A Systematic Review Analyzing the Prevalence and Circulation of Influenza Viruses in Swine Population Worldwide
Abstract
1. Introduction
2. Methods
2.1. Systematic Review Protocol and Search Strategy
2.2. Inclusion and Exclusion Criteria
2.3. Ethical Approvals
3. Results
3.1. Influenza Viruses in Swine in Africa
3.1.1. Cameroon
3.1.2. Nigeria
3.1.3. Egypt
3.1.4. Kenya
3.1.5. Other African Countries
3.2. Influenza Viruses in Swine in Asia
3.2.1. China
3.2.2. Hong Kong and Tibet
3.2.3. Bhutan
3.2.4. Cambodia
3.2.5. Japan
3.2.6. South Korea
3.2.7. Thailand
3.2.8. Vietnam
3.2.9. India
3.2.10. Lebanon
3.2.11. Malaysia
3.2.12. Laos
3.2.13. Russia
3.2.14. Taiwan
3.2.15. Indonesia
3.2.16. Sri Lanka
3.2.17. Kazakhstan
3.3. Influenza Viruses in Swine in Australia
3.4. Influenza Viruses in Swine in Europe
3.4.1. Belgium
3.4.2. Denmark
3.4.3. United Kingdom
3.4.4. Finland
3.4.5. France
3.4.6. Germany
3.4.7. Greece
3.4.8. Italy
3.4.9. Spain
3.4.10. Luxembourg
3.4.11. The Netherlands
3.4.12. Norway
3.4.13. Poland
3.4.14. Czechoslovakia
3.4.15. Hungary
3.4.16. Multi-National Surveillances in European Countries
3.5. Influenza Viruses in Swine in North America
3.5.1. Canada
3.5.2. United States
3.5.3. Mexico
3.5.4. Guatemala
3.5.5. Cuba
3.5.6. Trinidad and Tobago
3.6. South America
3.6.1. Argentina
3.6.2. Brazil
3.6.3. Colombia
3.6.4. Peru
3.6.5. Chile
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kawano, J.; Onta, T.; Kida, H.; Yanagawa, R. Distribution of antibodies in animals against influenza B and C viruses. Jpnj. Vet. Res. 1978, 26, 74–80. [Google Scholar]
- Osterhaus, A.D.; Rimmelzwaan, G.F.; Martina, B.E.; Bestebroer, T.M.; Fouchier, R.A. Influenza B virus in seals. Science 2000, 288, 1051–1053. [Google Scholar] [CrossRef] [PubMed]
- Ran, Z.; Shen, H.; Lang, Y.; Kolb, E.A.; Turan, N.; Zhu, L.; Ma, J.; Bawa, B.; Liu, Q.; Liu, H.; et al. Domestic pigs are susceptible to infection with influenza B viruses. J. Virol. 2015, 89, 4818–4826. [Google Scholar] [CrossRef] [PubMed]
- Romvary, J.; Meszaros, J.; Barb, K. Susceptibility of birds to type-B influenza virus. Acta. Microbiol. Acad. Sc.I Hung. 1980, 27, 279–287. [Google Scholar]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and Swine: Proposal for a new genus in the Orthomyxoviridae family. Mbio. 2014, 5, e00031-14. [Google Scholar] [CrossRef]
- Salem, E.; Cook, E.A.J.; Lbacha, H.A.; Oliva, J.; Awoume, F.; Aplogan, G.L.; Hymann, E.C.; Muloi, D.; Deem, S.L.; Alali, S.; et al. Serologic Evidence for Influenza C and D Virus among Ruminants and Camelids, Africa, 1991–2015. Emerg. Infect. Dis. 2017, 23, 1556–1559. [Google Scholar] [CrossRef]
- Scholtissek, C. Pigs as ‘Mixing Vessels’ for the creation of new pandemic influenza A viruses. Med Princ. Pract. 1990, 2, 65–71. [Google Scholar] [CrossRef]
- Nakatsu, S.; Murakami, S.; Shindo, K.; Horimoto, T.; Sagara, H.; Noda, T.; Kawaoka, Y. Influenza C and D Viruses Package Eight Organized Ribonucleoprotein Complexes. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Tao, H.; Li, L.; White, M.C.; Steel, J.; Lowen, A.C. Influenza A Virus Coinfection through Transmission Can Support High Levels of Reassortment. J. Virol. 2015, 89, 8453–8461. [Google Scholar] [CrossRef]
- Salomon, R.; Webster, R.G. The influenza virus enigma. Cell 2009, 136, 402–410. [Google Scholar] [CrossRef]
- Vincent, A.; Awada, L.; Brown, I.; Chen, H.; Claes, F.; Dauphin, G.; Donis, R.; Culhane, M.; Hamilton, K.; Lewis, N.; et al. Review of influenza A virus in swine worldwide: A call for increased surveillance and research. Zoonoses Public Health 2014, 61, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Lager, K.M.; Vincent, A.L.; Janke, B.H.; Gramer, M.R.; Richt, J.A. The role of swine in the generation of novel influenza viruses. Zoonoses Public Health 2009, 56, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Rajao, D.S.; Vincent, A.L.; Perez, D.R. Adaptation of Human Influenza Viruses to Swine. Front. Vet. Sci. 2018, 5, 347. [Google Scholar] [CrossRef] [PubMed]
- Kaverin, N.V.; Gambaryan, A.S.; Bovin, N.V.; Rudneva, I.A.; Shilov, A.A.; Khodova, O.M.; Varich, N.L.; Sinitsin, B.V.; Makarova, N.V.; Kropotkina, E.A. Postreassortment changes in influenza A virus hemagglutinin restoring HA-NA functional match. Virology 1998, 244, 315–321. [Google Scholar] [CrossRef]
- Jang, J.; Bae, S.E. Comparative Co-Evolution Analysis Between the HA and NA Genes of Influenza A Virus. Virol. (Auckl) 2018, 9. [Google Scholar] [CrossRef]
- Kosik, I.; Yewdell, J.W. Influenza Hemagglutinin and Neuraminidase: Yin(-)Yang Proteins Coevolving to Thwart Immunity. Viruses 2019, 11, 346. [Google Scholar] [CrossRef]
- Francis, T., Jr. A NEW TYPE OF VIRUS FROM EPIDEMIC INFLUENZA. Science 1940, 92, 405–408. [Google Scholar] [CrossRef]
- Langat, P.; Raghwani, J.; Dudas, G.; Bowden, T.A.; Edwards, S.; Gall, A.; Bedford, T.; Rambaut, A.; Daniels, R.S.; Russell, C.A.; et al. Genome-wide evolutionary dynamics of influenza B viruses on a global scale. PLoS. Pathog. 2017, 13, e1006749. [Google Scholar] [CrossRef]
- Rogers, G.N.; Paulson, J.C. Receptor determinants of human and animal influenza virus isolates: Differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 1983, 127, 361–373. [Google Scholar] [CrossRef]
- Rogers, G.N.; D’Souza, B.L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 1989, 173, 317–322. [Google Scholar] [CrossRef]
- Baum, L.G.; Paulson, J.C. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta. Histochem. Suppl. 1990, 40, 35–38. [Google Scholar] [PubMed]
- Suzuki, Y.; Ito, T.; Suzuki, T.; Holland, R.E., Jr.; Chambers, T.M.; Kiso, M.; Ishida, H.; Kawaoka, Y. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 2000, 74, 11825–11831. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Veit, M. Hemagglutinin-esterase-fusion (HEF) protein of influenza C virus. Protein Cell 2016, 7, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Herrler, G.; Durkop, I.; Becht, H.; Klenk, H.D. The glycoprotein of influenza C virus is the haemagglutinin, esterase and fusion factor. J. Gen. Virol. 1988, 69 Pt 4, 839–846. [Google Scholar] [CrossRef]
- Song, H.; Qi, J.; Khedri, Z.; Diaz, S.; Yu, H.; Chen, X.; Varki, A.; Shi, Y.; Gao, G.F. An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism. PLoS. Pathog. 2016, 12, e1005411. [Google Scholar] [CrossRef]
- Castrucci, M.R.; Donatelli, I.; Sidoli, L.; Barigazzi, G.; Kawaoka, Y.; Webster, R.G. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology 1993, 193, 503–506. [Google Scholar] [CrossRef]
- Ito, T.; Couceiro, J.N.; Kelm, S.; Baum, L.G.; Krauss, S.; Castrucci, M.R.; Donatelli, I.; Kida, H.; Paulson, J.C.; Webster, R.G.; et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998, 72, 7367–7373. [Google Scholar] [CrossRef]
- Koen, J.S. A practical method for field diagnosis of swine diseases. Am. J. Vet. Med. 1919, 14, 468–470. [Google Scholar]
- Taubenberger, J.K.; Morens, D.M. 1918 Influenza: The mother of all pandemics. Emerg. Infect. Dis. 2006, 12, 15–22. [Google Scholar] [CrossRef]
- Worobey, M.; Han, G.Z.; Rambaut, A. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc. Natl. Acad. Sci. USA 2014, 111, 8107–8112. [Google Scholar] [CrossRef]
- Saunders-Hastings, P.R.; Krewski, D. Reviewing the History of Pandemic Influenza: Understanding Patterns of Emergence and Transmission. Pathogens 2016, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.J.; Bahl, J.; Vijaykrishna, D.; Zhang, J.; Poon, L.L.; Chen, H.; Webster, R.G.; Peiris, J.S.; Guan, Y. Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. USA 2009, 106, 11709–11712. [Google Scholar] [CrossRef]
- Mena, I.; Nelson, M.I.; Quezada-Monroy, F.; Dutta, J.; Cortes-Fernandez, R.; Lara-Puente, J.H.; Castro-Peralta, F.; Cunha, L.F.; Trovao, N.S.; Lozano-Dubernard, B.; et al. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 2016, 5. [Google Scholar] [CrossRef]
- Shope, R.E. Swine Influenza: Iii. Filtration Experiments and Etiology. J. Exp. Med. 1931, 54, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.; Andrewes, C.H.; Laidlaw, P.P. A virus obtained from influenza patients. Lancet. 1933, 222, 66–68. [Google Scholar] [CrossRef]
- Janke, B.H. Influenza A virus infections in swine: Pathogenesis and diagnosis. Vet. Pathol. 2014, 51, 410–426. [Google Scholar] [CrossRef] [PubMed]
- Selvaraju, S.B.; Selvarangan, R. Evaluation of three influenza A and B real-time reverse transcription-PCR assays and a new 2009 H1N1 assay for detection of influenza viruses. J. Clin. Microbiol. 2010, 48, 3870–3875. [Google Scholar] [CrossRef]
- Guo, Y.J.; Jin, F.G.; Wang, P.; Wang, M.; Zhu, J.M. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J. Gen. Virol. 1983, 64 Pt 1, 177–182. [Google Scholar] [CrossRef]
- Manuguerra, J.C.; Hannoun, C.; Saenz Mdel, C.; Villar, E.; Cabezas, J.A. Sero-epidemiological survey of influenza C virus infection in Spain. Eurj. Epidemiol. 1994, 10, 91–94. [Google Scholar] [CrossRef]
- Nishimura, H.; Sugawara, K.; Kitame, F.; Nakamura, K.; Sasaki, H. Prevalence of the antibody to influenza C virus in a northern Luzon Highland Village, Philippines. Microbiol. Immunol. 1987, 31, 1137–1143. [Google Scholar] [CrossRef]
- Salez, N.; Melade, J.; Pascalis, H.; Aherfi, S.; Dellagi, K.; Charrel, R.N.; Carrat, F.; de Lamballerie, X. Influenza C virus high seroprevalence rates observed in 3 different population groups. J. Infect. 2014, 69, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Odagiri, T.; Matsuzaki, Y.; Okamoto, M.; Suzuki, A.; Saito, M.; Tamaki, R.; Lupisan, S.P.; Sombrero, L.T.; Hongo, S.; Oshitani, H. Isolation and characterization of influenza C viruses in the Philippines and Japan. J. Clin. Microbiol. 2015, 53, 847–858. [Google Scholar] [CrossRef]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog 2013, 9, e1003176. [Google Scholar] [CrossRef]
- Zhai, S.L.; Zhang, H.; Chen, S.N.; Zhou, X.; Lin, T.; Liu, R.; Lv, D.H.; Wen, X.H.; Wei, W.K.; Wang, D.; et al. Influenza D Virus in Animal Species in Guangdong Province, Southern China. Emerg. Infect. Dis. 2017, 23, 1392–1396. [Google Scholar] [CrossRef]
- Foni, E.; Chiapponi, C.; Baioni, L.; Zanni, I.; Merenda, M.; Rosignoli, C.; Kyriakis, C.S.; Luini, M.V.; Mandola, M.L.; Bolzoni, L.; et al. Influenza D in Italy: Towards a better understanding of an emerging viral infection in swine. Sci.. Rep. 2017, 7, 11660. [Google Scholar] [CrossRef]
- Snoeck, C.J.; Oliva, J.; Pauly, M.; Losch, S.; Wildschutz, F.; Muller, C.P.; Hubschen, J.M.; Ducatez, M.F. Influenza D Virus Circulation in Cattle and Swine, Luxembourg, 2012-2016. Emerg. Infect. Dis 2018, 24, 1388–1389. [Google Scholar] [CrossRef]
- Kyriakis, C.S.; Rose, N.; Foni, E.; Maldonado, J.; Loeffen, W.L.; Madec, F.; Simon, G.; Van Reeth, K. Influenza A virus infection dynamics in swine farms in Belgium, France, Italy and Spain, 2006–2008. Vet. Microbiol. 2013, 162, 543–550. [Google Scholar] [CrossRef]
