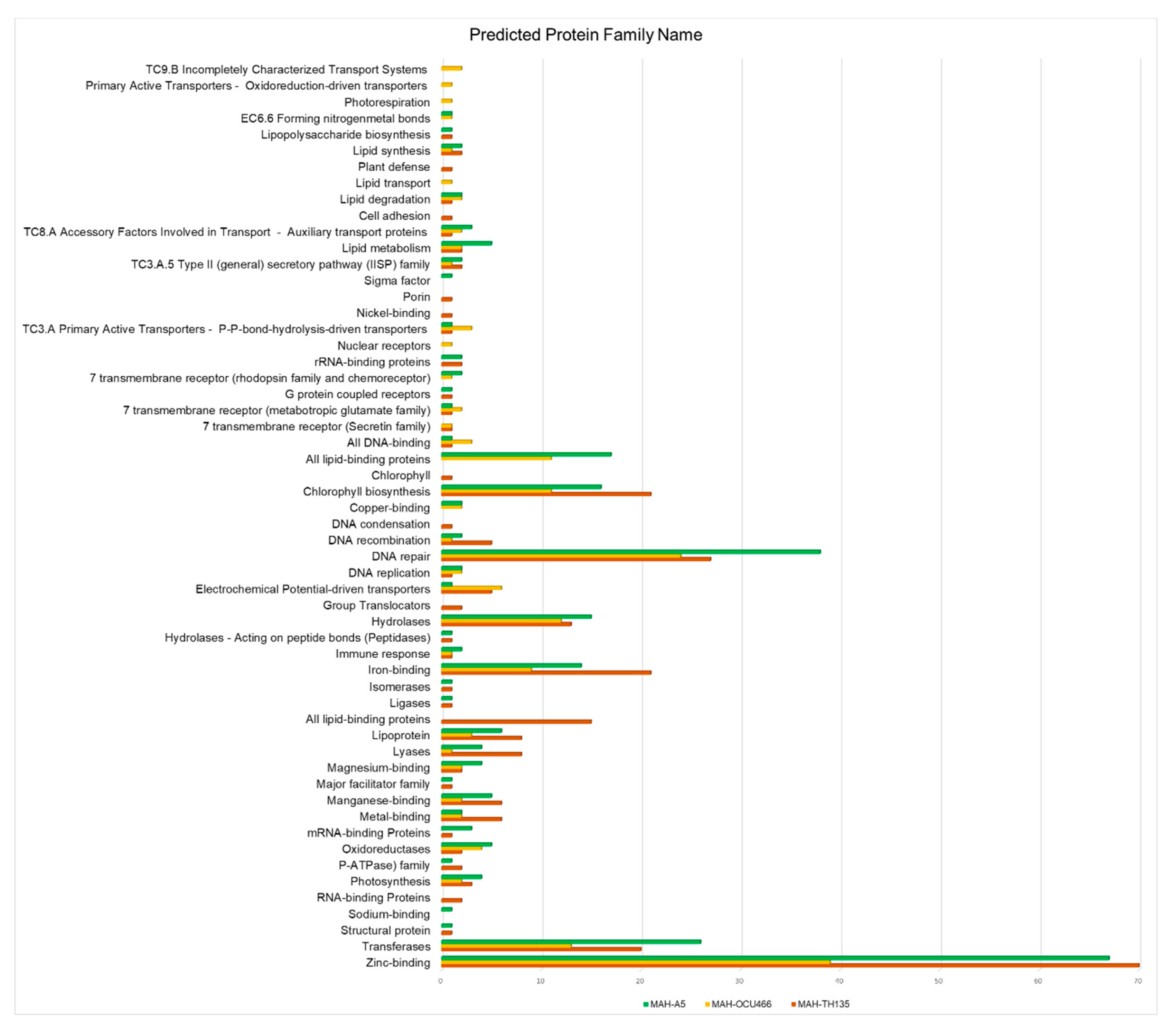
2.3.2. Functional Family Classification
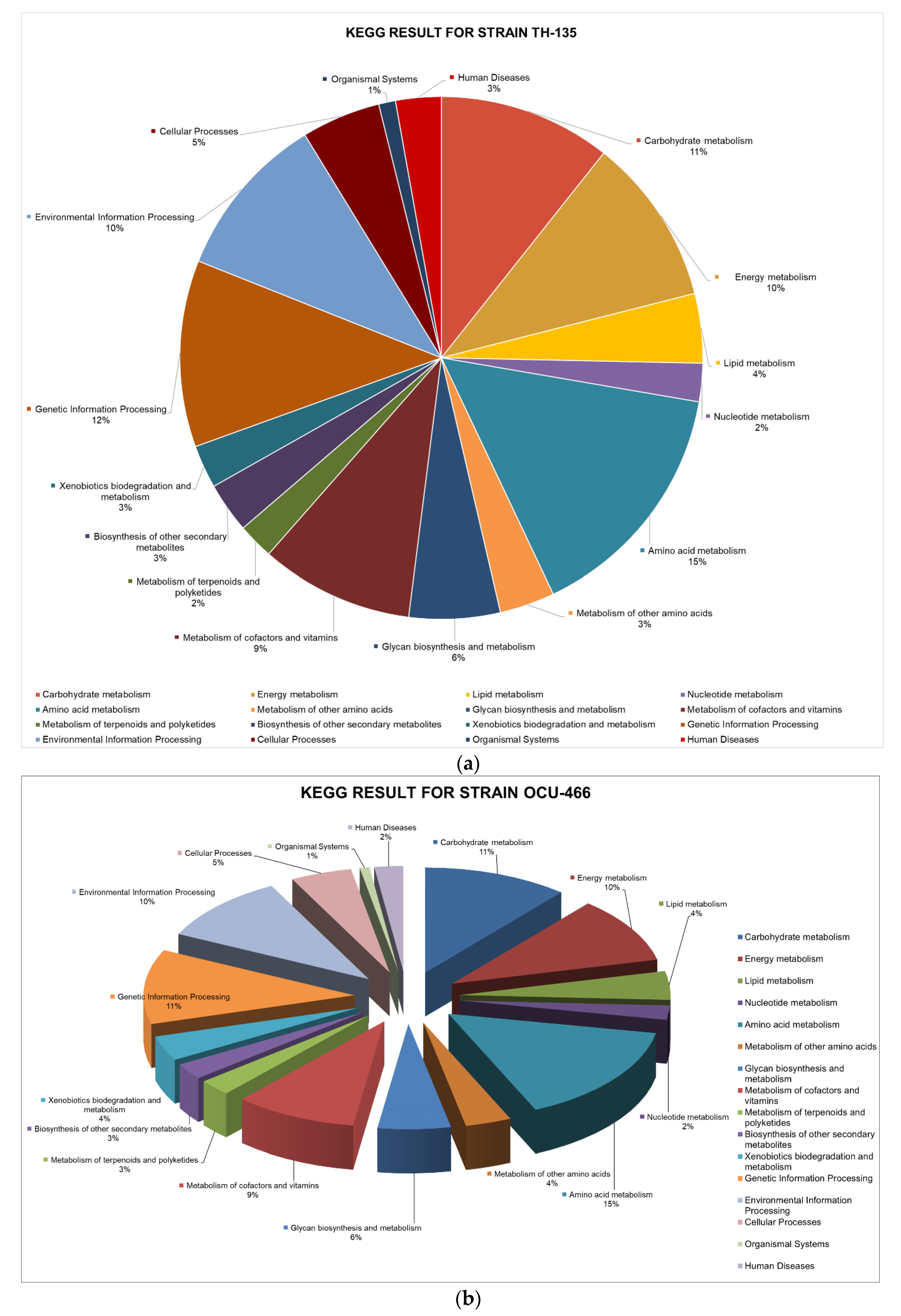
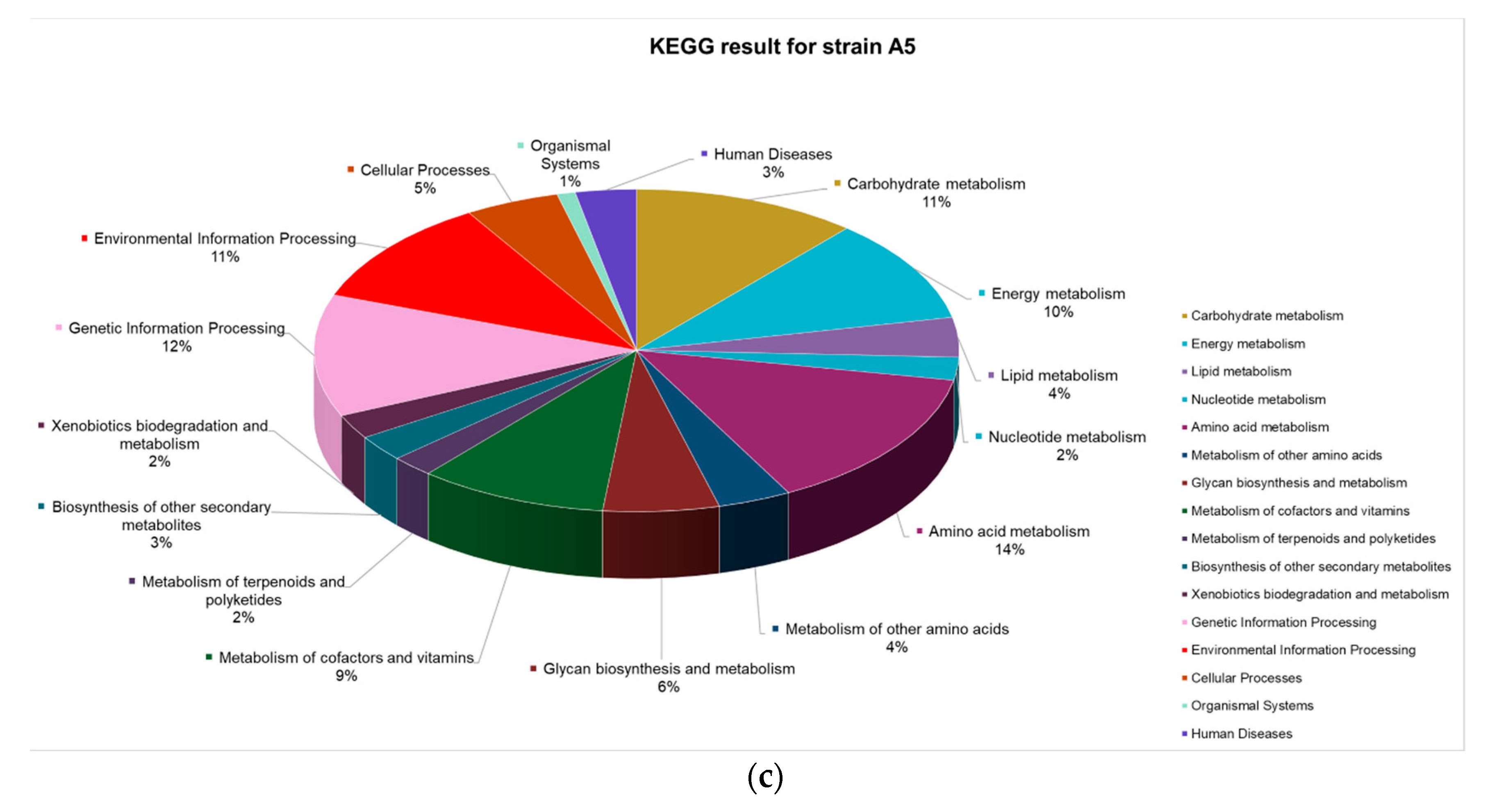
2.3.3. Metabolic Pathway Analysis via KEGG
2.4. Discussion of Significant Unique Metabolic Pathways (UMPs) of the Pathogens
2.4.1. Energy Metabolism
2.4.2. Biosynthesis of Secondary Metabolites
2.4.3. Amino Acid Metabolism
2.5. Shortlisting of Proteins Sequences as Druggable
3. Materials and Methods
3.1. Extraction of the Host–Pathogen Proteome
3.2. Grouping of Common Proteins in All Strains
3.3. Identification of Non-Homologous Proteins
3.4. Finding of Essential Genes
3.5. Information about Metabolic Pathways
3.6. Annotation of the Curated Proteins
3.7. Druggability of the Shortlisted Sequences
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Daley, C. Mycobacterium avium Complex Disease. Microbiol. Spectr. 2017, 5, 663–701. [Google Scholar] [CrossRef]
- Iwamoto, T.; Nakajima, C.; Nishiuchi, Y.; Kato, T.; Yoshida, S.; Nakanishi, N.; Tamaru, A.; Tamura, Y.; Suzuki, Y.; Nasu, M. Genetic diversity of Mycobacterium avium subsp. hominissuis strains isolated from humans, pigs, and human living environment. Infect. Genet. Evol. 2012, 12, 846–852. [Google Scholar] [CrossRef]
- Uchiya, K.-I.; Takahashi, H.; Yagi, T.; Moriyama, M.; Inagaki, T.; Ichikawa, K.; Nakagawa, T.; Nikai, T.; Ogawa, K. Comparative genome analysis of Mycobacterium avium revealed genetic diversity in strains that cause pulmonary and disseminated disease. PLoS ONE 2013, 8, e71831. [Google Scholar] [CrossRef] [PubMed]
- Mijs, W.; De Haas, P.; Rossau, R.; Van Der Laan, T.; Rigouts, L.; Portaels, F.; Van Soolingen, D. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and ‘M. avium subsp. hominissuis’ for the human/porcine type of M. avium. Int. J. Syst. Evol. Micr. 2002, 52, 1505–1518. [Google Scholar]
- Porvaznik, I.; Solovič, I.; Mokrý, J. Non-tuberculous mycobacteria: Classification, diagnostics, and therapy. In Respiratory Treatment and Prevention; Springer: Berlin/Heidelberg, Germany, 2016; pp. 19–25. [Google Scholar]
