Genomics and High-Resolution Typing Confirm Predominant Clonal Evolution Down to a Microevolutionary Scale in Trypanosoma cruzi
Abstract
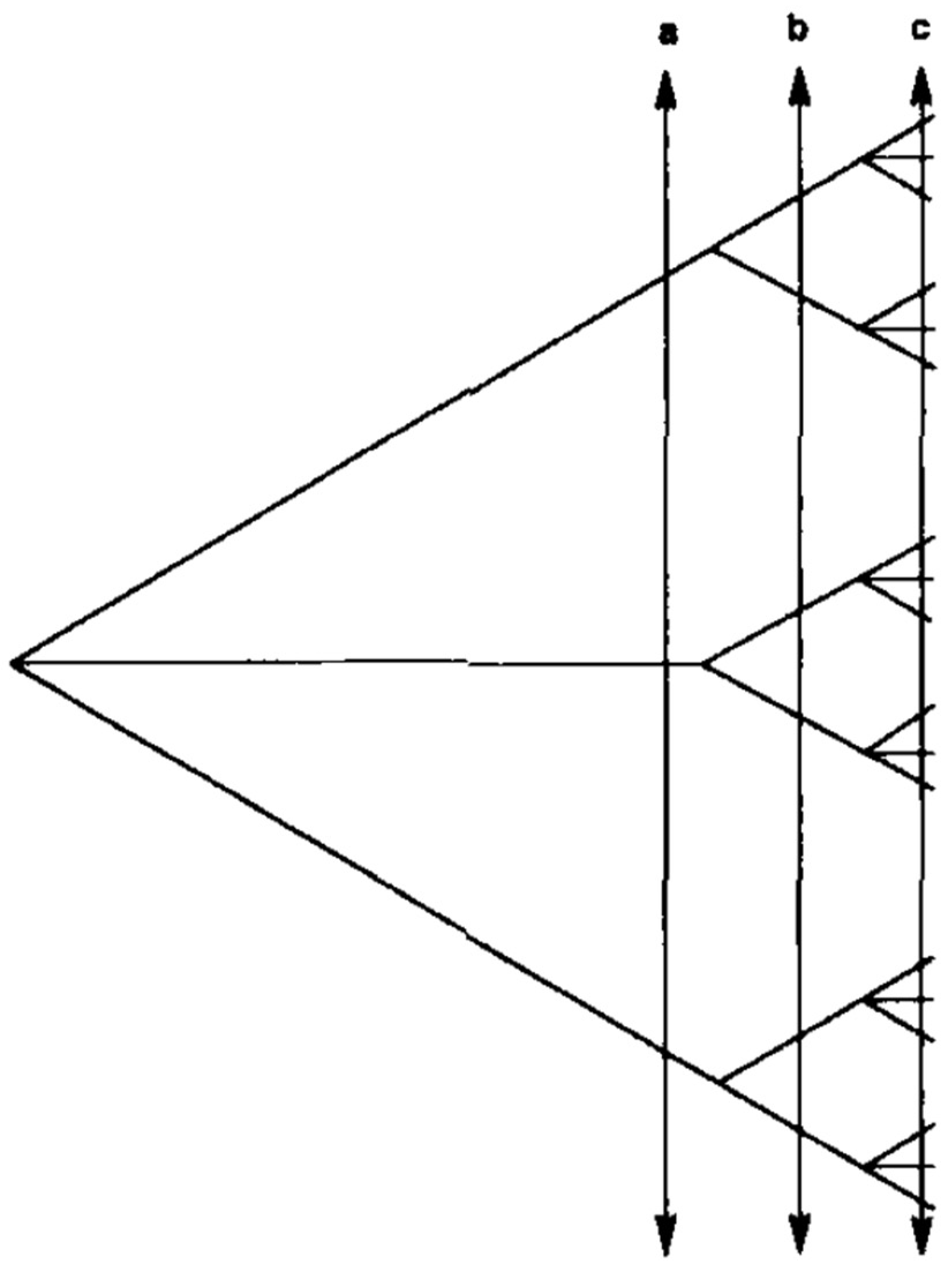
1. Preliminary Recalls about the Predominant Clonal Evolution (PCE) Model
2. Trypanosoma cruzi and the PCE Model
3. T. cruzi PCE Challengers
- (a)
- Biological speciation—each of the near-clades correspond to cryptic species that are genetically isolated from each other, but within which genetic exchange is random, except for physical obstacles (time and/or space) to this random gene flow (see Figure 3). This hypothesis of speciation has been invoked to claim that the main subdivisions (Savannah, Killifi, Forest) within