- Poljak, Z.; Carman, S.; McEwen, B. Assessment of seasonality of influenza in swine using field submissions to a diagnostic laboratory in Ontario between 2007 and 2012. Influenza Other Respir. Viruses 2014, 8, 482–492. [Google Scholar] [CrossRef]
- Vincent, A.L.; Ma, W.; Lager, K.M.; Janke, B.H.; Richt, J.A. Swine influenza viruses a North American perspective. Adv. Virus. Res. 2008, 72, 127–154. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gotzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Health Care Interventions: Explanation and Elaboration. PLoS. Med. 2009, 6. [Google Scholar] [CrossRef]
- Njabo, K.Y.; Fuller, T.L.; Chasar, A.; Pollinger, J.P.; Cattoli, G.; Terregino, C.; Monne, I.; Reynes, J.M.; Njouom, R.; Smith, T.B. Pandemic A/H1N1/2009 influenza virus in swine, Cameroon, 2010. Vet. Microbiol. 2012, 156, 189–192. [Google Scholar] [CrossRef]
- Snoeck, C.J.; Abiola, O.J.; Sausy, A.; Okwen, M.P.; Olubayo, A.G.; Owoade, A.A.; Muller, C.P. Serological evidence of pandemic (H1N1) 2009 virus in pigs, West and Central Africa. Vet. Microbiol. 2015, 176, 165–171. [Google Scholar] [CrossRef]
- Larison, B.; Njabo, K.Y.; Chasar, A.; Fuller, T.; Harrigan, R.J.; Smith, T.B. Spillover of pH1N1 to swine in Cameroon: An investigation of risk factors. Bmc Vet. Res. 2014, 10, 55. [Google Scholar] [CrossRef]
- Adeola, O.A.; Adeniji, J.A.; Olugasa, B.O. Detection of haemagglutination-inhibiting antibodies against human H1 and H3 strains of influenza A viruses in pigs in Ibadan, Nigeria. Zoonoses Public Health 2010, 57, e89–e94. [Google Scholar] [CrossRef]
- Meseko, C.A.; Odaibo, G.N.; Olaleye, D.O. Detection and isolation of 2009 pandemic influenza A/H1N1 virus in commercial piggery, Lagos Nigeria. Vet. Microbiol 2014, 168, 197–201. [Google Scholar] [CrossRef]
- Adeola, O.A.; Olugasa, B.O.; Emikpe, B.O. Molecular detection of influenza A(H1N1)pdm09 viruses with M genes from human pandemic strains among Nigerian pigs, 2013-2015: Implications and associated risk factors. Epidemio.L Infect. 2017, 145, 3345–3360. [Google Scholar] [CrossRef]
- Adeola, O.A.; Olugasa, B.O.; Emikpe, B.O. Antigenic Detection of Human Strain of Influenza Virus A (H3N2) in Swine Populations at Three Locations in Nigeria and Ghana during the Dry Early Months of 2014. Zoonoses Public Health 2016, 63, 106–111. [Google Scholar] [CrossRef]
- Awosanya, E.J.; Ogundipe, G.; Babalobi, O.; Omilabu, S. Prevalence and correlates of influenza-A in piggery workers and pigs in two communities in Lagos, Nigeria. Pan. Afr. Med. J. 2013, 16, 102. [Google Scholar] [CrossRef]
- Meseko, C.A.; Globig, A.; Ijomanta, J.; Joannis, T.; Nwosuh, C.; Shamaki, D.; Harder, T.; Hoffman, D.; Pohlmann, A.; Beer, M.; et al. Evidence of exposure of domestic pigs to Highly Pathogenic Avian Influenza H5N1 in Nigeria. Sc.I Rep. 2018, 8, 5900. [Google Scholar] [CrossRef]
- El-Sayed, A.; Awad, W.; Fayed, A.; Hamann, H.P.; Zschock, M. Avian influenza prevalence in pigs, Egypt. Emerg. Infect. Dis. 2010, 16, 726–727. [Google Scholar] [CrossRef]
- Gomaa, M.R.; Kandeil, A.; El-Shesheny, R.; Shehata, M.M.; McKenzie, P.P.; Webby, R.J.; Ali, M.A.; Kayali, G. Evidence of infection with avian, human, and swine influenza viruses in pigs in Cairo, Egypt. Arch. Virol. 2018, 163, 359–364. [Google Scholar] [CrossRef]
- Munyua, P.; Onyango, C.; Mwasi, L.; Waiboci, L.W.; Arunga, G.; Fields, B.; Mott, J.A.; Cardona, C.J.; Kitala, P.; Nyaga, P.N.; et al. Identification and characterization of influenza A viruses in selected domestic animals in Kenya, 2010-2012. PLoS ONE 2018, 13, e0192721. [Google Scholar] [CrossRef]
- Osoro, E.M.; Lidechi, S.; Marwanga, D.; Nyaundi, J.; Mwatondo, A.; Muturi, M.; Ng’ang’a, Z.; Njenga, K. Seroprevalence of influenza A virus in pigs and low risk of acute respiratory illness among pig workers in Kenya. Environ. Health Prev. Med. 2019, 24, 53. [Google Scholar] [CrossRef]
- Osoro, E.M.; Lidechi, S.; Nyaundi, J.; Marwanga, D.; Mwatondo, A.; Muturi, M.; Ng’ang’a, Z.; Njenga, K. Detection of pandemic influenza A/H1N1/pdm09 virus among pigs but not in humans in slaughterhouses in Kenya, 2013-2014. Bmc Res. Notes 2019, 12, 628. [Google Scholar] [CrossRef]
- Couacy-Hymann, E.; Kouakou, V.A.; Aplogan, G.L.; Awoume, F.; Kouakou, C.K.; Kakpo, L.; Sharp, B.R.; McClenaghan, L.; McKenzie, P.; Webster, R.G.; et al. Surveillance for influenza viruses in poultry and swine, west Africa, 2006-2008. Emerg. Infect. Dis. 2012, 18, 1446–1452. [Google Scholar] [CrossRef]
- Cardinale, E.; Pascalis, H.; Temmam, S.; Herve, S.; Saulnier, A.; Turpin, M.; Barbier, N.; Hoarau, J.; Queguiner, S.; Gorin, S.; et al. Influenza A(H1N1)pdm09 virus in pigs, Reunion Island. Emerg. Infect. Dis. 2012, 18, 1665–1668. [Google Scholar] [CrossRef]
- Ducatez, M.F.; Awoume, F.; Webby, R.J. Influenza A(H1N1)pdm09 virus in pigs, Togo, 2013. Vet. Microbiol. 2015, 177, 201–205. [Google Scholar] [CrossRef]
- Dione, M.; Masembe, C.; Akol, J.; Amia, W.; Kungu, J.; Lee, H.S.; Wieland, B. The importance of on-farm biosecurity: Sero-prevalence and risk factors of bacterial and viral pathogens in smallholder pig systems in Uganda. Acta. Trop. 2018, 187, 214–221. [Google Scholar] [CrossRef]
- Shortridge, K.F.; Stuart-Harris, C.H. An influenza epicentre? Lancet. 1982, 2, 812–813. [Google Scholar] [CrossRef]
- Ninomiya, A.; Takada, A.; Okazaki, K.; Shortridge, K.F.; Kida, H. Seroepidemiological evidence of avian H4, H5, and H9 influenza A virus transmission to pigs in southeastern China. Vet. Microbiol. 2002, 88, 107–114. [Google Scholar] [CrossRef]
- Qi, X.; Lu, C.P. Genetic characterization of novel reassortant H1N2 influenza A viruses isolated from pigs in southeastern China. Arch. Virol. 2006, 151, 2289–2299. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.L.; Wang, C.F.; Yan, C.M.; Peng, J.S.; Jiang, Z.L.; Liu, J.H. Swine infection with H9N2 influenza viruses in China in 2004. Virus Genes 2008, 36, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, G.H.; Hua, R.H.; Zhang, Q.; Liu, T.Q.; Liao, M.; Tong, G.Z. Isolation and genetic analysis of human origin H1N1 and H3N2 influenza viruses from pigs in China. Biochem. Biophys. Res. Commun. 2007, 356, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Zhou, H.; Jiang, T.; Li, C.; Zhang, A.; Guo, X.; Zou, W.; Chen, H.; Jin, M. Isolation and molecular characterization of equine H3N8 influenza viruses from pigs in China. Arch. Virol 2009, 154, 887–890. [Google Scholar] [CrossRef]
- Yu, H.; Hua, R.H.; Wei, T.C.; Zhou, Y.J.; Tian, Z.J.; Li, G.X.; Liu, T.Q.; Tong, G.Z. Isolation and genetic characterization of avian origin H9N2 influenza viruses from pigs in China. Vet. Microbiol 2008, 131, 82–92. [Google Scholar] [CrossRef]
- Bi, Y.; Fu, G.; Chen, J.; Peng, J.; Sun, Y.; Wang, J.; Pu, J.; Zhang, Y.; Gao, H.; Ma, G.; et al. Novel swine influenza virus reassortants in pigs, China. Emerg. Infect. Dis. 2010, 16, 1162–1164. [Google Scholar] [CrossRef]
- Hou, D.; Bi, Y.; Sun, H.; Yang, J.; Fu, G.; Sun, Y.; Liu, J.; Pu, J. Identification of swine influenza A virus and Stenotrophomonas maltophilia co-infection in Chinese pigs. Virolj. 2012, 9, 169. [Google Scholar] [CrossRef]
- He, L.; Zhao, G.; Zhong, L.; Liu, Q.; Duan, Z.; Gu, M.; Wang, X.; Liu, X.; Liu, X. Isolation and characterization of two H5N1 influenza viruses from swine in Jiangsu Province of China. Arch. Virol. 2013, 158, 2531–2541. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhu, W.; Chen, Y.; Zhou, P.; Cao, Z.; Xie, J.; Zhang, C.; Ke, C.; Qi, W.; Su, S.; et al. Serological surveillance of H5 and H9 avian influenza A viral infections among pigs in Southern China. Microb Pathog 2013, 64, 39–42. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, X.; Li, S.; Guo, X.; Yang, Y.; Jin, M. Complete genome sequence of a novel H4N1 influenza virus isolated from a pig in central China. J. Virol 2012, 86, 13879. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, J.; Qiao, C.; Yang, H.; Zhang, Y.; Xin, X.; Chen, H. Co-circulation of pandemic 2009 H1N1, classical swine H1N1 and avian-like swine H1N1 influenza viruses in pigs in China. Infect. Genet. Evol. 2013, 13, 331–338. [Google Scholar] [CrossRef]
- Zhao, G.; Fan, Q.; Zhong, L.; Li, Y.; Liu, W.; Liu, X.; Gao, S.; Peng, D.; Liu, X. Isolation and phylogenetic analysis of pandemic H1N1/09 influenza virus from swine in Jiangsu province of China. Res. Vet. Sci. 2012, 93, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhou, B.; Fan, X.; Lam, T.T.; Wang, J.; Chen, A.; Chen, X.; Chen, H.; Webster, R.G.; Webby, R.; et al. Novel reassortment of Eurasian avian-like and pandemic/2009 influenza viruses in swine: Infectious potential for humans. J. Virol. 2011, 85, 10432–10439. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.L.; Huang, L.Z.; Qi, H.T.; Cao, N.; Zhang, L.Q.; Wang, H.; Guan, S.S.; Qi, W.B.; Jiao, P.R.; Liao, M.; et al. Genetic characterization of H1N2 influenza a virus isolated from sick pigs in Southern China in 2010. Virolj. 2011, 8, 469. [Google Scholar] [CrossRef]
- Yan, J.H.; Xiong, Y.; Yi, C.H.; Sun, X.X.; He, Q.S.; Fu, W.; Xu, X.K.; Jiang, J.X.; Ma, L.; Liu, Q. Pandemic (H1N1) 2009 virus circulating in pigs, Guangxi, China. Emerg. Infect. Dis. 2012, 18, 357–359. [Google Scholar] [CrossRef]
- Liang, H.; Lam, T.T.; Fan, X.; Chen, X.; Zeng, Y.; Zhou, J.; Duan, L.; Tse, M.; Chan, C.H.; Li, L.; et al. Expansion of genotypic diversity and establishment of 2009 H1N1 pandemic-origin internal genes in pigs in China. J. Virol. 2014, 88, 10864–10874. [Google Scholar] [CrossRef]
- Sun, Y.F.; Wang, X.H.; Li, X.L.; Zhang, L.; Li, H.H.; Lu, C.; Yang, C.L.; Feng, J.; Han, W.; Ren, W.K.; et al. Novel triple-reassortant H1N1 swine influenza viruses in pigs in Tianjin, Northern China. Vet. Microbiol. 2016, 183, 85–91. [Google Scholar] [CrossRef]
- Ma, M.J.; Wang, G.L.; Anderson, B.D.; Bi, Z.Q.; Lu, B.; Wang, X.J.; Wang, C.X.; Chen, S.H.; Qian, Y.H.; Song, S.X.; et al. Evidence for Cross-species Influenza A Virus Transmission Within Swine Farms, China: A One Health, Prospective Cohort Study. Clin. Infect. Dis. 2018, 66, 533–540. [Google Scholar] [CrossRef]
- He, P.; Wang, G.; Mo, Y.; Yu, Q.; Xiao, X.; Yang, W.; Zhao, W.; Guo, X.; Chen, Q.; He, J.; et al. Novel triple-reassortant influenza viruses in pigs, Guangxi, China. Emerg. Microbes. Infect. 2018, 7, 85. [Google Scholar] [CrossRef]
- Peng, X.; Wu, H.; Xu, L.; Peng, X.; Cheng, L.; Jin, C.; Xie, T.; Lu, X.; Wu, N. Molecular characterization of a novel reassortant H1N2 influenza virus containing genes from the 2009 pandemic human H1N1 virus in swine from eastern China. Virus Genes 2016, 52, 405–410. [Google Scholar] [CrossRef]
- Anderson, B.D.; Ma, M.J.; Wang, G.L.; Bi, Z.Q.; Lu, B.; Wang, X.J.; Wang, C.X.; Chen, S.H.; Qian, Y.H.; Song, S.X.; et al. Prospective surveillance for influenza. virus in Chinese swine farms. Emerg. Microbes. Infect. 2018, 7, 87. [Google Scholar] [CrossRef]