- Bruffaerts, N.; Vluggen, C.; Roupie, V.; Duytschaever, L.; Van den Poel, C.; Denoël, J.; Wattiez, R.; Letesson, J.-J.; Fretin, D.; Rigouts, L. Virulence and immunogenicity of genetically defined human and porcine isolates of M. avium subsp. hominissuis in an experimental mouse infection. PLoS ONE 2017, 12, e0171895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchiya, K.-I.; Takahashi, H.; Nakagawa, T.; Yagi, T.; Moriyama, M.; Inagaki, T.; Ichikawa, K.; Nikai, T.; Ogawa, K. Characterization of a novel plasmid, pMAH135, from Mycobacterium avium subsp. hominissuis. PLoS ONE 2015, 10, e0117797. [Google Scholar] [CrossRef] [Green Version]
- Maekawa, K.; Ito, Y.; Hirai, T.; Kubo, T.; Imai, S.; Tatsumi, S.; Fujita, K.; Takakura, S.; Niimi, A.; Iinuma, Y. Environmental risk factors for pulmonary Mycobacterium avium-intracellulare complex disease. Chest 2011, 140, 723–729. [Google Scholar] [CrossRef] [Green Version]
- Weiss, C.; Glassroth, J. Pulmonary disease caused by nontuberculous mycobacteria. Expert. Rev. Respir. Med. 2012, 6, 597–613. [Google Scholar] [CrossRef]
- Uchiya, K.-I.; Asahi, S.; Futamura, K.; Hamaura, H.; Nakagawa, T.; Nikai, T.; Ogawa, K. Antibiotic susceptibility and genotyping of Mycobacterium avium strains that cause pulmonary and disseminated infection. Antimicrob. Agents Chemother. 2018, 62, e02035-17. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, J.D.; Elias, V.; Cipolla, D.; Gonda, I.; Bermudez, L.E. Effective Treatment of Mycobacterium avium subsp. hominissuis and Mycobacterium abscessus Species Infections in Macrophages, Biofilm, and Mice by Using Liposomal Ciprofloxacin. Antimicrob. Agents Chemother. 2018, 62, e00440-18. [Google Scholar] [CrossRef] [Green Version]
- Griffith, D.E.; Brown-Elliott, B.A.; Langsjoen, B.; Zhang, Y.; Pan, X.; Girard, W.; Nelson, K.; Caccitolo, J.; Alvarez, J.; Shepherd, S. Clinical and molecular analysis of macrolide resistance in Mycobacterium avium complex lung disease. Am. J. Respir. Crit. Care Med. 2006, 174, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Nicolle, L. Community-acquired MRSA: A practitioner's guide. CMAJ 2006, 175, 145. [Google Scholar] [CrossRef] [PubMed]