Trypanosoma congolense are not evidence for PCE, because they could correspond to cryptic “species”. However, the authors did not clearly refer to a model of biological speciation [17]; and
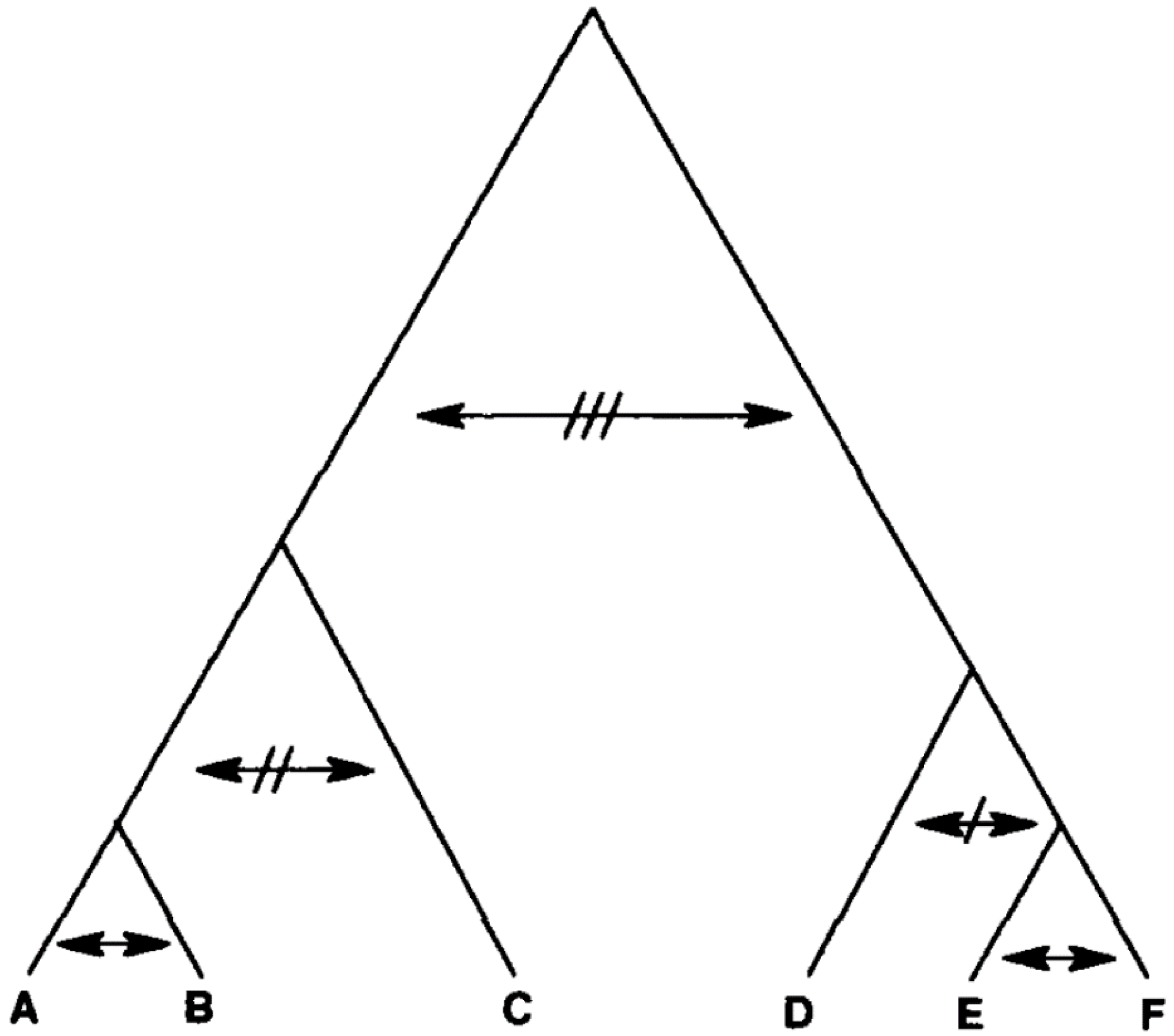
- (b)
- Progressive clonality—this situation refers to the case where the amount of genetic exchange is inversely proportional to the evolutionary distance between any two given genotypes [16]. If the genotypes are either identical or very similar, genetic exchange is abundant (homogamy, selfing). If they are distantly related, genetic exchange is either severely limited or lacking (Figure 4). Such an evolutionary model is believed to be frequent in bacteria [19].