- Xu, M.; Huang, Y.; Chen, J.; Huang, Z.; Zhang, J.; Zhu, Y.; Xie, S.; Chen, Q.; Wei, W.; Yang, D.; et al. Isolation and genetic analysis of a novel triple-reassortant H1N1 influenza virus from a pig in China. Vet. Microbiol. 2011, 147, 403–409. [Google Scholar] [CrossRef]
- Fan, X.; Zhu, H.; Zhou, B.; Smith, D.K.; Chen, X.; Lam, T.T.; Poon, L.L.; Peiris, M.; Guan, Y. Emergence and dissemination of a swine H3N2 reassortant influenza virus with 2009 pandemic H1N1 genes in pigs in China. J. Virol 2012, 86, 2375–2378. [Google Scholar] [CrossRef][Green Version]
- Yang, H.; Qiao, C.; Tang, X.; Chen, Y.; Xin, X.; Chen, H. Human infection from avian-like influenza A (H1N1) viruses in pigs, China. Emerg Infect. Dis 2012, 18, 1144–1146. [Google Scholar] [CrossRef]
- Wang, N.; Zou, W.; Yang, Y.; Guo, X.; Hua, Y.; Zhang, Q.; Zhao, Z.; Jin, M. Complete genome sequence of an H10N5 avian influenza virus isolated from pigs in central China. J. Virol 2012, 86, 13865–13866. [Google Scholar] [CrossRef]
- Su, S.; Qi, W.; Chen, J.; Zhu, W.; Huang, Z.; Xie, J.; Zhang, G. Seroepidemiological evidence of avian influenza A virus transmission to pigs in southern China. J. Clin. Microbiol. 2013, 51, 601–602. [Google Scholar] [CrossRef][Green Version]
- Zhou, P.; Hong, M.; Merrill, M.M.; He, H.; Sun, L.; Zhang, G. Serological report of influenza A (H7N9) infections among pigs in Southern China. Bmc Vet. Res. 2014, 10, 203. [Google Scholar] [CrossRef]
- Cao, N.; Zhu, W.; Chen, Y.; Tan, L.; Zhou, P.; Cao, Z.; Ke, C.; Li, Y.; Wu, J.; Qi, W.; et al. Avian influenza A (H5N1) virus antibodies in pigs and residents of swine farms, southern China. J. Clin. Virol. 2013, 58, 647–651. [Google Scholar] [CrossRef]
- Zhou, H.; Cao, Z.; Tan, L.; Fu, X.; Lu, G.; Qi, W.; Ke, C.; Wang, H.; Sun, L.; Zhang, G. Avian-like A (H1N1) swine influenza virus antibodies among swine farm residents and pigs in southern China. Jpnj. Infect. Dis 2014, 67, 184–190. [Google Scholar] [CrossRef][Green Version]
- Li, S.; Zhou, Y.; Zhao, Y.; Li, W.; Song, W.; Miao, Z. Avian influenza H9N2 seroprevalence among pig population and pig farm staff in Shandong, China. Virolj. 2015, 12, 34. [Google Scholar] [CrossRef]
- Liang, Q.L.; Nie, L.B.; Zou, Y.; Hou, J.L.; Chen, X.Q.; Bai, M.J.; Gao, Y.H.; Hu, G.X.; Zhu, X.Q. Serological evidence of hepatitis E virus and influenza A virus infection in farmed wild boars in China. Acta. Trop. 2019, 192, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.M.; Wang, S.C.; Peng, C.; Yu, J.M.; Zhuang, Q.Y.; Hou, G.Y.; Liu, S.; Li, J.P.; Chen, J.M. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes 2014, 49, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Shortridge, K.F.; Krauss, S.; Li, P.H.; Kawaoka, Y.; Webster, R.G. Emergence of avian H1N1 influenza viruses in pigs in China. J. Virol. 1996, 70, 8041–8046. [Google Scholar] [CrossRef] [PubMed]
- Connor, R.J.; Kawaoka, Y.; Webster, R.G.; Paulson, J.C. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 1994, 205, 17–23. [Google Scholar] [CrossRef]
- Matrosovich, M.N.; Krauss, S.; Webster, R.G. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 2001, 281, 156–162. [Google Scholar] [CrossRef]
- Liu, G.H.; Zhou, D.H.; Cong, W.; Zhang, X.X.; Shi, X.C.; Danba, C.; Huang, S.Y.; Zhu, X.Q. First report of seroprevalence of swine influenza A virus in Tibetan pigs in Tibet, China. Trop. Anim. Health Prod. 2014, 46, 257–259. [Google Scholar] [CrossRef]
- Monger, V.R.; Stegeman, J.A.; Koop, G.; Dukpa, K.; Tenzin, T.; Loeffen, W.L. Seroprevalence and associated risk factors of important pig viral diseases in Bhutan. Prev. Vet. Med. 2014, 117, 222–232. [Google Scholar] [CrossRef]
- Rith, S.; Netrabukkana, P.; Sorn, S.; Mumford, E.; Mey, C.; Holl, D.; Goutard, F.; Fenwick, S.; Robertson, I.; Roger, F.; et al. Serologic evidence of human influenza virus infections in swine populations, Cambodia. Influenza Other Respir. Viruses 2013, 7, 271–279. [Google Scholar] [CrossRef]
- Osbjer, K.; Berg, M.; Sokerya, S.; Chheng, K.; San, S.; Davun, H.; Magnusson, U.; Olsen, B.; Zohari, S. Influenza A Virus in Backyard Pigs and Poultry in Rural Cambodia. Transbound Emerg. Dis. 2017, 64, 1557–1568. [Google Scholar] [CrossRef]
- Sugimura, T.; Kataoka, T.; Kita, E.; Nishino, K.; Sasahara, J. An outbreak of influenza in the swine with Hong Kong influenza virus (author’s transl). Uirusu 1975, 25, 19–24. [Google Scholar] [CrossRef]
- Nerome, K.; Ishida, M.; Nakayama, M.; Oya, A.; Kanai, C.; Suwicha, K. Antigenic and genetic analysis of A/Hong Kong (H3N2) influenza viruses isolated from swine and man. J. Gen. Virol. 1981, 56, 441–445. [Google Scholar] [CrossRef]
- Arikawa, J.; Yamane, N.; Totsukawa, K.; Ishida, N. The follow-up study of swine and Hong Kong influenza virus infection among Japanese hogs. Tohokuj. Exp. Med. 1982, 136, 353–358. [Google Scholar] [CrossRef]
- Sugimura, T.; Yonemochi, H.; Ogawa, T.; Tanaka, Y.; Kumagai, T. Isolation of a recombinant influenza virus (Hsw 1 N2) from swine in Japan. Arch. Virol. 1980, 66, 271–274. [Google Scholar] [CrossRef]
- Ogawa, Y.; Goto, H.; Hirano, T.; Shimizu, K.; Ohno, Y.; Kuroda, H.; Hongo, H. Sero-epizootiological study on swine influenza in a prefecture of Japan. J. Hyg. (Lond) 1983, 90, 403–406. [Google Scholar] [CrossRef]
- Yasuhara, H.; Hirahara, T.; Nakai, M.; Sasaki, N.; Kato, J.; Watanabe, T.; Morikawa, M. Further isolation of a recombinant virus (H1N2, formerly Hsw1N2) from a pig in Japan in 1980. Microbiol. Immunol. 1983, 27, 43–50. [Google Scholar] [CrossRef]
- Nerome, K.; Ishida, M.; Oya, A.; Oda, K. The possible origin H1N1 (Hsw1N1) virus in the swine population of Japan and antigenic analysis of the isolates. J. Gen. Virol 1982, 62 Pt 1, 171–175. [Google Scholar] [CrossRef]
- Yamaoka, M.; Hotta, H.; Itoh, M.; Homma, M. Prevalence of antibody to influenza C virus among pigs in Hyogo Prefecture, Japan. J. Gen. Virol. 1991, 72 Pt 3, 711–714. [Google Scholar] [CrossRef]
- Ohwada, K.; Kitame, F.; Sugawara, K.; Nishimura, H.; Homma, M.; Nakamura, K. Distribution of the antibody to influenza C virus in dogs and pigs in Yamagata Prefecture, Japan. Microbiol Immunol 1987, 31, 1173–1180. [Google Scholar] [CrossRef]
- Ito, T.; Kawaoka, Y.; Vines, A.; Ishikawa, H.; Asai, T.; Kida, H. Continued circulation of reassortant H1N2 influenza viruses in pigs in Japan. Arch. Virol. 1998, 143, 1773–1782. [Google Scholar] [CrossRef]
- Kobayashi, M.; Takayama, I.; Kageyama, T.; Tsukagoshi, H.; Saitoh, M.; Ishioka, T.; Yokota, Y.; Kimura, H.; Tashiro, M.; Kozawa, K. Novel reassortant influenza A(H1N2) virus derived from A(H1N1)pdm09 virus isolated from swine, Japan, 2012. Emerg. Infect. Dis. 2013, 19, 1972–1974. [Google Scholar] [CrossRef]
- Yoneyama, S.; Hayashi, T.; Kojima, H.; Usami, Y.; Kubo, M.; Takemae, N.; Uchida, Y.; Saito, T. Occurrence of a pig respiratory disease associated with swine influenza A (H1N2) virus in Tochigi Prefecture, Japan. J. Vet. Med. Sci. 2010, 72, 481–488. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Katsuda, K.; Sato, S.; Imai, M.; Shirahata, T.; Goto, H. Prevalence of antibodies to type A influenza viruses in swine sera 1990-1994. J. Vet. Med. Sci 1995, 57, 773–775. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Katsuda, K.; Sato, S.; Shirahata, T.; Lindstrom, S.; Nerome, R.; Ishida, M.; Nerome, K.; Goto, H. Antigenic and genetic characteristics of H1N1 human influenza virus isolated from pigs in Japan. J. Gen. Virol. 1995, 76 Pt 5, 1247–1249. [Google Scholar] [CrossRef]
- Kirisawa, R.; Ogasawara, Y.; Yoshitake, H.; Koda, A.; Furuya, T. Genomic reassortants of pandemic A (H1N1) 2009 virus and endemic porcine H1 and H3 viruses in swine in Japan. J. Vet. Med. Sci. 2014, 76, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, H.; Van Nguyen, D.; Yonemitsu, K.; Minami, S.; Nagata, N.; Hara, N.; Kuwata, R.; Murakami, S.; Kodera, Y.; Takeda, T.; et al. Influenza A virus infection in Japanese wild boars (Sus scrofa leucomystax). J. Vet. Med. Sci. 2017, 79, 848–851. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Inoue, H.; Ozawa, M.; Matsuu, A. Serological survey of influenza A virus infection in Japanese wild boars (Sus scrofa leucomystax). Microbiol. Immunol. 2019. [Google Scholar] [CrossRef]
- Song, D.S.; Lee, J.Y.; Oh, J.S.; Lyoo, K.S.; Yoon, K.J.; Park, Y.H.; Park, B.K. Isolation of H3N2 swine influenza virus in South Korea. J. Vet. Diagn. Invest. 2003, 15, 30–34. [Google Scholar] [CrossRef]
- Kwon, T.Y.; Lee, S.S.; Kim, C.Y.; Shin, J.Y.; Sunwoo, S.Y.; Lyoo, Y.S. Genetic characterization of H7N2 influenza virus isolated from pigs. Vet. Microbiol. 2011, 153, 393–397. [Google Scholar] [CrossRef]
- Choi, C.; Ha, S.K.; Chae, C. Detection and isolation of H1N1 influenza virus from pigs in Korea. Vet. Rec 2004, 154, 274–275. [Google Scholar] [CrossRef]
- Jung, T.; Choi, C.; Chae, C. Localization of swine influenza virus in naturally infected pigs. Vet. Pathol. 2002, 39, 10–16. [Google Scholar] [CrossRef]
- Jung, K.; Chae, C. First outbreak of respiratory disease associated with swine influenza H1N2 virus in pigs in Korea. J. Vet. Diagn. Invest. 2005, 17, 176–178. [Google Scholar] [CrossRef]
- Jung, K.; Song, D.S.; Kang, B.K.; Oh, J.S.; Park, B.K. Serologic surveillance of swine H1 and H3 and avian H5 and H9 influenza A virus infections in swine population in Korea. Prev. Vet. Med. 2007, 79, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.K.; Kim, H.S.; Cho, S.W.; Seo, S.H. Genetic and antigenic characterization of swine H1N2 influenza viruses isolated from Korean pigs. J. Microbiol. Biotechnol. 2007, 17, 868–872. [Google Scholar] [PubMed]
- Kim, S.H.; Kim, H.J.; Jin, Y.H.; Yeoul, J.J.; Lee, K.K.; Oem, J.K.; Lee, M.H.; Park, C.K. Isolation of influenza A(H3N2)v virus from pigs and characterization of its biological properties in pigs and mice. Arch. Virol. 2013, 158, 2351–2357. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Pascua, P.N.; Song, M.S.; Baek, Y.H.; Kim, C.J.; Choi, H.W.; Sung, M.H.; Webby, R.J.; Webster, R.G.; Poo, H.; et al. Isolation and genetic characterization of H5N2 influenza viruses from pigs in Korea. J. Virol. 2009, 83, 4205–4215. [Google Scholar] [CrossRef] [PubMed]
- Song, D.S.; Lee, C.S.; Jung, K.; Kang, B.K.; Oh, J.S.; Yoon, Y.D.; Lee, J.H.; Park, B.K. Isolation and phylogenetic analysis of H1N1 swine influenza virus isolated in Korea. Virus Res. 2007, 125, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Kang, B.K.; Kim, H.K.; Park, S.J.; Park, B.K.; Jung, K.; Song, D.S. Phylogenetic analysis of swine influenza viruses recently isolated in Korea. Virus Genes 2008, 37, 168–176. [Google Scholar] [CrossRef]
- Shin, J.Y.; Song, M.S.; Lee, E.H.; Lee, Y.M.; Kim, S.Y.; Kim, H.K.; Choi, J.K.; Kim, C.J.; Webby, R.J.; Choi, Y.K. Isolation and characterization of novel H3N1 swine influenza viruses from pigs with respiratory diseases in Korea. J. Clin. Microbiol. 2006, 44, 3923–3927. [Google Scholar] [CrossRef]