- Rathi, B.; Sarangi, A.N.; Trivedi, N. Genome subtraction for novel target definition in Salmonella typhi. Bioinformation 2009, 4, 143–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butt, A.M.; Tahir, S.; Nasrullah, I.; Idrees, M.; Lu, J.; Tong, Y. Mycoplasma genitalium: A comparative genomics study of metabolic pathways for the identification of drug and vaccine targets. Infect. Genet. Evol. 2012, 12, 53–62. [Google Scholar] [CrossRef]
- Barh, D.; Tiwari, S.; Jain, N.; Ali, A.; Santos, A.R.; Misra, A.N.; Azevedo, V.; Kumar, A. In silico subtractive genomics for target identification in human bacterial pathogens. Drug Develop. Res. 2011, 72, 162–177. [Google Scholar] [CrossRef]
- Bottacini, F.; Motherway, M.O.C.; Kuczynski, J.; O’Connell, K.J.; Serafini, F.; Duranti, S.; Milani, C.; Turroni, F.; Lugli, G.A.; Zomer, A. Comparative genomics of the Bifidobacterium breve taxon. BMC Genom. 2014, 15, 170. [Google Scholar] [CrossRef] [Green Version]
- Uddin, R.; Saeed, K. Identification and characterization of potential drug targets by subtractive genome analyses of methicillin resistant Staphylococcus aureus. Comput. Biol. Chem. 2014, 48, 55–63. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Koonin, E.V. Searching for drug targets in microbial genomes. Curr. Opin. Biotechnol. 1999, 10, 571–578. [Google Scholar] [CrossRef]
- Uddin, R.; Masood, F.; Azam, S.S.; Wadood, A. Identification of putative non-host essential genes and novel drug targets against Acinetobacter baumannii by in silico comparative genome analysis. Microb. Pathog. 2019, 128, 28–35. [Google Scholar] [CrossRef]
- Dutta, A.; Singh, S.K.; Ghosh, P.; Mukherjee, R.; Mitter, S.; Bandyopadhyay, D. In silico identification of potential therapeutic targets in the human pathogen Helicobacter pylori. Silico Biol. 2006, 6, 43–47. [Google Scholar]
- Marri, P.R.; Bannantine, J.P.; Golding, G.B. Comparative genomics of metabolic pathways in Mycobacterium species: Gene duplication, gene decay and lateral gene transfer. FEMS Microbiol. Rev. 2006, 30, 906–925. [Google Scholar] [CrossRef] [PubMed]
- Uddin, R.; Jamil, F. Prioritization of potential drug targets against P. aeruginosa by core proteomic analysis using computational subtractive genomics and protein-Protein interaction network. Comput. Biol. Chem. 2018, 74, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Navid, A.; Akhtar, A.S.; Azam, S.S.; Wadood, A.; Pérez-Sánchez, H. Subtractive Genomics, Molecular Docking and Molecular Dynamics Simulation Revealed LpxC as a Potential Drug Target Against Multi-Drug Resistant Klebsiella pneumoniae. Interdiscipl. Sci. Comput. Life Sci. 2019, 11, 508–526. [Google Scholar] [CrossRef] [PubMed]