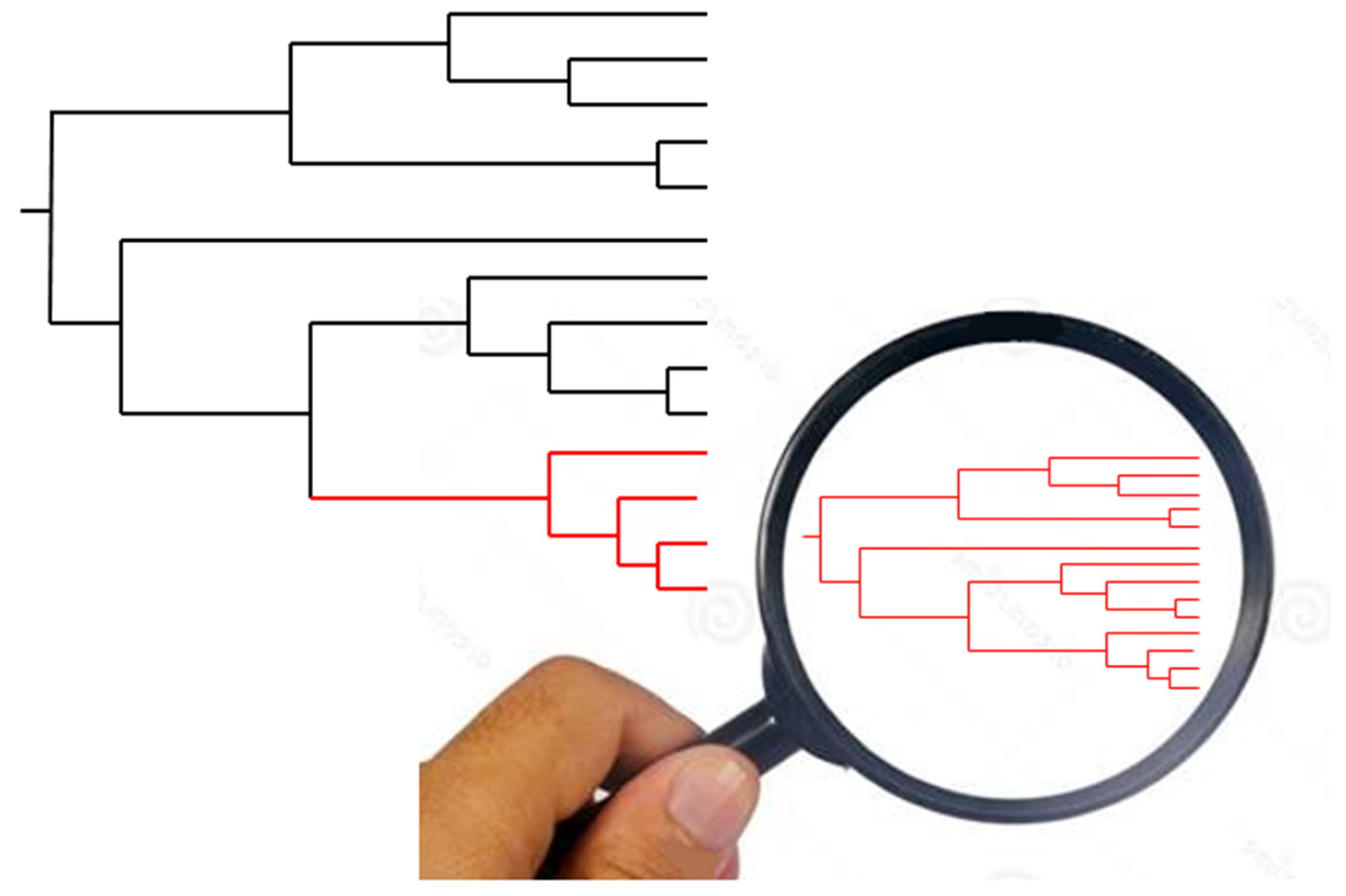
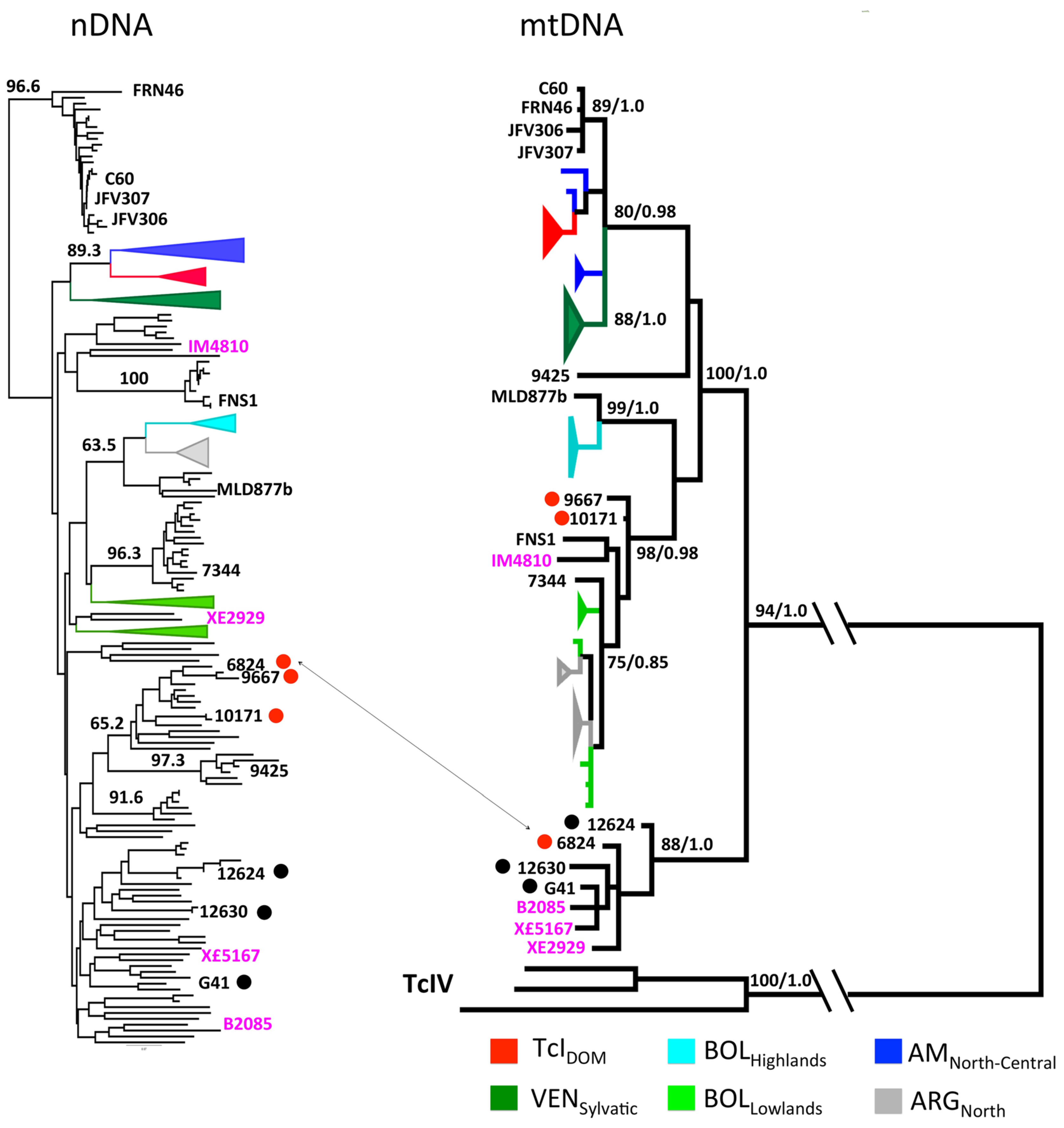
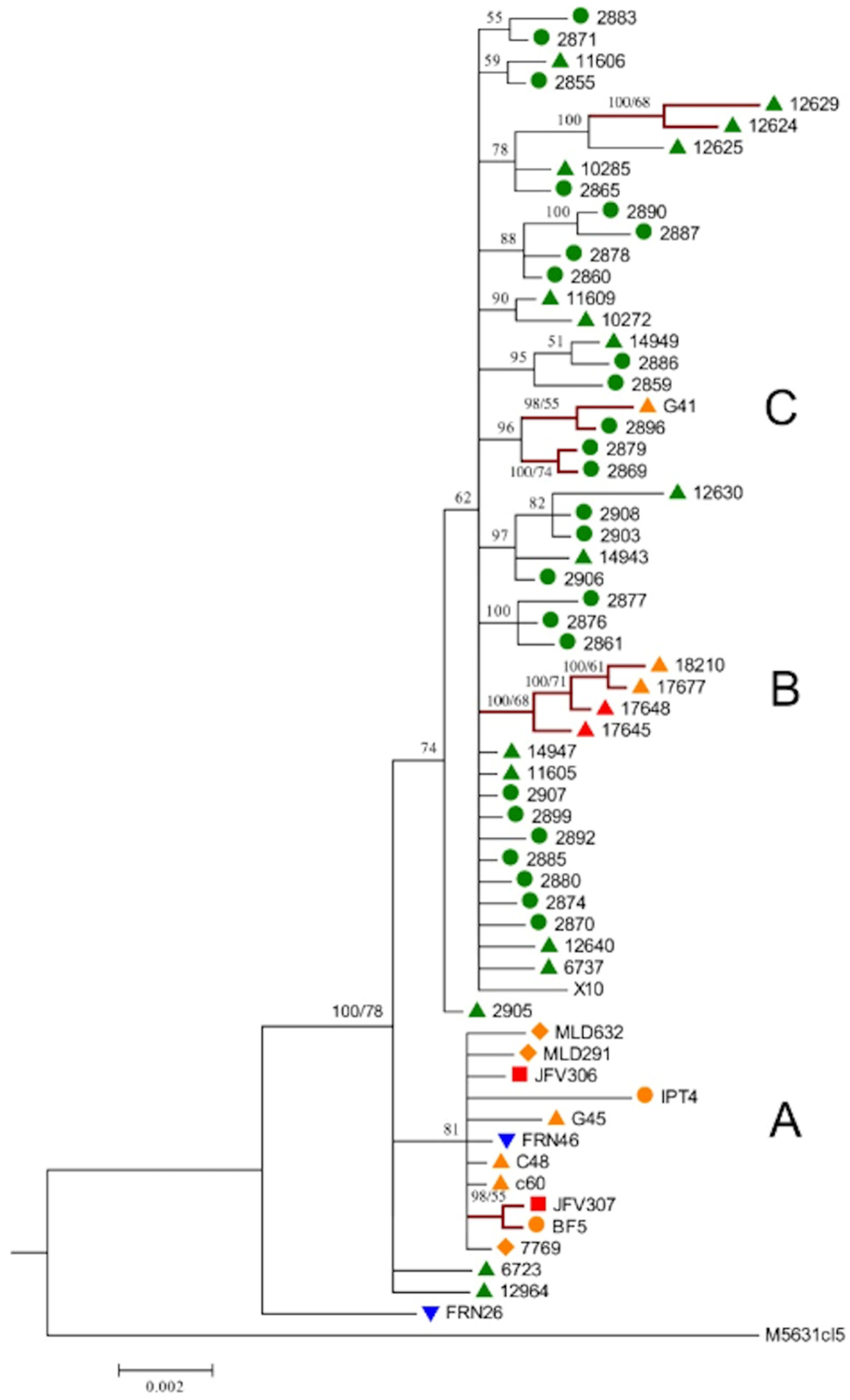
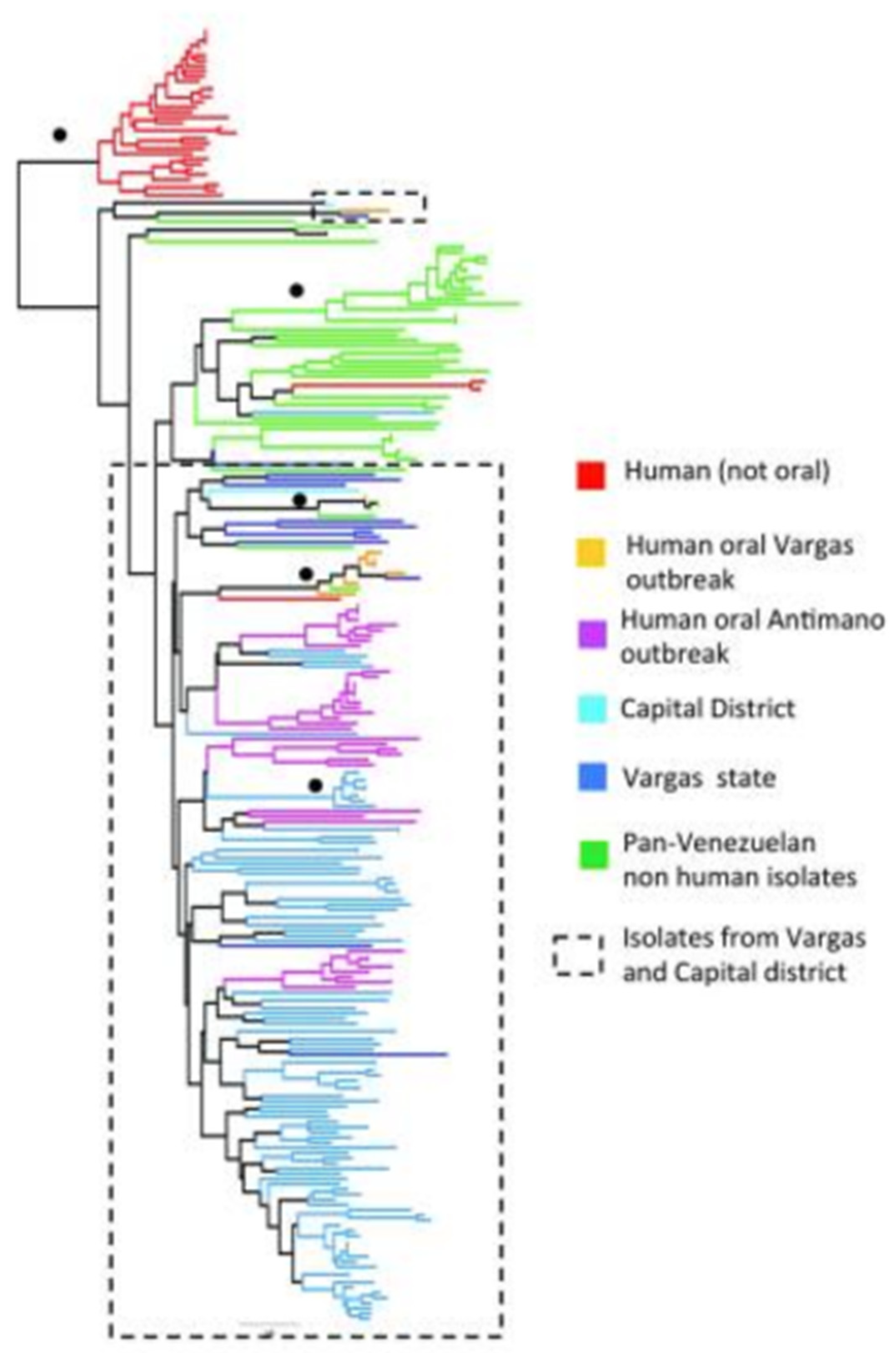
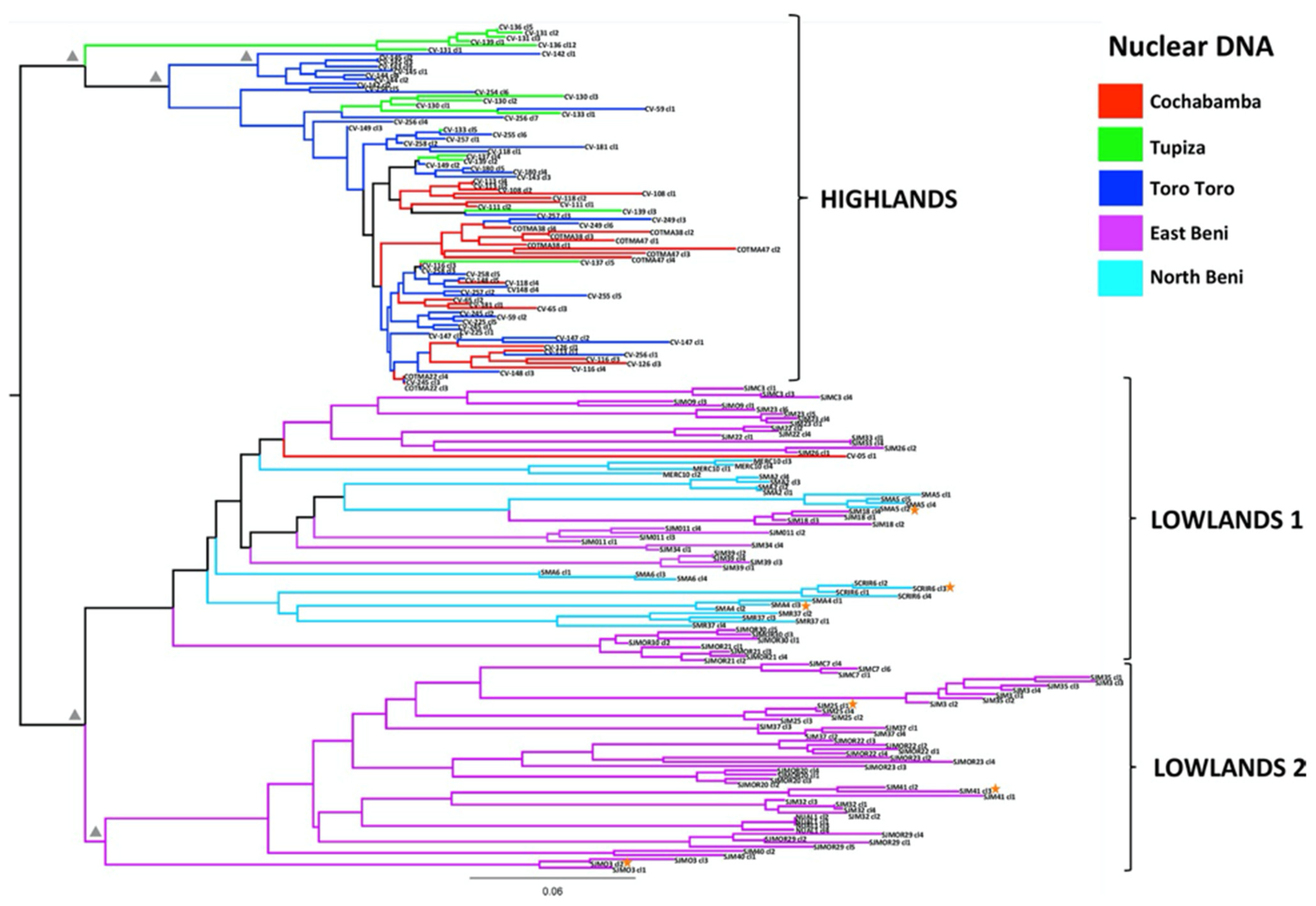
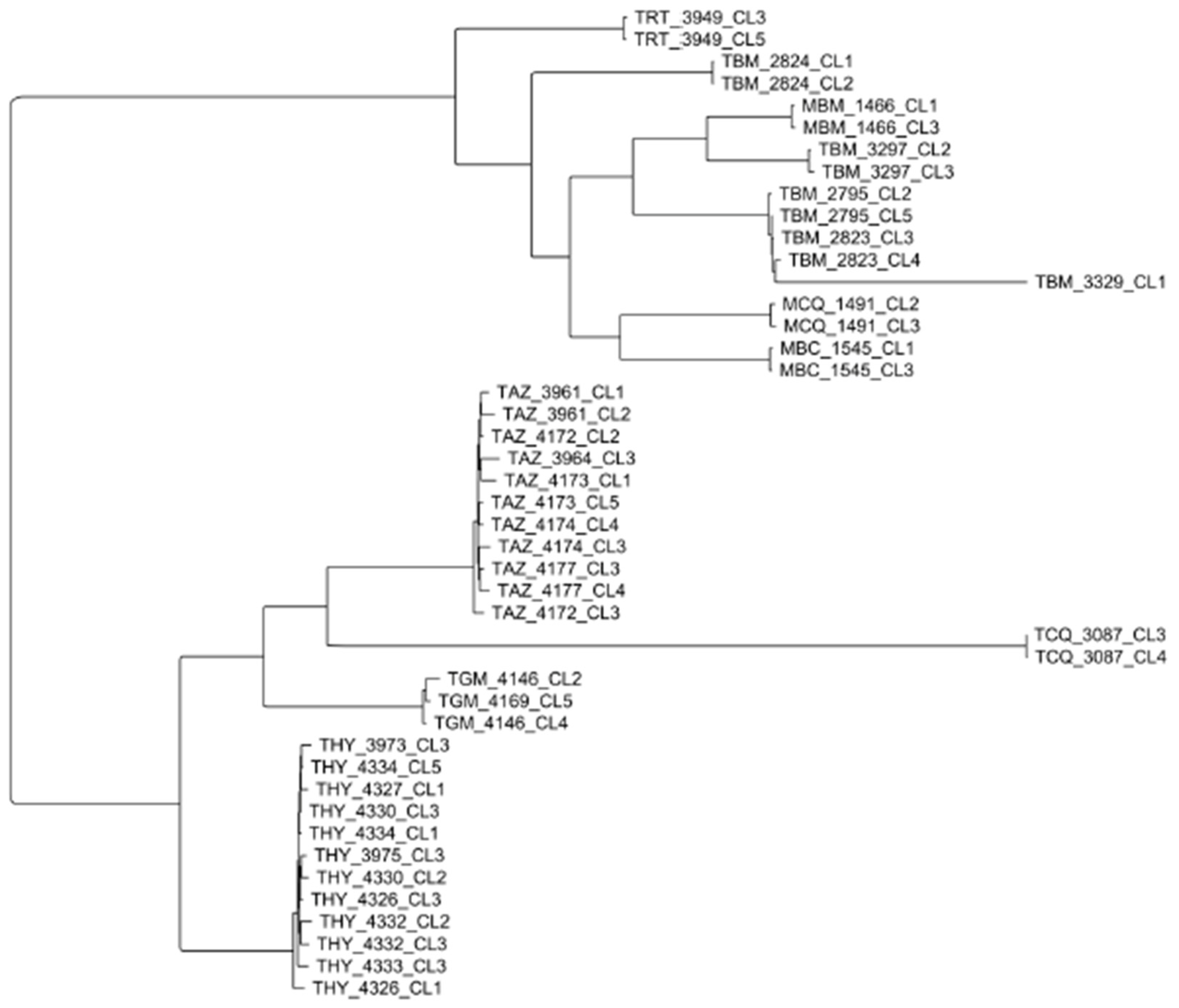
4. New Analyses with High-Resolution Typing Challenge the Challengers
5. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Miles, M.A.; Llewellyn, M.S.; Lewis, M.D.; Yeo, M.; Baleela, R.; Fitzpatrick, S.; Gaunt, M.W.; Mauricio, I.L. The molecular epidemiology and phylogeography of Trypanosoma cruzi and parallel research on Leishmania: Looking back and to the future. Parasitology 2009, 136, 1509–1528. [Google Scholar] [CrossRef] [PubMed]
- Tibayrenc, M.; Ayala, F.J. Is predominant clonal evolution a common evolutionary adaptation to parasitisms in parasitic protozoa, fungi, bacteria and viruses? Adv. Parasitol. 2017, 96, 243–325. [Google Scholar]
- Tibayrenc, M.; Ayala, F.J. Relevant units of analysis for applied and basic research dealing with neglected transmissible diseases: The predominant clonal evolution model of pathogenic microorganisms. PLoS Neglect. Trop. Dis. 2017, 11, e0005293. [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.M.; Bull, J.J. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Messenger, L.A.; Llewellyn, M.S.; Bhattacharyya, T.; Franzén, O.; Lewis, M.D.; Ramírez, J.D.; Carrasco, H.J.; Andersson, B.; Miles, M.A. Multiple Mitochondrial Introgression Events and Heteroplasmy in Trypanosoma cruzi Revealed by Maxicircle MLST and Next Generation Sequencing. PLoS Neglect. Trop. Dis. 2012, 6, e1584. [Google Scholar] [CrossRef]
- Schwabl, P.; Imamura, H.; Van den Broeck, F.; Costales, J.A.; Maiguashca-Sánchez, J.; Miles, M.A.; Andersson, B.; Grijalva, M.J.; Llewellyn, M.S. Meiotic sex in Chagas disease parasite Trypanosoma cruzi. Nat. Commun. 2019, 10, 3972. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Ayala, F.J. A misleading description of the predominant clonal evolution model in Trypanosoma cruzi. Acta Trop. 2018, 187, 13–14. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Ayala, F.J. Reproductive clonality of pathogens: A perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc. Nat. Acad. Sci. USA 2012, 109, E3305–E3313. [Google Scholar] [CrossRef]
- Miles, M.A.; Souza, A.; Povoa, M.; Shaw, J.J.; Lainson, R.; Toyé, P.J. Isozymic heterogeneity of Trypanosoma cruzi in the first autochtonous patients with Chagas’disease in Amazonian Brazil. Nature 1978, 272, 819–821. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Ward, P.; Moya, A.; Ayala, F.J. Natural populations of Trypanosoma cruzi, the agent of Chagas’disease, have a complex multiclonal structure. Proc. Nat. Acad. Sci. USA 1986, 83, 115–119. [Google Scholar] [CrossRef]