- Song, M.S.; Lee, J.H.; Pascua, P.N.; Baek, Y.H.; Kwon, H.I.; Park, K.J.; Choi, H.W.; Shin, Y.K.; Song, J.Y.; Kim, C.J.; et al. Evidence of human-to-swine transmission of the pandemic (H1N1) 2009 influenza virus in South Korea. J. Clin. Microbiol. 2010, 48, 3204–3211. [Google Scholar] [CrossRef]
- Han, J.Y.; Park, S.J.; Kim, H.K.; Rho, S.; Nguyen, G.V.; Song, D.; Kang, B.K.; Moon, H.J.; Yeom, M.J.; Park, B.K. Identification of reassortant pandemic H1N1 influenza virus in Korean pigs. J. Microbiol. Biotechnol. 2012, 22, 699–707. [Google Scholar] [CrossRef]
- Kim, J.I.; Lee, I.; Park, S.; Lee, S.; Hwang, M.W.; Bae, J.Y.; Heo, J.; Kim, D.; Jang, S.I.; Kim, K.; et al. Phylogenetic analysis of a swine influenza A(H3N2) virus isolated in Korea in 2012. PLoS ONE 2014, 9, e88782. [Google Scholar] [CrossRef]
- Cho, Y.Y.; Lim, S.I.; Jeoung, H.Y.; Kim, Y.K.; Song, J.Y.; Lee, J.B.; An, D.J. Serological evidence for influenza virus infection in Korean wild boars. J. Vet. Med. Sci. 2015, 77, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Ouh, I.O.; Cho, S.D.; Cho, I.S.; Park, C.K.; Song, J.Y. Complete Genome Sequence of H1N1 Swine Influenza Virus from Pigs in the Republic of Korea in 2016. Microbiol. Resour. Announc. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Kanai, C.; Suwicha, K.; Nadhirat, S.; Nerome, K.; Nakayama, M.; Oya, A. Isolation and serological characterization of influenza A virus from a pig in Thailand. Jpnj. Med. Sci. Biol. 1981, 34, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Kupradinun, S.; Peanpijit, P.; Bhodhikosoom, C.; Yoshioka, Y.; Endo, A.; Nerome, K. The first isolation of swine H1N1 influenza viruses from pigs in Thailand. Arch. Virol. 1991, 118, 289–297. [Google Scholar] [CrossRef]
- Takemae, N.; Parchariyanon, S.; Damrongwatanapokin, S.; Uchida, Y.; Ruttanapumma, R.; Watanabe, C.; Yamaguchi, S.; Saito, T. Genetic diversity of swine influenza viruses isolated from pigs during 2000 to 2005 in Thailand. Influenza Other Respir. Viruses 2008, 2, 181–189. [Google Scholar] [CrossRef]
- Thippamom, N.; Sreta, D.; Kitikoon, P.; Thanawongnuwech, R.; Poovorawan, Y.; Theamboonlers, A.; Suwannakarn, K.; Parchariyanon, S.; Damrongwatanapokin, S.; Amonsin, A. Genetic variations of nucleoprotein gene of influenza A viruses isolated from swine in Thailand. Virol. J. 2010, 7, 185. [Google Scholar] [CrossRef]
- Chutinimitkul, S.; Thippamom, N.; Damrongwatanapokin, S.; Payungporn, S.; Thanawongnuwech, R.; Amonsin, A.; Boonsuk, P.; Sreta, D.; Bunpong, N.; Tantilertcharoen, R.; et al. Genetic characterization of H1N1, H1N2 and H3N2 swine influenza virus in Thailand. Arch. Virol. 2008, 153, 1049–1056. [Google Scholar] [CrossRef]
- Hiromoto, Y.; Parchariyanon, S.; Ketusing, N.; Netrabukkana, P.; Hayashi, T.; Kobayashi, T.; Takemae, N.; Saito, T. Isolation of the pandemic (H1N1) 2009 virus and its reassortant with an H3N2 swine influenza virus from healthy weaning pigs in Thailand in 2011. Virus. Res. 2012, 169, 175–181. [Google Scholar] [CrossRef]
- Mine, J.; Abe, H.; Parchariyanon, S.; Boonpornprasert, P.; Ubonyaem, N.; Nuansrichay, B.; Takemae, N.; Tanikawa, T.; Tsunekuni, R.; Uchida, Y.; et al. Genetic and antigenic dynamics of influenza A viruses of swine on pig farms in Thailand. Arch. Virol. 2019, 164, 457–472. [Google Scholar] [CrossRef]
- Nonthabenjawan, N.; Chanvatik, S.; Chaiyawong, S.; Jairak, W.; Boonyapisusopha, S.; Tuanudom, R.; Thontiravong, A.; Bunpapong, N.; Amonsin, A. Genetic diversity of swine influenza viruses in Thai swine farms, 2011-2014. Virus Genes 2015, 50, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Charoenvisal, N.; Keawcharoen, J.; Sreta, D.; Chaiyawong, S.; Nonthabenjawan, N.; Tantawet, S.; Jittimanee, S.; Arunorat, J.; Amonsin, A.; Thanawongnuwech, R. Genetic characterization of Thai swine influenza viruses after the introduction of pandemic H1N1 2009. Virus Genes 2013, 47, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Nguyen, T.D.; Ozaki, H.; Webby, R.J.; Puthavathana, P.; Buranathal, C.; Chaisingh, A.; Auewarakul, P.; Hanh, N.T.; Ma, S.K.; et al. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J. Virol. 2005, 79, 10821–10825. [Google Scholar] [CrossRef] [PubMed]
- Takemae, N.; Parchariyanon, S.; Ruttanapumma, R.; Hiromoto, Y.; Hayashi, T.; Uchida, Y.; Saito, T. Swine influenza virus infection in different age groups of pigs in farrow-to-finish farms in Thailand. Virol. J. 2011, 8, 537. [Google Scholar] [CrossRef]
- Loeffen, W.L.; Nodelijk, G.; Heinen, P.P.; van Leengoed, L.A.; Hunneman, W.A.; Verheijden, J.H. Estimating the incidence of influenza-virus infections in Dutch weaned piglets using blood samples from a cross-sectional study. Vet. Microbiol 2003, 91, 295–308. [Google Scholar] [CrossRef]
- Kitikoon, P.; Sreta, D.; Tuanudom, R.; Amonsin, A.; Suradhat, S.; Oraveerakul, K.; Poovorawan, Y.; Thanawongnuwech, R. Serological evidence of pig-to-human influenza virus transmission on Thai swine farms. Vet. Microbiol. 2011, 148, 413–418. [Google Scholar] [CrossRef]
- Sreta, D.; Tantawet, S.; Na Ayudhya, S.N.; Thontiravong, A.; Wongphatcharachai, M.; Lapkuntod, J.; Bunpapong, N.; Tuanudom, R.; Suradhat, S.; Vimolket, L.; et al. Pandemic (H1N1) 2009 virus on commercial swine farm, Thailand. Emerg. Infect. Dis. 2010, 16, 1587–1590. [Google Scholar] [CrossRef]
- Kitikoon, P.; Sreta, D.; Na Ayudhya, S.N.; Wongphatcharachai, M.; Lapkuntod, J.; Prakairungnamthip, D.; Bunpapong, N.; Suradhat, S.; Thanawongnuwech, R.; Amonsin, A. Brief report: Molecular characterization of a novel reassorted pandemic H1N1 2009 in Thai pigs. Virus Genes 2011, 43, 1–5. [Google Scholar] [CrossRef]
- Abe, H.; Mine, J.; Parchariyanon, S.; Takemae, N.; Boonpornprasert, P.; Ubonyaem, N.; Patcharasinghawut, P.; Nuansrichay, B.; Tanikawa, T.; Tsunekuni, R.; et al. Co-infection of influenza A viruses of swine contributes to effective shuffling of gene segments in a naturally reared pig. Virology 2015, 484, 203–212. [Google Scholar] [CrossRef]
- Trevennec, K.; Leger, L.; Lyazrhi, F.; Baudon, E.; Cheung, C.Y.; Roger, F.; Peiris, M.; Garcia, J.M. Transmission of pandemic influenza H1N1 (2009) in Vietnamese swine in 2009-2010. Influenza Other Respir Viruses 2012, 6, 348–357. [Google Scholar] [CrossRef]
- Ngo, L.T.; Hiromoto, Y.; Pham, V.P.; Le, H.T.; Nguyen, H.T.; Le, V.T.; Takemae, N.; Saito, T. Isolation of novel triple-reassortant swine H3N2 influenza viruses possessing the hemagglutinin and neuraminidase genes of a seasonal influenza virus in Vietnam in 2010. Influenza Other Respir Viruses 2012, 6, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Takemae, N.; Shobugawa, Y.; Nguyen, P.T.; Nguyen, T.; Nguyen, T.N.; To, T.L.; Thai, P.D.; Nguyen, T.D.; Nguyen, D.T.; Nguyen, D.K.; et al. Effect of herd size on subclinical infection of swine in Vietnam with influenza A viruses. Bmc Vet. Res. 2016, 12, 227. [Google Scholar] [CrossRef] [PubMed]
- Baudon, E.; Poon, L.L.; Dao, T.D.; Pham, N.T.; Cowling, B.J.; Peyre, M.; Nguyen, K.V.; Peiris, M. Detection of Novel Reassortant Influenza A (H3N2) and H1N1 2009 Pandemic Viruses in Swine in Hanoi, Vietnam. Zoonoses Public Health 2015, 62, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Mukherjee, K.K.; Mondal, M.C.; Chakravarti, S.K.; Chakraborty, M.S. A serological survey of influenza a antibody in human and pig sera in Calcutta. Folia. Microbiol. (Praha) 1995, 40, 345–348. [Google Scholar] [CrossRef]
- Nagarajan, K.; Saikumar, G.; Arya, R.S.; Gupta, A.; Somvanshi, R.; Pattnaik, B. Influenza A H1N1 virus in Indian pigs & its genetic relatedness with pandemic human influenza A 2009 H1N1. Indianj. Med. Res. 2010, 132, 160–167. [Google Scholar]
- Barbour, E.K.; Sagherian, V.K.; Sagherian, N.K.; Dankar, S.K.; Jaber, L.S.; Usayran, N.N.; Farran, M.T. Avian influenza outbreak in poultry in the Lebanon and transmission to neighbouring farmers and swine. Vet. Ital. 2006, 42, 77–85. [Google Scholar]
- Suriya, R.; Hassan, L.; Omar, A.R.; Aini, I.; Tan, C.G.; Lim, Y.S.; Kamaruddin, M.I. Seroprevalence and risk factors for influenza A viruses in pigs in Peninsular Malaysia. Zoonoses Public Health 2008, 55, 342–351. [Google Scholar] [CrossRef]
- Conlan, J.V.; Vongxay, K.; Jarman, R.G.; Gibbons, R.V.; Lunt, R.A.; Fenwick, S.; Thompson, R.C.A.; Blacksell, S.D. Serologic study of pig-associated viral zoonoses in Laos. Am. J. Trop Med. Hyg. 2012, 86, 1077–1084. [Google Scholar] [CrossRef]
- Sobolev, I.; Kurskaya, O.; Murashkina, T.; Leonov, S.; Sharshov, K.; Kabilov, M.; Alikina, T.; Tolstykh, N.; Gorodov, V.; Alekseev, A.; et al. Genome Sequence of an Unusual Reassortant H1N1 Swine Influenza Virus Isolated from a Pig in Russia, 2016. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Kundin, W.D. Hong Kong A-2 influenza virus infection among swine during a human epidemic in Taiwan. Nature 1970, 228, 857. [Google Scholar] [CrossRef]
- Tsai, C.P.; Tsai, H.J. Influenza B viruses in pigs, Taiwan. Influenza Other Respir. Viruses 2019, 13, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Nidom, C.A.; Takano, R.; Yamada, S.; Sakai-Tagawa, Y.; Daulay, S.; Aswadi, D.; Suzuki, T.; Suzuki, Y.; Shinya, K.; Iwatsuki-Horimoto, K.; et al. Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Infect. Dis 2010, 16, 1515–1523. [Google Scholar] [CrossRef] [PubMed]
- Perera, H.K.K.; Wickramasinghe, G.; Cheung, C.L.; Nishiura, H.; Smith, D.K.; Poon, L.L.M.; Perera, A.K.C.; Ma, S.K.; Sunil-Chandra, N.P.; Guan, Y.; et al. Swine influenza in Sri Lanka. Emerg. Infect. Dis. 2013, 19, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Klivleyeva, N.G.; Ongarbayeva, N.S.; Saktaganov, N.T.; Glebova, T.I.; Lukmanova, G.V.; Shamenova, M.G.; Sayatov, M.K.; Berezin, V.E.; Nusupbaeva, G.E.; Aikimbayev, A.M.; et al. Circulation of influenza viruses among humans and swine in the territory of Kazakhstan during 2017- 2018. Bull. Natl. Acad. Sci. Repub. Kazakhstan 2019, 2, 6–13. [Google Scholar] [CrossRef]
- Holyoake, P.K.; Kirkland, P.D.; Davis, R.J.; Arzey, K.E.; Watson, J.; Lunt, R.A.; Wang, J.; Wong, F.; Moloney, B.J.; Dunn, S.E. The first identified case of pandemic H1N1 influenza in pigs in Australia. Aust. Vet. J. 2011, 89, 427–431. [Google Scholar] [CrossRef]
- Deng, Y.M.; Iannello, P.; Smith, I.; Watson, J.; Barr, I.G.; Daniels, P.; Komadina, N.; Harrower, B.; Wong, F.Y. Transmission of influenza A(H1N1) 2009 pandemic viruses in Australian swine. Influenza Other Respir Viruses 2012, 6, e42–e47. [Google Scholar] [CrossRef]
- Smith, D.W.; Barr, I.G.; Loh, R.; Levy, A.; Tempone, S.; O’Dea, M.; Watson, J.; Wong, F.Y.K.; Effler, P.V. Respiratory Illness in a Piggery Associated with the First Identified Outbreak of Swine Influenza in Australia: Assessing the Risk to Human Health and Zoonotic Potential. Trop. Med. Infect. Dis. 2019, 4, 96. [Google Scholar] [CrossRef]