- Asalone, K.C.; Nelson, M.M.; Bracht, J.R. Novel Sequence Discovery by Subtractive Genomics. J. Vis. Exp. 2019, 143, e58877. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.; Pradhan, D.; Singh, H.; Reddy, M.S. Computational screening of potential drug targets for pathogens causing bacterial pneumonia. Microb. Pathog. 2019, 130, 271–282. [Google Scholar] [CrossRef]
- Prabha, R.; Singh, D.P.; Ahmad, K.; Kumar, S.P.J.; Kumar, P. Subtractive genomics approach for identification of putative antimicrobial targets in Xanthomonas oryzae pv. oryzae KACC10331. Arch. Phytopath. Plant Protect. 2019, 52, 863–872. [Google Scholar] [CrossRef]
- Auster, L.; Sutton, M.; Gwin, M.C.; Nitkin, C.; Bonfield, T.L. Optimization of In Vitro Mycobacterium avium and Mycobacterium intracellulare Growth Assays for Therapeutic Development. Microorganisms 2019, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Shoukat, K.; Rasheed, N.; Sajid, M. Subtractive genome analysis for In silico identification and characterization of novel drug targets IN C. trachomatis STRAIN D/UW-3/Cx. Int. J. Curr. Res. 2012, 4, 017–021. [Google Scholar]
- Koteswara, R.G.; Nagamalleswara, R.K.; Phani, R.; Krishna, B.; Aravind, S. In silico identification of potential therapeutic targets inclostridium botulinum by the approach subtractive genomics. Int. J. Bioinform. Res. 2010, 2, 12–16. [Google Scholar]
- Sharma, V.; Gupta, P.; Dixit, A. In silico identification of putative drug targets from different metabolic pathways of Aeromonas hydrophila. Silico Biol. 2008, 8, 331–338. [Google Scholar]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalton, H. Structure and Mechanism of Action of the Enzyme(s) Involved in Methane Oxidation. In Applications of Enzyme Biotechnology; Kelly, J.W., Baldwin, T.O., Eds.; Springer: Boston, MA, USA, 1991; pp. 55–68. [Google Scholar]