- Brisse, S.; Barnabé, C.; Tibayrenc, M. Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. Int. J. Parasitol. 2000, 30, 35–44. [Google Scholar] [CrossRef]
- Zingales, B.; Miles, M.A.; Campbell, D.A.; Tibayrenc, M.; Macedo, A.M.; Teixeira, M.M.; Schijman, A.G.; Llewellyn, M.S.; Lages-Silva, E.; Machado, C.R.; et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect. Genet. Evol. 2012, 12, 240–253. [Google Scholar] [CrossRef] [PubMed]
- Rougeron, V.; De Meeûs, T.; Kako Ouraga, S.; Hide, M.; Bañuls, A.L. ‘‘Everything You Always Wanted to Know about Sex (but Were Afraid to Ask)’’ in Leishmania after Two Decades of Laboratory and Field Analyses. PLoS Pathog. 2010, 6, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, J.D.; Llewellyn, M.S. Response to Tibayrenc and Ayala: Reproductive clonality in protozoan pathogens–truth or artefact? Mol. Ecol. 2015, 24, 5782–5784. [Google Scholar] [CrossRef]
- Tibayrenc, M.; Ayala, F.J. How clonal are Trypanosoma and Leishmania? Trends Parasitol. 2013, 29, 264–269. [Google Scholar] [CrossRef]
- Tibayrenc, M. Population Genetics of Parasitic Protozoa and other Microorganisms. Adv. Parasitol. 1995, 36, 47–115. [Google Scholar]