- Wong, F.Y.K.; Donato, C.; Deng, Y.M.; Teng, D.; Komadina, N.; Baas, C.; Modak, J.; O’Dea, M.; Smith, D.W.; Effler, P.V.; et al. Divergent Human-Origin Influenza Viruses Detected in Australian Swine Populations. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Ottis, K.; Bachmann, P.A. Occurrence of Hsw 1 N 1 subtype influenza A viruses in wild ducks in Europe. Arch. Virol. 1980, 63, 185–190. [Google Scholar] [CrossRef]
- Pensaert, M.; Ottis, K.; Vandeputte, J.; Kaplan, M.M.; Bachmann, P.A. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull. World Health Organ. 1981, 59, 75–78. [Google Scholar]
- Van Reeth, K.; Brown, I.H.; Pensaert, M. Isolations of H1N2 influenza A virus from pigs in Belgium. Vet. Rec. 2000, 146, 588–589. [Google Scholar] [CrossRef] [PubMed]
- Breum, S.O.; Hjulsager, C.K.; Trebbien, R.; Larsen, L.E. Influenza a virus with a human-like n2 gene is circulating in pigs. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [PubMed]
- Ryt-Hansen, P.; Larsen, I.; Kristensen, C.S.; Krog, J.S.; Wacheck, S.; Larsen, L.E. Longitudinal field studies reveal early infection and persistence of influenza A virus in piglets despite the presence of maternally derived antibodies. Vet. Res. 2019, 50, 36. [Google Scholar] [CrossRef] [PubMed]
- Krog, J.S.; Hjulsager, C.K.; Larsen, M.A.; Larsen, L.E. Triple-reassortant influenza A virus with H3 of human seasonal origin, NA of swine origin, and internal A(H1N1) pandemic 2009 genes is established in Danish pigs. Influenza Other Respir. Viruses 2017, 11, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.S.; Lamont, P.H.; Harkness, J.W. Serological evidence of continuing infection of swine in Great Britain with an influenza A virus (H3N2). J. Hyg (Lond) 1978, 80, 415–422. [Google Scholar] [CrossRef]
- Brown, I.H.; Harris, P.A.; Alexander, D.J. Serological studies of influenza viruses in pigs in Great Britain 1991-2. Epidemiol. Infect. 1995, 114, 511–520. [Google Scholar] [CrossRef]
- Brown, I.H.; Hill, M.L.; Harris, P.A.; Alexander, D.J.; McCauley, J.W. Genetic characterisation of an influenza A virus of unusual subtype (H1N7) isolated from pigs in England. Arch. Virol. 1997, 142, 1045–1050. [Google Scholar] [CrossRef]
- Brown, I.H.; Alexander, D.J.; Chakraverty, P.; Harris, P.A.; Manvell, R.J. Isolation of an influenza A virus of unusual subtype (H1N7) from pigs in England, and the subsequent experimental transmission from pig to pig. Vet. Microbiol. 1994, 39, 125–134. [Google Scholar] [CrossRef]
- Welsh, M.D.; Baird, P.M.; Guelbenzu-Gonzalo, M.P.; Hanna, A.; Reid, S.M.; Essen, S.; Russell, C.; Thomas, S.; Barrass, L.; McNeilly, F.; et al. Initial incursion of pandemic (H1N1) 2009 influenza A virus into European pigs. Vet. Rec. 2010, 166, 642–645. [Google Scholar] [CrossRef]
- Williamson, S.M.; Tucker, A.W.; McCrone, I.S.; Bidewell, C.A.; Brons, N.; Habernoll, H.; Essen, S.C.; Brown, I.H.; Wood, J.L. Descriptive clinical and epidemiological characteristics of influenza A H1N1 2009 virus infections in pigs in England. Vet. Rec. 2012, 171, 271. [Google Scholar] [CrossRef]
- Howard, W.A.; Essen, S.C.; Strugnell, B.W.; Russell, C.; Barass, L.; Reid, S.M.; Brown, I.H. Reassortant Pandemic (H1N1) 2009 virus in pigs, United Kingdom. Emerg. Infect. Dis. 2011, 17, 1049–1052. [Google Scholar] [CrossRef] [PubMed]
- Gerber, P.F.; Dawson, L.; Strugnell, B.; Burgess, R.; Brown, H.; Opriessnig, T. Using oral fluids samples for indirect influenza A virus surveillance in farmed UK pigs. Vet. Med. Sci. 2017, 3, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Nokireki, T.; Laine, T.; London, L.; Ikonen, N.; Huovilainen, A. The first detection of influenza in the Finnish pig population: A retrospective study. Acta. Vet. Scand. 2013, 55, 69. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haimi-Hakala, M.; Halli, O.; Laurila, T.; Raunio-Saarnisto, M.; Nokireki, T.; Laine, T.; Nykasenoja, S.; Pelkola, K.; Segales, J.; Sibila, M.; et al. Etiology of acute respiratory disease in fattening pigs in Finland. Porc. Health Manag 2017, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- Aymard, M.; Douglas, A.R.; Fontaine, M.; Gourreau, J.M.; Kaiser, C.; Million, J.; Skehel, J.J. Antigenic characterization of influenza A (H1N1) viruses recently isolated from pigs and turkeys in France. Bull. World Health Organ. 1985, 63, 537–542. [Google Scholar] [PubMed]
- Gourreau, J.M.; Kaiser, C.; Valette, M.; Douglas, A.R.; Labie, J.; Aymard, M. Isolation of two H1N2 influenza viruses from swine in France. Arch. Virol. 1994, 135, 365–382. [Google Scholar] [CrossRef]
- Marozin, S.; Gregory, V.; Cameron, K.; Bennett, M.; Valette, M.; Aymard, M.; Foni, E.; Barigazzi, G.; Lin, Y.; Hay, A. Antigenic and genetic diversity among swine influenza A H1N1 and H1N2 viruses in Europe. J. Gen. Virol. 2002, 83, 735–745. [Google Scholar] [CrossRef]
- Vittecoq, M.; Grandhomme, V.; Simon, G.; Herve, S.; Blanchon, T.; Renaud, F.; Thomas, F.; Gauthier-Clerc, M.; van der Werf, S. Study of influenza A virus in wild boars living in a major duck wintering site. Infect. Genet. Evol. 2012, 12, 483–486. [Google Scholar] [CrossRef]
- Chastagner, A.; Enouf, V.; Peroz, D.; Herve, S.; Lucas, P.; Queguiner, S.; Gorin, S.; Beven, V.; Behillil, S.; Leneveu, P.; et al. Bidirectional Human-Swine Transmission of Seasonal Influenza A(H1N1)pdm09 Virus in Pig Herd, France, 2018. Emerg. Infect. Dis. 2019, 25, 1940–1943. [Google Scholar] [CrossRef]
- Kaden, V.; Lange, E.; Starick, E.; Bruer, W.; Krakowski, W.; Klopries, M. Epidemiological survey of swine influenza A virus in selected wild boar populations in Germany. Vet. Microbiol. 2008, 131, 123–132. [Google Scholar] [CrossRef]
- Starick, E.; Lange, E.; Fereidouni, S.; Bunzenthal, C.; Hoveler, R.; Kuczka, A.; grosse Beilage, E.; Hamann, H.P.; Klingelhofer, I.; Steinhauer, D.; et al. Reassorted pandemic (H1N1) 2009 influenza A virus discovered from pigs in Germany. J. Gen. Virol. 2011, 92, 1184–1188. [Google Scholar] [CrossRef] [PubMed]
- Lange, J.; Groth, M.; Schlegel, M.; Krumbholz, A.; Wieczorek, K.; Ulrich, R.; Koppen, S.; Schulz, K.; Appl, D.; Selbitz, H.J.; et al. Reassortants of the pandemic (H1N1) 2009 virus and establishment of a novel porcine H1N2 influenza virus, lineage in Germany. Vet. Microbiol. 2013, 167, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Pippig, J.; Ritzmann, M.; Buttner, M.; Neubauer-Juric, A. Influenza A Viruses Detected in Swine in Southern Germany after the H1N1 Pandemic in 2009. Zoonoses Public Health 2016, 63, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Kyriakis, C.S.; Papatsiros, V.G.; Athanasiou, L.V.; Valiakos, G.; Brown, I.H.; Simon, G.; Van Reeth, K.; Tsiodras, S.; Spyrou, V.; Billinis, C. Serological Evidence of Pandemic H1N1 Influenza Virus Infections in Greek Swine. Zoonoses Public Health 2016, 63, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Ottis, K.; Sidoli, L.; Bachmann, P.A.; Webster, R.G.; Kaplan, M.M. Human influenza A viruses in pigs: Isolation of a H3N2 strain antigenically related to A/England/42/72 and evidence for continuous circulation of human viruses in the pig population. Arch. Virol. 1982, 73, 103–108. [Google Scholar] [CrossRef]
- Donatelli, I.; Campitelli, L.; Castrucci, M.R.; Ruggieri, A.; Sidoli, L.; Oxford, J.S. Detection of two antigenic subpopulations of A(H1N1) influenza viruses from pigs: Antigenic drift or interspecies transmission? J. Med. Virol. 1991, 34, 248–257. [Google Scholar] [CrossRef]
- Mancini, G.; Donatelli, I.; Rozera, C.; Arangio Ruiz, G.; Butto, S. Antigenic and biochemical analysis of influenza “A” H3N2 viruses isolated from pigs. Arch. Virol 1985, 83, 157–167. [Google Scholar] [CrossRef]
- Campitelli, L.; Donatelli, I.; Foni, E.; Castrucci, M.R.; Fabiani, C.; Kawaoka, Y.; Krauss, S.; Webster, R.G. Continued evolution of H1N1 and H3N2 influenza viruses in pigs in Italy. Virology 1997, 232, 310–318. [Google Scholar] [CrossRef]
- Foti, M.; Bottari, T.; Daidone, A.; Rinaldo, D.; De Leo, F.; Foti, S.; Giacopello, C. Serological survey on Aujeszky’s disease, swine influenza and porcine reproductive and respiratory syndrome virus infections in Italian pigs. Polj. Vet. Sci. 2008, 11, 323–325. [Google Scholar]
- Moreno, A.; Di Trani, L.; Alborali, L.; Vaccari, G.; Barbieri, I.; Falcone, E.; Sozzi, E.; Puzelli, S.; Ferri, G.; Cordioli, P. First Pandemic H1N1 Outbreak from a Pig Farm in Italy. Open Virolj. 2010, 4, 52–56. [Google Scholar] [CrossRef]
- Guercio, A.; Purpari, G.; Conaldi, P.G.; Pagano, V.; Moreno, A.; Giambruno, P.; Di Trani, L.; Vaccari, G.; Falcone, E.; Istituto, A.B.; et al. Pandemic influenza A/H1N1 virus in a swine farm house in Sicily, Italy. J. Environ. Biol 2012, 33, 155–157. [Google Scholar] [PubMed]
- Boni, A.; Vaccari, G.; Di Trani, L.; Zaccaria, G.; Alborali, G.L.; Lelli, D.; Cordioli, P.; Moreno, A.M. Genetic characterization and evolution of H1N1pdm09 after circulation in a swine farm. Biomed. Res. Int. 2014, 2014, 598732. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moreno, A.; Di Trani, L.; Faccini, S.; Vaccari, G.; Nigrelli, D.; Boniotti, M.B.; Falcone, E.; Boni, A.; Chiapponi, C.; Sozzi, E.; et al. Novel H1N2 swine influenza reassortant strain in pigs derived from the pandemic H1N1/2009 virus. Vet. Microbiol. 2011, 149, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Beato, M.S.; Tassoni, L.; Milani, A.; Salviato, A.; Di Martino, G.; Mion, M.; Bonfanti, L.; Monne, I.; Watson, S.J.; Fusaro, A. Circulation of multiple genotypes of H1N2 viruses in a swine farm in Italy over a two-month period. Vet. Microbiol. 2016, 195, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Delogu, M.; Cotti, C.; Vaccari, G.; Raffini, E.; Frasnelli, M.; Nicoloso, S.; Biacchessi, V.; Boni, A.; Foni, E.; Castrucci, M.R.; et al. Serologic and Virologic Evidence of Influenza A Viruses in Wild Boars ( Sus scrofa) from Two Different Locations in Italy. J. Wildl. Dis. 2019, 55, 158–163. [Google Scholar] [CrossRef]
- Foni, E.; Garbarino, C.; Chiapponi, C.; Baioni, L.; Zanni, I.; Cordioli, P. Epidemiological survey of swine influenza A virus in the wild boar population of two Italian provinces. Influenza Other Respir. Viruses 2013, 7 (Suppl. 4), 16–20. [Google Scholar] [CrossRef]
- Chiapponi, C.; Faccini, S.; De Mattia, A.; Baioni, L.; Barbieri, I.; Rosignoli, C.; Nigrelli, A.; Foni, E. Detection of Influenza D Virus among Swine and Cattle, Italy. Emerg. Infect. Dis. 2016, 22, 352–354. [Google Scholar] [CrossRef]
- Maldonado, J.; Van Reeth, K.; Riera, P.; Sitja, M.; Saubi, N.; Espuna, E.; Artigas, C. Evidence of the concurrent circulation of H1N2, H1N1 and H3N2 influenza A viruses in densely populated pig areas in Spain. Vet. J. 2006, 172, 377–381. [Google Scholar] [CrossRef]