- Beste, D.J.; Noh, K.; Niedenfuhr, S.; Mendum, T.A.; Hawkins, N.D.; Ward, J.L.; Beale, M.H.; Wiechert, W.; McFadden, J. 13C-flux spectral analysis of host-pathogen metabolism reveals a mixed diet for intracellular Mycobacterium tuberculosis. Chem. Biol. 2013, 20, 1012–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouzy, A.; Larrouy-Maumus, G.; Bottai, D.; Levillain, F.; Dumas, A.; Wallach, J.B.; Caire-Brandli, I.; De Chastellier, C.; Wu, T.D.; Poincloux, R.; et al. Mycobacterium tuberculosis exploits asparagine to assimilate nitrogen and resist acid stress during infection. PLoS Pathog. 2014, 10, e1003928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouzy, A.; Larrouy-Maumus, G.; Wu, T.D.; Peixoto, A.; Levillain, F.; Lugo-Villarino, G.; Guerquin-Kern, J.L.; De Carvalho, L.P.; Poquet, Y.; Neyrolles, O. Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate. Nat. Chem. Biol. 2013, 9, 674–676. [Google Scholar] [CrossRef] [Green Version]
- Tullius, M.V.; Harth, G.; Horwitz, M.A. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun. 2003, 71, 3927–3936. [Google Scholar] [CrossRef] [Green Version]
- Gillner, D.M.; Becker, D.P.; Holz, R.C. Lysine biosynthesis in bacteria: A metallodesuccinylase as a potential antimicrobial target. J. Biol. Inorg. Chem 2013, 18, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Mandal, R.S.; Das, S. In silico approach towards identification of potential inhibitors of Helicobacter pylori DapE. J. Biomol. Struct. Dyn. 2015, 33, 1460–1473. [Google Scholar] [CrossRef]
- Halouska, S.; Fenton, R.J.; Zinniel, D.K.; Marshall, D.D.; Barletta, R.G.; Powers, R. Metabolomics analysis identifies D-Alanine-D-Alanine ligase as the primary lethal target of D-Cycloserine in mycobacteria. J. Proteome Res. 2014, 13, 1065–1076. [Google Scholar] [CrossRef] [Green Version]
- Qiu, W.; Zheng, X.; Wei, Y.; Zhou, X.; Zhang, K.; Wang, S.; Cheng, L.; Li, Y.; Ren, B.; Xu, X.; et al. D-Alanine metabolism is essential for growth and biofilm formation of Streptococcus mutans. Mol. Oral Microbiol. 2016, 31, 435–444. [Google Scholar] [CrossRef]
- Silver, L.L. Appropriate Targets for Antibacterial Drugs. Cold Spring Harb. Perspect. Med. 2016, 6, a030239. [Google Scholar] [CrossRef] [Green Version]