- Van den Broeck, F.; Tavernier, L.J.M.; Vermeiren, L.; Dujardin, J.C.; Van Den Abbeele, J. Mitonuclear genomics challenges the theory of clonality in Trypanosoma congolense: Reply to Tibayrenc and Ayala. Mol. Ecol. 2018, 27, 3425–3431. [Google Scholar] [CrossRef]
- Maynard Smith, J.; Smith, N.H.; O’Rourke, M.; Spratt, B.G. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 1993, 90, 4384–4388. [Google Scholar] [CrossRef]
- Hauck, S.; Maiden, M.C. Clonally Evolving Pathogenic Bacteria. In Molecular Mechanisms of Microbial Evolution; Rampelotto, P.H., Ed.; Grand Challenges in Biology and Biotechnology Springer International Publishing AG, Part of Springer Nature: Berlin, Germany, 2018; pp. 307–325. [Google Scholar]
- Lima, V.S.; Jansen, A.M.; Messenger, L.A.; Miles, M.A.; Llewellyn, M.S. Wild Trypanosoma cruzi I genetic diversity in Brazil suggests admixture and disturbance in parasite populations from the Atlantic Forest region. Parasite Vector 2014, 7, 263. [Google Scholar] [CrossRef][Green Version]
- Roman, F.; das Chagas Xavier, S.; Messenger, L.A.; Pavan, M.G.; Miles, M.A.; Jansen, A.M.; Yeo, M. Dissecting the phyloepidemiology of Trypanosoma cruzi I (TcI) in Brazil by the use of high resolution genetic markers. PLoS Neglect. Trop. D 2018, 12, e0006466. [Google Scholar] [CrossRef]