- Baratelli, M.; Cordoba, L.; Perez, L.J.; Maldonado, J.; Fraile, L.; Nunez, J.I.; Montoya, M. Genetic characterization of influenza A viruses circulating in pigs and isolated in north-east Spain during the period 2006-2007. Res. Vet. Sci. 2014, 96, 380–388. [Google Scholar] [CrossRef]
- Martin-Valls, G.E.; Simon-Grife, M.; van Boheemen, S.; de Graaf, M.; Bestebroer, T.M.; Busquets, N.; Martin, M.; Casal, J.; Fouchier, R.A.; Mateu, E. Phylogeny of Spanish swine influenza viruses isolated from respiratory disease outbreaks and evolution of swine influenza virus within an endemically infected farm. Vet. Microbiol. 2014, 170, 266–277. [Google Scholar] [CrossRef]
- Simon-Grife, M.; Martin-Valls, G.E.; Vilar, M.J.; Busquets, N.; Mora-Salvatierra, M.; Bestebroer, T.M.; Fouchier, R.A.; Martin, M.; Mateu, E.; Casal, J. Swine influenza virus infection dynamics in two pig farms; results of a longitudinal assessment. Vet. Res. 2012, 43, 24. [Google Scholar] [CrossRef] [PubMed]
- Loeffen, W.L.; Hunneman, W.A.; Quak, J.; Verheijden, J.H.; Stegeman, J.A. Population dynamics of swine influenza virus in farrow-to-finish and specialised finishing herds in the Netherlands. Vet. Microbiol 2009, 137, 45–50. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hofshagen, M.; Gjerset, B.; Er, C.; Tarpai, A.; Brun, E.; Dannevig, B.; Bruheim, T.; Fostad, I.G.; Iversen, B.; Hungnes, O.; et al. Pandemic influenza A(H1N1)v: Human to pig transmission in Norway? Euro Surveill 2009, 14. [Google Scholar] [CrossRef]
- Grontvedt, C.A.; Er, C.; Gjerset, B.; Germundsson, A.; Framstad, T.; Brun, E.; Jorgensen, A.; Lium, B. Clinical Impact of Infection with Pandemic Influenza (H1N1) 2009 Virus in Naive Nucleus and Multiplier Pig Herds in Norway. Influenza Res. Treat. 2011, 2011, 163745. [Google Scholar] [CrossRef] [PubMed]
- Gjerset, B.; Er, C.; Lotvedt, S.; Jorgensen, A.; Hungnes, O.; Lium, B.; Germundsson, A. Experiences after Twenty Months with Pandemic Influenza A (H1N1) 2009 Infection in the Naive Norwegian Pig Population. Influenza Res. Treat. 2011, 2011, 206975. [Google Scholar] [CrossRef]
- Er, C.; Lium, B.; Tavornpanich, S.; Hofmo, P.O.; Forberg, H.; Hauge, A.G.; Grontvedt, C.A.; Framstad, T.; Brun, E. Adverse effects of Influenza A(H1N1)pdm09 virus infection on growth performance of Norwegian pigs - a longitudinal study at a boar testing station. Bmc Vet. Res. 2014, 10, 284. [Google Scholar] [CrossRef]
- Biernacka, K.; Karbowiak, P.; Wrobel, P.; Chareza, T.; Czopowicz, M.; Balka, G.; Goodell, C.; Rauh, R.; Stadejek, T. Detection of porcine reproductive and respiratory syndrome virus (PRRSV) and influenza A virus (IAV) in oral fluid of pigs. Res. Vet. Sci. 2016, 109, 74–80. [Google Scholar] [CrossRef]
- Lepek, K.; Pajak, B.; Rabalski, L.; Urbaniak, K.; Kucharczyk, K.; Markowska-Daniel, I.; Szewczyk, B. Analysis of Coinfections with A/H1N1 Strain Variants among Pigs in Poland by Multitemperature Single-Strand Conformational Polymorphism. Biomed. Res. Int. 2015, 2015, 535908. [Google Scholar] [CrossRef]
- Czyzewska-Dors, E.; Dors, A.; Kwit, K.; Pejsak, Z.; Pomorska-Mol, M. Serological Survey of the Influenza a Virus in Polish Farrow-to-finish Pig Herds in 2011-2015. J. Vet. Res. 2017, 61, 157–161. [Google Scholar] [CrossRef]
- Tumova, B.; Mensik, J.; Stumpa, A.; Fedova, D.; Pospisil, Z. Serological evidence and isolation of a virus closely related to the human A/Hong Kong/68 (H3N2) strain in swine populations in Czechoslovakia in 1969–1972. Zent. Vet. B 1976, 23, 590–603. [Google Scholar] [CrossRef]
- Banyai, K.; Kovacs, E.; Toth, A.; Biksi, I.; Szentpali-Gavaller, K.; Balint, A.; Dencso, L.; Dan, A. Genome sequence of a monoreassortant H1N1 swine influenza virus isolated from a pig in Hungary. J. Virol. 2012, 86, 13133. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van Reeth, K.; Brown, I.H.; Durrwald, R.; Foni, E.; Labarque, G.; Lenihan, P.; Maldonado, J.; Markowska-Daniel, I.; Pensaert, M.; Pospisil, Z.; et al. Seroprevalence of H1N1, H3N2 and H1N2 influenza viruses in pigs in seven European countries in 2002-2003. Influenza Other Respir. Viruses 2008, 2, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Kyriakis, C.S.; Brown, I.H.; Foni, E.; Kuntz-Simon, G.; Maldonado, J.; Madec, F.; Essen, S.C.; Chiapponi, C.; Van Reeth, K. Virological surveillance and preliminary antigenic characterization of influenza viruses in pigs in five European countries from 2006 to 2008. Zoonoses Public Health 2011, 58, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Van Dreumel, A.A.; Ditchfield, W.J. Case report. An outbreak of swine influenza in Manitoba. Can. Vet. J. 1968, 9, 275–278. [Google Scholar] [PubMed]
- Arora, D.J.; N’Diaye, M.; Dea, S. Genomic study of hemagglutinins of swine influenza (H1N1) viruses associated with acute and chronic respiratory diseases in pigs. Arch. Virol. 1997, 142, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Sanford, S.E.; Josephson, G.K.; Key, D.W. An epizootic of Swine influenza in ontario. Can. Vet. J. 1983, 24, 167–171. [Google Scholar] [PubMed]
- Bikour, M.H.; Frost, E.H.; Deslandes, S.; Talbot, B.; Elazhary, Y. Persistence of a 1930 swine influenza A (H1N1) virus in Quebec. J. Gen. Virol. 1995, 76 Pt 10, 2539–2547. [Google Scholar] [CrossRef]
- Bikour, M.H.; Cornaglia, E.; Weber, J.M.; Elazhary, Y. Antigenic characterization of an H3N2 swine influenza virus isolated from pigs with proliferative and necrotizing pneumonia in Quebec. Can. J. Vet. Res. 1994, 58, 287–290. [Google Scholar]
- Carman, S.; Stansfield, C.; Weber, J.; Bildfell, R.; Van Dreumel, T. H3N2 influenza A virus recovered from a neonatal pig in Ontario--1997. Can. Vet. J. 1999, 40, 889–890. [Google Scholar]
- Ferreira, J.B.; Grgic, H.; Friendship, R.; Nagy, E.; Poljak, Z. Influence of microclimate conditions on the cumulative exposure of nursery pigs to swine influenza A viruses. Transbound Emerg. Dis. 2018, 65, e145–e154. [Google Scholar] [CrossRef]
- Grgic, H.; Costa, M.; Friendship, R.M.; Carman, S.; Nagy, E.; Wideman, G.; Weese, S.; Poljak, Z. Molecular characterization of H3N2 influenza A viruses isolated from Ontario swine in 2011 and 2012. Virolj. 2014, 11, 194. [Google Scholar] [CrossRef] [PubMed]
- Grgic, H.; Costa, M.; Friendship, R.M.; Carman, S.; Nagy, E.; Poljak, Z. Genetic Characterization of H1N1 and H1N2 Influenza A Viruses Circulating in Ontario Pigs in 2012. PLoS ONE 2015, 10, e0127840. [Google Scholar] [CrossRef]
- Nelson, M.I.; Culhane, M.R.; Trovao, N.S.; Patnayak, D.P.; Halpin, R.A.; Lin, X.; Shilts, M.H.; Das, S.R.; Detmer, S.E. The emergence and evolution of influenza A (H1alpha) viruses in swine in Canada and the United States. J. Gen. Virol. 2017, 98, 2663–2675. [Google Scholar] [CrossRef] [PubMed]
- Dea, S.; Bilodeau, R.; Sauvageau, R.; Montpetit, C.; Martineau, G.P. Antigenic variant of swine influenza virus causing proliferative and necrotizing pneumonia in pigs. J. Vet. Diagn. Invest. 1992, 4, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Larochelle, R.; Sauvageau, R.; Magar, R. Immunohistochemical detection of swine influenza virus and porcine reproductive and respiratory syndrome virus in porcine proliferative and necrotizing pneumonia cases from Quebec. Can. Vet. J. 1994, 35, 513–515. [Google Scholar] [PubMed]
- Karasin, A.I.; Brown, I.H.; Carman, S.; Olsen, C.W. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J. Virol 2000, 74, 9322–9327. [Google Scholar] [CrossRef]
- Karasin, A.I.; West, K.; Carman, S.; Olsen, C.W. Characterization of avian H3N3 and H1N1 influenza A viruses isolated from pigs in Canada. J. Clin. Microbiol. 2004, 42, 4349–4354. [Google Scholar] [CrossRef]
- Karasin, A.I.; Carman, S.; Olsen, C.W. Identification of human H1N2 and human-swine reassortant H1N2 and H1N1 influenza A viruses among pigs in Ontario, Canada (2003 to 2005). J. Clin. Microbiol. 2006, 44, 1123–1126. [Google Scholar] [CrossRef]
- Olsen, C.W.; Karasin, A.I.; Carman, S.; Li, Y.; Bastien, N.; Ojkic, D.; Alves, D.; Charbonneau, G.; Henning, B.M.; Low, D.E.; et al. Triple reassortant H3N2 influenza A viruses, Canada, 2005. Emerg. Infect. Dis. 2006, 12, 1132–1135. [Google Scholar] [CrossRef]
- Nfon, C.; Berhane, Y.; Zhang, S.; Handel, K.; Labrecque, O.; Pasick, J. Molecular and antigenic characterization of triple-reassortant H3N2 swine influenza viruses isolated from pigs, turkey and quail in Canada. Transbound Emerg. Dis. 2011, 58, 394–401. [Google Scholar] [CrossRef]
- Nfon, C.K.; Berhane, Y.; Hisanaga, T.; Zhang, S.; Handel, K.; Kehler, H.; Labrecque, O.; Lewis, N.S.; Vincent, A.L.; Copps, J.; et al. Characterization of H1N1 swine influenza viruses circulating in Canadian pigs in 2009. J. Virol. 2011, 85, 8667–8679. [Google Scholar] [CrossRef] [PubMed]
- Forgie, S.E.; Keenliside, J.; Wilkinson, C.; Webby, R.; Lu, P.; Sorensen, O.; Fonseca, K.; Barman, S.; Rubrum, A.; Stigger, E.; et al. Swine outbreak of pandemic influenza A virus on a Canadian research farm supports human-to-swine transmission. Clin. Infect. Dis. 2011, 52, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Howden, K.J.; Brockhoff, E.J.; Caya, F.D.; McLeod, L.J.; Lavoie, M.; Ing, J.D.; Bystrom, J.M.; Alexandersen, S.; Pasick, J.M.; Berhane, Y.; et al. An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can. Vet. J. 2009, 50, 1153–1161. [Google Scholar] [PubMed]
- Weingartl, H.M.; Berhane, Y.; Hisanaga, T.; Neufeld, J.; Kehler, H.; Emburry-Hyatt, C.; Hooper-McGreevy, K.; Kasloff, S.; Dalman, B.; Bystrom, J.; et al. Genetic and pathobiologic characterization of pandemic H1N1 2009 influenza viruses from a naturally infected swine herd. J. Virol. 2010, 84, 2245–2256. [Google Scholar] [CrossRef] [PubMed]
- Pasma, T.; Joseph, T. Pandemic (H1N1) 2009 infection in swine herds, Manitoba, Canada. Emerg Infect. Dis 2010, 16, 706–708. [Google Scholar] [CrossRef]
- Lange, E.; Kalthoff, D.; Blohm, U.; Teifke, J.P.; Breithaupt, A.; Maresch, C.; Starick, E.; Fereidouni, S.; Hoffmann, B.; Mettenleiter, T.C.; et al. Pathogenesis and transmission of the novel swine-origin influenza virus A/H1N1 after experimental infection of pigs. J. Gen. Virol. 2009, 90, 2119–2123. [Google Scholar] [CrossRef]
- Tremblay, D.; Allard, V.; Doyon, J.F.; Bellehumeur, C.; Spearman, J.G.; Harel, J.; Gagnon, C.A. Emergence of a new swine H3N2 and pandemic (H1N1) 2009 influenza A virus reassortant in two Canadian animal populations, mink and swine. J. Clin. Microbiol. 2011, 49, 4386–4390. [Google Scholar] [CrossRef]
- Yang, W.; Elankumaran, S.; Marr, L.C. Relationship between humidity and influenza A viability in droplets and implications for influenza’s seasonality. PLoS ONE 2012, 7, e46789. [Google Scholar] [CrossRef]
- Shope, R.E. Serological evidence for the occurrence of infection with human influenza virus in swine. J. Exp. Med. 1938, 67, 739–748. [Google Scholar] [CrossRef]