- Caffrey, C.R.; Rohwer, A.; Oellien, F.; Marhöfer, R.J.; Braschi, S.; Oliveira, G.; McKerrow, J.H.; Selzer, P.M. A comparative chemogenomics strategy to predict potential drug targets in the metazoan pathogen, Schistosoma mansoni. PLoS ONE 2009, 4, e4413. [Google Scholar] [CrossRef]
- Consortium, U. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Tao, T. Standalone BLAST Setup for Unix; National Center for Biotechnology Information: Bethesda, MD, USA, 2008. [Google Scholar]
- Kerfeld, C.A.; Scott, K.M. Using BLAST to teach “E-value-tionary” concepts. PLoS Biol. 2011, 9, e1001014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, F.; Luo, H.; Zhang, C.-T.; Zhang, R. Gene essentiality analysis based on DEG 10, an updated database of essential genes. In Gene Essentiality; Springer: Berlin/Heidelberg, Germany, 2015; pp. 219–233. [Google Scholar]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2016, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Xu, J.Y.; Tao, L.; Li, X.F.; Li, S.; Zeng, X.; Chen, S.Y.; Zhang, P.; Qin, C.; Zhang, C. SVM-Prot 2016: A web-server for machine learning prediction of protein functional families from sequence irrespective of similarity. PLoS ONE 2016, 11, e0155290. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2017, 46, D1074–D1082. [Google Scholar] [CrossRef]
| UNIPROT STRAIN ID MAH-TH135 | ||||
| S. No. | Protein ID | DrugBank target name | DrugBank ID | Localization Site |
| 1. | T2GUW6 | DNA polymerase III subunit epsilon (DB01643) | P03007 | Cytoplasmic |
| UNIPROT STRAIN ID MAH-OCU466 | ||||
| S. No. | Protein ID | DrugBank target name | DrugBank ID | Localization Site |
| 1. | A0A2A3L1J8 | Inter-alpha-trypsin inhibitor heavy chain H4 (DB01593; DB14487; DB14533) Inter-alpha-trypsin inhibitor heavy chain H4 (DB01593; DB14487; DB14533) | Q14624 Q06033 | Cytoplasmic |
| 2. | A0A2A3L805 | O67040 Exopolyphosphatase (DB03382) | O67040 | Cytoplasmic |
| 3. | A0A2A3L3Y2 | DNA polymerase III subunit epsilon (DB01643) | P03007 | Cytoplasmic |
| 4. | A0A2A3LDY9 | Mannoside ABC transport system, sugar-binding protein (DB01942) | Q9X0V0 | Unknown |
| UNIPROT STRAIN ID MAH-A5 | ||||
| S. No. | Protein ID | DrugBank target name | DrugBank ID | Localization Site |
| 1. | A0A0E2W125 | Exopolyphosphatase (DB03382) | O67040 | Cytoplasmic |