- Segovia, M.; Carrasco, H.J.; Martínez, C.E.; Messenger, L.A.; Nessi, A.; Londoño, J.C.; Espinosa, R.; Martínez, C.; Alfredo, M.; Bonfante-Cabarcas, R.; et al. Molecular Epidemiologic Source Tracking of Orally Transmitted Chagas Disease, Venezuela. Emerg. Infect. Dis. 2013, 19, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Messenger, L.A.; Garcia, L.; Vanhove, M.; Huaranca, C.; Bustamante, M.; Torrico, M.; Torrico, F.; Miles, M.A.; Llewellyn, M.S. Ecological host fitting of Trypanosoma cruzi TcI in Bolivia: Mosaic population structure, hybridization and a role for humans in Andean parasite dispersal. Mol. Ecol. 2015, 24, 2406–2422. [Google Scholar] [CrossRef] [PubMed]
- Reis-Cunha, J.L.; Baptista, R.P.; Rodrigues-Luiz, G.F.; Coqueiro-dos-Santos, A.; Valdivia, H.O.; de Almeida, L.V.; Cardoso, M.S.; D’Ávila, D.A.; Dias, F.H.C.; Fujiwara, R.T.; et al. Whole genome sequencing of Trypanosoma cruzi field isolates reveals extensive genomic variability and complex aneuploidy patterns within TcII DTU. BMC Genom. 2018, 191, 816. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, J.C.; Torres, C.; Curto, M.; Schijman, A.G. New insights into Trypanosoma cruzi evolution, genotyping and molecular diagnostics from satellite DNA sequence analysis. PLoS Neglect. Trop. D 2017, 11, e0006139. [Google Scholar] [CrossRef] [PubMed]