- Young, G.A., Jr.; Underdahl, N.R. Swine influenza as a possible factor in suckling pig mortalities. 4. Relationship of passive swine infiuenzal immunity in suckling pits to rate of weight gam. Cornell Vet. 1950, 40, 201–205. [Google Scholar]
- Young, G.A.; Underdahl, N.R. An evaluation of influenza in midwestern swine. Am. J. Vet. Res. 1955, 16, 545–552. [Google Scholar] [PubMed]
- Nakamura, R.M.; Easterday, B.C.; Pawlisch, R.; Walker, G.L. Swine Influenza: Epizootiological and Serological Studies; World Health Organization: Geneva, Switzerland, 1972; pp. 481–487. [Google Scholar]
- Hinshaw, V.S.; Bean, W.J.; Webster, R.G.; Easterday, B.C. The prevalence of influenza viruses in swine and the antigenic and genetic relatedness of influenza viruses from man and swine. Virology 1978, 84, 51–62. [Google Scholar] [CrossRef]
- Gaydos, J.C.; Top, F.H., Jr.; Hodder, R.A.; Russell, P.K. Swine influenza a outbreak, Fort Dix, New Jersey, 1976. Emerg Infect. Dis. 2006, 12, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Gaydos, J.C.; Hodder, R.A.; Top, F.H., Jr.; Soden, V.J.; Allen, R.G.; Bartley, J.D.; Zabkar, J.H.; Nowosiwsky, T.; Russell, P.K. Swine influenza A at Fort Dix, New Jersey (January–February 1976). I. Case finding and clinical study of cases. J. Infect. Dis 1977, 136 (Suppl. 3), S356–S362. [Google Scholar] [CrossRef]
- Gaydos, J.C.; Hodder, R.A.; Top, F.H., Jr.; Allen, R.G.; Soden, V.J.; Nowosiwsky, T.; Russell, P.K. Swine influenza A at Fort Dix, New Jersey (January–February 1976). II. Transmission and morbidity in units with cases. J. Infect. Dis. 1977, 136 (Suppl. 3), S363–S368. [Google Scholar] [CrossRef]
- Olsen, C.W.; Carey, S.; Hinshaw, L.; Karasin, A.I. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch. Virol. 2000, 145, 1399–1419. [Google Scholar] [CrossRef]
- Webby, R.J.; Swenson, S.L.; Krauss, S.L.; Gerrish, P.J.; Goyal, S.M.; Webster, R.G. Evolution of swine H3N2 influenza viruses in the United States. J. Virol. 2000, 74, 8243–8251. [Google Scholar] [CrossRef]
- Choi, Y.K.; Goyal, S.M.; Joo, H.S. Prevalence of swine influenza virus subtypes on swine farms in the United States. Arch. Virol. 2002, 147, 1209–1220. [Google Scholar] [CrossRef]
- Choi, Y.K.; Goyal, S.M.; Joo, H.S. Retrospective analysis of etiologic agents associated with respiratory diseases in pigs. Can. Vet. J. 2003, 44, 735–737. [Google Scholar]
- Beaudoin, A.; Johnson, S.; Davies, P.; Bender, J.; Gramer, M. Characterization of influenza a outbreaks in Minnesota swine herds and measures taken to reduce the risk of zoonotic transmission. Zoonoses Public Health 2012, 59, 96–106. [Google Scholar] [CrossRef]
- Karasin, A.I.; Olsen, C.W.; Anderson, G.A. Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana. J. Clin. Microbiol. 2000, 38, 2453–2456. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Gramer, M.; Rossow, K.; Yoon, K.J. Isolation and genetic characterization of new reassortant H3N1 swine influenza virus from pigs in the midwestern United States. J. Virol. 2006, 80, 5092–5096. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Vincent, A.L.; Gramer, M.R.; Brockwell, C.B.; Lager, K.M.; Janke, B.H.; Gauger, P.C.; Patnayak, D.P.; Webby, R.J.; Richt, J.A. Identification of H2N3 influenza A viruses from swine in the United States. Proc. Natl. Acad. Sci. USA 2007, 104, 20949–20954. [Google Scholar] [CrossRef] [PubMed]
- Gray, G.C.; Bender, J.B.; Bridges, C.B.; Daly, R.F.; Krueger, W.S.; Male, M.J.; Heil, G.L.; Friary, J.A.; Derby, R.B.; Cox, N.J. Influenza A(H1N1)pdm09 virus among healthy show pigs, United States. Emerg. Infect. Dis. 2012, 18, 1519–1521. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Mantilla, J.; Alvarez, J.; Culhane, M.; Nirmala, J.; Cano, J.P.; Torremorell, M. Comparison of individual, group and environmental sampling strategies to conduct influenza surveillance in pigs. Bmc Vet. Res. 2019, 15, 61. [Google Scholar] [CrossRef] [PubMed]
- Bose, M.E.; Beck, E.T.; Ledeboer, N.; Kehl, S.C.; Jurgens, L.A.; Patitucci, T.; Witt, L.; LaGue, E.; Darga, P.; He, J.; et al. Rapid semiautomated subtyping of influenza virus species during the 2009 swine origin influenza A H1N1 virus epidemic in Milwaukee, Wisconsin. J. Clin. Microbiol. 2009, 47, 2779–2786. [Google Scholar] [CrossRef]
- Bowman, A.S.; Nolting, J.M.; Nelson, S.W.; Slemons, R.D. Subclinical influenza virus A infections in pigs exhibited at agricultural fairs, Ohio, USA, 2009-2011. Emerg. Infect. Dis. 2012, 18, 1945–1950. [Google Scholar] [CrossRef]
- Ducatez, M.F.; Hause, B.; Stigger-Rosser, E.; Darnell, D.; Corzo, C.; Juleen, K.; Simonson, R.; Brockwell-Staats, C.; Rubrum, A.; Wang, D.; et al. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg. Infect. Dis. 2011, 17, 1624–1629. [Google Scholar] [CrossRef]
- Ali, A.; Khatri, M.; Wang, L.; Saif, Y.M.; Lee, C.W. Identification of swine H1N2/pandemic H1N1 reassortant influenza virus in pigs, United States. Vet. Microbiol. 2012, 158, 60–68. [Google Scholar] [CrossRef]
- Corzo, C.A.; Culhane, M.; Juleen, K.; Stigger-Rosser, E.; Ducatez, M.F.; Webby, R.J.; Lowe, J.F. Active surveillance for influenza A virus among swine, midwestern United States, 2009-2011. Emerg. Infect. Dis. 2013, 19, 954–960. [Google Scholar] [CrossRef]
- Allerson, M.W.; Davies, P.R.; Gramer, M.R.; Torremorell, M. Infection dynamics of pandemic 2009 H1N1 influenza virus in a two-site swine herd. Transbound Emerg. Dis. 2014, 61, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Perez, A.; Sreevatsan, S.; Davies, P.; Culhane, M.; Torremorell, M. Association between Influenza A Virus Infection and Pigs Subpopulations in Endemically Infected Breeding Herds. PLoS ONE 2015, 10, e0129213. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, B.S.; DeBeauchamp, J.; Stigger-Rosser, E.; Franks, J.; Crumpton, J.C.; Turner, J.; Darnell, D.; Jeevan, T.; Kayali, G.; Harding, A.; et al. Influenza Virus Surveillance in Coordinated Swine Production Systems, United States. Emerg. Infect. Dis. 2015, 21, 1834–1836. [Google Scholar] [CrossRef] [PubMed]
- Panyasing, Y.; Goodell, C.; Kittawornrat, A.; Wang, C.; Levis, I.; Desfresne, L.; Rauh, R.; Gauger, P.C.; Zhang, J.; Lin, X.; et al. Influenza A Virus Surveillance Based on Pre-Weaning Piglet Oral Fluid Samples. Transbound Emerg. Dis. 2016, 63, e328–e338. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.L.; Nelson, S.W.; Workman, J.D.; Slemons, R.D.; Szablewski, C.M.; Nolting, J.M.; Bowman, A.S. Utility of snout wipe samples for influenza A virus surveillance in exhibition swine populations. Influenza Other Respir Viruses 2014, 8, 574–579. [Google Scholar] [CrossRef]
- Kyriakis, C.S.; Zhang, M.; Wolf, S.; Jones, L.P.; Shim, B.S.; Chocallo, A.H.; Hanson, J.M.; Jia, M.; Liu, D.; Tripp, R.A. Molecular epidemiology of swine influenza A viruses in the Southeastern United States, highlights regional differences in circulating strains. Vet. Microbiol. 2017, 211, 174–179. [Google Scholar] [CrossRef]
- Bliss, N.; Stull, J.W.; Moeller, S.J.; Rajala-Schultz, P.J.; Bowman, A.S. Movement patterns of exhibition swine and associations of influenza A virus infection with swine management practices. J. Am. Vet. Med. Assoc. 2017, 251, 706–713. [Google Scholar] [CrossRef]
- Bowman, A.S.; Walia, R.R.; Nolting, J.M.; Vincent, A.L.; Killian, M.L.; Zentkovich, M.M.; Lorbach, J.N.; Lauterbach, S.E.; Anderson, T.K.; Davis, C.T.; et al. Influenza A(H3N2) Virus in Swine at Agricultural Fairs and Transmission to Humans, Michigan and Ohio, USA, 2016. Emerg. Infect. Dis. 2017, 23, 1551–1555. [Google Scholar] [CrossRef]
- Clavijo, A.; Nikooienejad, A.; Esfahani, M.S.; Metz, R.P.; Schwartz, S.; Atashpaz-Gargari, E.; Deliberto, T.J.; Lutman, M.W.; Pedersen, K.; Bazan, L.R.; et al. Identification and analysis of the first 2009 pandemic H1N1 influenza virus from U.S. feral swine. Zoonoses Public Health 2013, 60, 327–335. [Google Scholar] [CrossRef]
- Martin, B.E.; Sun, H.; Carrel, M.; Cunningham, F.L.; Baroch, J.A.; Hanson-Dorr, K.C.; Young, S.G.; Schmit, B.; Nolting, J.M.; Yoon, K.J.; et al. Feral Swine in the United States Have Been Exposed to both Avian and Swine Influenza A Viruses. Appl Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Weller, C.B.; Cadmus, K.J.; Ehrhart, E.J.; Powers, B.E.; Pabilonia, K.L. Detection and isolation of Influenza A virus subtype H1N1 from a small backyard swine herd in Colorado. J. Vet. Diagn. Invest. 2013, 25, 782–784. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.S.; Minnis, R.B.; Campbell, T.A.; Barras, S.; Deyoung, R.W.; Pabilonia, K.; Avery, M.L.; Sullivan, H.; Clark, L.; McLean, R.G. Influenza exposure in United States feral swine populations. J. Wildl Dis 2008, 44, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Baroch, J.A.; Long, L.P.; Xu, Y.; Cunningham, F.L.; Pedersen, K.; Lutman, M.W.; Schmit, B.S.; Bowman, A.S.; Deliberto, T.J.; et al. Influenza A subtype H3 viruses in feral swine, United States, 2011–2012. Emerg. Infect. Dis. 2014, 20, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.; Luo, K.; Olivier, A.K.; Cunningham, F.L.; Blackmon, S.; Hanson-Dorr, K.; Sun, H.; Baroch, J.; Lutman, M.W.; Quade, B.; et al. Influenza D Virus Infection in Feral Swine Populations, United States. Emerg. Infect. Dis. 2018, 24, 1020–1028. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.B.; Grgic, H.; Friendship, R.; Wideman, G.; Nagy, E.; Poljak, Z. Longitudinal study of influenza A virus circulation in a nursery swine barn. Vet. Res. 2017, 48, 63. [Google Scholar] [CrossRef]
- Lamb, R.A.; Krung, R.M. Orthomyxoviridae: The Viruses and Their Replication, 4th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2001. [Google Scholar]
- Decorte, I.; Steensels, M.; Lambrecht, B.; Cay, A.B.; De Regge, N. Detection and Isolation of Swine Influenza A Virus in Spiked Oral Fluid and Samples from Individually Housed, Experimentally Infected Pigs: Potential Role of Porcine Oral Fluid in Active Influenza A Virus Surveillance in Swine. PLoS ONE 2015, 10, e0139586. [Google Scholar] [CrossRef] [PubMed]
- Goodell, C.K.; Prickett, J.; Kittawornrat, A.; Johnson, J.; Zhang, J.; Wang, C.; Zimmerman, J.J. Evaluation of Screening Assays for the Detection of Influenza A Virus Serum Antibodies in Swine. Transbound Emerg. Dis. 2016, 63, 24–35. [Google Scholar] [CrossRef]
- Hause, B.M.; Duff, J.W.; Scheidt, A.; Anderson, G. Virus detection using metagenomic sequencing of swine nasal and rectal swabs. J. Swine Health Prod. 2016, 24, 304–308. [Google Scholar]
- Abente, E.J.; Gauger, P.C.; Walia, R.R.; Rajao, D.S.; Zhang, J.; Harmon, K.M.; Killian, M.L.; Vincent, A.L. Detection and characterization of an H4N6 avian-lineage influenza A virus in pigs in the Midwestern United States. Virology 2017, 511, 56–65. [Google Scholar] [CrossRef]
- Chamba Pardo, F.O.; Schelkopf, A.; Allerson, M.; Morrison, R.; Culhane, M.; Perez, A.; Torremorell, M. Breed-to-wean farm factors associated with influenza A virus infection in piglets at weaning. Prev. Vet. Med. 2018, 161, 33–40. [Google Scholar] [CrossRef]