| 2. | A0A0E2W9K2 | Inter-alpha-trypsin inhibitor heavy chain H4 (DB01593; DB14487; DB14533) Inter-alpha-trypsin inhibitor heavy chain H4 (DB01593; DB14487; DB14533) | Q14624 Q06033 | Cytoplasmic |
| 3. | A0A0E2W6U1 | Diacylglycerol acyltransferase/mycolyltransferase Ag85C (DB02811; DB08558) | P9WQN9 | Unknown (This protein may have multiple localization sites.) |
| 4. | A0A0E2W8I5 | Diacylglycerol acyltransferase/mycolyltransferase Ag85C (DB02811; DB08558) | P9WQN9 | Extracellular |
| 5. | A0A0E2W8U0 | DNA polymerase III subunit epsilon (DB01643) | P03007 | Cytoplasmic |
| 6. | A0A0E2WAR7 | Mannoside ABC transport system, sugar-binding protein (DB01942) Nickel-binding periplasmic protein (DB03374) | Q9X0V0 P33590 | Unknown |
| 7. | A0A0E2WQA2 | Mannoside ABC transport system, sugar-binding protein (DB01942) Nickel-binding periplasmic protein (DB03374) Periplasmic oligopeptide-binding protein (DB07365) ABC transporter, periplasmic substrate-binding protein (DB02078) | Q9X0V0 P33590 P06202 Q5LRQ9 | Periplasmic |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uddin, R.; Siraj, B.; Rashid, M.; Khan, A.; Ahsan Halim, S.; Al-Harrasi, A. Genome Subtraction and Comparison for the Identification of Novel Drug Targets against Mycobacterium avium subsp. hominissuis. Pathogens 2020, 9, 368. https://doi.org/10.3390/pathogens9050368
Uddin R, Siraj B, Rashid M, Khan A, Ahsan Halim S, Al-Harrasi A. Genome Subtraction and Comparison for the Identification of Novel Drug Targets against Mycobacterium avium subsp. hominissuis. Pathogens. 2020; 9(5):368. https://doi.org/10.3390/pathogens9050368
Chicago/Turabian StyleUddin, Reaz, Bushra Siraj, Muhammad Rashid, Ajmal Khan, Sobia Ahsan Halim, and Ahmed Al-Harrasi. 2020. "Genome Subtraction and Comparison for the Identification of Novel Drug Targets against Mycobacterium avium subsp. hominissuis" Pathogens 9, no. 5: 368. https://doi.org/10.3390/pathogens9050368
APA StyleUddin, R., Siraj, B., Rashid, M., Khan, A., Ahsan Halim, S., & Al-Harrasi, A. (2020). Genome Subtraction and Comparison for the Identification of Novel Drug Targets against Mycobacterium avium subsp. hominissuis. Pathogens, 9(5), 368. https://doi.org/10.3390/pathogens9050368