- Brisse, S.; Henriksson, J.; Barnabé, C.; Douzery, E.J.; Berkvens, D.; Serrano, M.; De Carvalho, M.R.C.; Buck, G.A.; Dujardin, J.C.; Tibayrenc, M. Evidence for genetic exchange and hybridization in Trypanosoma cruzi based on nucleotide sequences and molecular karyotype. Infect. Genet Evol. 2003, 2, 173–183. [Google Scholar] [CrossRef]
- Westenberger, S.J.; Barnabé, C.; Campbell, D.A.; Sturm, N.R. Two Hybridization Events Define the Population Structure of Trypanosoma cruzi. Genetics 2005, 171, 527–543. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tibayrenc, M.; Ayala, F.J. Genomics and High-Resolution Typing Confirm Predominant Clonal Evolution Down to a Microevolutionary Scale in Trypanosoma cruzi. Pathogens 2020, 9, 356. https://doi.org/10.3390/pathogens9050356
Tibayrenc M, Ayala FJ. Genomics and High-Resolution Typing Confirm Predominant Clonal Evolution Down to a Microevolutionary Scale in Trypanosoma cruzi. Pathogens. 2020; 9(5):356. https://doi.org/10.3390/pathogens9050356
Chicago/Turabian StyleTibayrenc, Michel, and Francisco J. Ayala. 2020. "Genomics and High-Resolution Typing Confirm Predominant Clonal Evolution Down to a Microevolutionary Scale in Trypanosoma cruzi" Pathogens 9, no. 5: 356. https://doi.org/10.3390/pathogens9050356
APA StyleTibayrenc, M., & Ayala, F. J. (2020). Genomics and High-Resolution Typing Confirm Predominant Clonal Evolution Down to a Microevolutionary Scale in Trypanosoma cruzi. Pathogens, 9(5), 356. https://doi.org/10.3390/pathogens9050356