- Zeller, M.A.; Li, G.; Harmon, K.M.; Zhang, J.; Vincent, A.L.; Anderson, T.K.; Gauger, P.C. Complete Genome Sequences of Two Novel Human-Like H3N2 Influenza A Viruses, A/swine/Oklahoma/65980/2017 (H3N2) and A/Swine/Oklahoma/65260/2017 (H3N2), Detected in Swine in the United States. Microbiol. Resour. Announc. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Ayora-Talavera, G.; Cadavieco-Burgos, J.M.; Canul-Armas, A.B. Serologic evidence of human and swine influenza in Mayan persons. Emerg. Infect. Dis. 2005, 11, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Saavedra-Montanez, M.; Carrera-Aguirre, V.; Castillo-Juarez, H.; Rivera-Benitez, F.; Rosas-Estrada, K.; Pulido-Camarillo, E.; Mercado-Garcia, C.; Carreon-Napoles, R.; Haro-Tirado, M.; Rosete, D.P.; et al. Retrospective serological survey of influenza viruses in backyard pigs from Mexico City. Influenza Other Respir. Viruses. 2013, 7, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Robles, G.; Montalvo-Corral, M.; Burgara-Estrella, A.; Hernandez, J. Serological and molecular prevalence of swine influenza virus on farms in northwestern Mexico. Vet. Microbiol. 2014, 172, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Escalera-Zamudio, M.; Cobian-Guemes, G.; de los Dolores Soto-del Rio, M.; Isa, P.; Sanchez-Betancourt, I.; Parissi-Crivelli, A.; Martinez-Cazares, M.T.; Romero, P.; Velazquez-Salinas, L.; Huerta-Lozano, B.; et al. Characterization of an influenza A virus in Mexican swine that is related to the A/H1N1/2009 pandemic clade. Virology 2012, 433, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.I.; Souza, C.K.; Trovao, N.S.; Diaz, A.; Mena, I.; Rovira, A.; Vincent, A.L.; Torremorell, M.; Marthaler, D.; Culhane, M.R. Human-Origin Influenza A(H3N2) Reassortant Viruses in Swine, Southeast Mexico. Emerg. Infect. Dis. 2019, 25, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Betancourt, J.I.; Cervantes-Torres, J.B.; Saavedra-Montanez, M.; Segura-Velazquez, R.A. Complete genome sequence of a novel influenza A H1N2 virus circulating in swine from Central Bajio region, Mexico. Transbound Emerg. Dis. 2017, 64, 2083–2092. [Google Scholar] [CrossRef]
- Gonzalez-Reiche, A.S.; Ramirez, A.L.; Muller, M.L.; Orellana, D.; Sosa, S.M.; Ola, P.; Paniagua, J.; Ortiz, L.; Hernandez, J.; Cordon-Rosales, C.; et al. Origin, distribution, and potential risk factors associated with influenza A virus in swine in two production systems in Guatemala. Influenza Other Respir. Viruses 2017, 11, 182–192. [Google Scholar] [CrossRef]
- Perez, L.J.; Perera, C.L.; Vega, A.; Frias, M.T.; Rouseaux, D.; Ganges, L.; Nunez, J.I.; Diaz de Arce, H. Isolation and complete genomic characterization of pandemic H1N1/2009 influenza viruses from Cuban swine herds. Res. Vet. Sc.I 2013, 94, 781–788. [Google Scholar] [CrossRef]
- Perez, L.J.; Perera, C.L.; Coronado, L.; Rios, L.; Vega, A.; Frias, M.T.; Ganges, L.; Nunez, J.I.; Diaz de Arce, H. Molecular epidemiology study of swine influenza virus revealing a reassorted virus H1N1 in swine farms in Cuba. Prev. Vet. Med. 2015, 119, 172–178. [Google Scholar] [CrossRef]
- Sookhoo, J.R.V.; Brown-Jordan, A.; Blake, L.; Holder, R.B.; Brookes, S.M.; Essen, S.; Carrington, C.V.F.; Brown, I.H.; Oura, C.A.L. Seroprevalence of economically important viral pathogens in swine populations of Trinidad and Tobago, West Indies. Trop. Anim. Health Prod. 2017, 49, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Cappuccio, J.A.; Pena, L.; Dibarbora, M.; Rimondi, A.; Pineyro, P.; Insarralde, L.; Quiroga, M.A.; Machuca, M.; Craig, M.I.; Olivera, V.; et al. Outbreak of swine influenza in Argentina reveals a non-contemporary human H3N2 virus highly transmissible among pigs. J. Gen. Virol. 2011, 92, 2871–2878. [Google Scholar] [CrossRef] [PubMed]
- Pereda, A.; Cappuccio, J.; Quiroga, M.A.; Baumeister, E.; Insarralde, L.; Ibar, M.; Sanguinetti, R.; Cannilla, M.L.; Franzese, D.; Escobar Cabrera, O.E.; et al. Pandemic (H1N1) 2009 outbreak on pig farm, Argentina. Emerg. Infect. Dis. 2010, 16, 304–307. [Google Scholar] [CrossRef] [PubMed]
- Pereda, A.; Rimondi, A.; Cappuccio, J.; Sanguinetti, R.; Angel, M.; Ye, J.; Sutton, T.; Dibarbora, M.; Olivera, V.; Craig, M.I.; et al. Evidence of reassortment of pandemic H1N1 influenza virus in swine in Argentina: Are we facing the expansion of potential epicenters of influenza emergence? Influenza Other Respir. Viruses 2011, 5, 409–412. [Google Scholar] [CrossRef]
- Cappuccio, J.; Dibarbora, M.; Lozada, I.; Quiroga, A.; Olivera, V.; Dangelo, M.; Perez, E.; Barrales, H.; Perfumo, C.; Pereda, A.; et al. Two years of surveillance of influenza a virus infection in a swine herd. Results of virological, serological and pathological studies. Comp. Immunol. Microbiol. Infect. Dis. 2017, 50, 110–115. [Google Scholar] [CrossRef]
- Dibarbora, M.; Cappuccio, J.; Olivera, V.; Quiroga, M.; Machuca, M.; Perfumo, C.; Perez, D.; Pereda, A. Swine influenza: Clinical, serological, pathological, and virological cross-sectional studies in nine farms in Argentina. Influenza Other Respir. Viruses 2013, 7 (Suppl. 4), 10–15. [Google Scholar] [CrossRef]
- Rajao, D.S.; Alves, F.; Del Puerto, H.L.; Braz, G.F.; Oliveira, F.G.; Ciacci-Zanella, J.R.; Schaefer, R.; dos Reis, J.K.; Guedes, R.M.; Lobato, Z.I.; et al. Serological evidence of swine influenza in Brazil. Influenza Other Respir. Viruses 2013, 7, 109–112. [Google Scholar] [CrossRef]
- Rajao, D.S.; Couto, D.H.; Gasparini, M.R.; Costa, A.T.; Reis, J.K.; Lobato, Z.I.; Guedes, R.M.; Leite, R.C. Diagnosis and clinic-pathological findings of influenza virus infection in Brazilian pigs. Pesqui. Veterinária Bras. 2013, 33, 30–36. [Google Scholar] [CrossRef]
- Amorim, A.R.; Fornells, L.A.; Reis Fda, C.; Rezende, D.J.; Mendes Gda, S.; Couceiro, J.N.; Santos, N.S. Influenza A virus infection of healthy piglets in an abattoir in Brazil: Animal-human interface and risk for interspecies transmission. Mem. Inst. Oswaldo Cruz 2013, 108, 548–553. [Google Scholar] [CrossRef]
- Ciacci-Zanella, J.R.; Schaefer, R.; Gava, D.; Haach, V.; Cantao, M.E.; Coldebella, A. Influenza A virus infection in Brazilian swine herds following the introduction of pandemic 2009 H1N1. Vet. Microbiol. 2015, 180, 118–122. [Google Scholar] [CrossRef]
- Souza, C.K.; Oldiges, D.P.; Poeta, A.P.S.; Vaz, I.D.S., Jr.; Schaefer, R.; Gava, D.; Ciacci-Zanella, J.R.; Canal, C.W.; Corbellini, L.G. Serological surveillance and factors associated with influenza A virus in backyard pigs in Southern Brazil. Zoonoses Public Health 2019, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Cibulski, S.P.; Andrade, C.P.; Teixeira, T.F.; Varela, A.P.; Scheffer, C.M.; Franco, A.C.; de Almeida, L.L.; Roehe, P.M. Swine Influenza Virus and Association with the Porcine Respiratory Disease Complex in Pig Farms in Southern Brazil. Zoonoses Public Health 2016, 63, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.I.; Schaefer, R.; Gava, D.; Cantao, M.E.; Ciacci-Zanella, J.R. Influenza A Viruses of Human Origin in Swine, Brazil. Emerg. Infect. Dis. 2015, 21, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Rajao, D.S.; Costa, A.T.; Brasil, B.S.; Del Puerto, H.L.; Oliveira, F.G.; Alves, F.; Braz, G.F.; Reis, J.K.; Guedes, R.M.; Lobato, Z.I.; et al. Genetic characterization of influenza virus circulating in Brazilian pigs during 2009 and 2010 reveals a high prevalence of the pandemic H1N1 subtype. Influenza Other Respir. Viruses 2013, 7, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, R.; Rech, R.R.; Gava, D.; Cantao, M.E.; da Silva, M.C.; Silveira, S.; Zanella, J.R. A human-like H1N2 influenza virus detected during an outbreak of acute respiratory disease in swine in Brazil. Arch. Virol. 2015, 160, 29–38. [Google Scholar] [CrossRef]
- Serafini Poeta Silva, A.P.; de Freitas Costa, E.; Sousa, E.S.G.; Souza, C.K.; Schaefer, R.; da Silva Vaz, I., Jr.; Corbellini, L.G. Biosecurity practices associated with influenza A virus seroprevalence in sows from southern Brazilian breeding herds. Prev. Vet. Med. 2019, 166, 1–7. [Google Scholar] [CrossRef]
- Schmidt, C.; Cibulski, S.P.; Muterle Varela, A.P.; Mengue Scheffer, C.; Wendlant, A.; Quoos Mayer, F.; Lopes de Almeida, L.; Franco, A.C.; Roehe, P.M. Full-Genome Sequence of a Reassortant H1N2 Influenza A Virus Isolated from Pigs in Brazil. Genome Announc. 2014, 2. [Google Scholar] [CrossRef]
- Jiménez, L.F.M.; Nieto, G.R.; Alfonso, V.V.; Correa, J.J. Association of swine infl uenza H1N1 pandemic virus (SIVH1N1p) with porcine respiratory disease complex in sows from commercial pig farms in Colombia. Virol. Sin. 2014, 29, 242–249. [Google Scholar] [CrossRef]
- Tinoco, Y.O.; Montgomery, J.M.; Kasper, M.R.; Nelson, M.I.; Razuri, H.; Guezala, M.C.; Azziz-Baumgartner, E.; Widdowson, M.A.; Barnes, J.; Gilman, R.H.; et al. Transmission dynamics of pandemic influenza A(H1N1)pdm09 virus in humans and swine in backyard farms in Tumbes, Peru. Influenza Other Respir. Viruses 2016, 10, 47–56. [Google Scholar] [CrossRef]
- Jimenez-Bluhm, P.; Di Pillo, F.; Bahl, J.; Osorio, J.; Schultz-Cherry, S.; Hamilton-West, C. Circulation of influenza in backyard productive systems in central Chile and evidence of spillover from wild birds. Prev. Vet. Med. 2018, 153, 1–6. [Google Scholar] [CrossRef]
- Bravo-Vasquez, N.; Di Pillo, F.; Lazo, A.; Jimenez-Bluhm, P.; Schultz-Cherry, S.; Hamilton-West, C. Presence of influenza viruses in backyard poultry and swine in El Yali wetland, Chile. Prev. Vet. Med. 2016, 134, 211–215. [Google Scholar] [CrossRef]
- Bravo-Vasquez, N.; Baumberger, C.; Jimenez-Bluhm, P.; Di Pillo, F.; Lazo, A.; Sanhueza, J.; Schultz-Cherry, S.; Hamilton-West, C. Risk factors and spatial relative risk assessment for influenza a virus in poultry and swine in backyard production systems of central Chile. Vet. Med. Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, T.; Dotti, S.; Renzi, S.; Ferrari, M. Susceptibility of different cell lines to Avian and Swine Influenza viruses. J. Virol. Methods 2012, 185, 82–88. [Google Scholar] [CrossRef]
- Cazacu, A.C.; Greer, J.; Taherivand, M.; Demmler, G.J. Comparison of lateral-flow immunoassay and enzyme immunoassay with viral culture for rapid detection of influenza virus in nasal wash specimens from children. J. Clin. Microbiol.. 2003, 41, 2132–2134. [Google Scholar] [CrossRef][Green Version]
- Koski, R.R.; Klepser, M.E. A systematic review of rapid diagnostic tests for influenza: Considerations for the community pharmacist. J. Am. Pharm Assoc. (2003) 2017, 57, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Hartley, J.; Dunn, H.; Williams, R.; Williams, C.A.; Breuer, J. Whole-genome Sequencing Provides Data for Stratifying Infection Prevention and Control Management of Nosocomial Influenza, A. Clin. Infect. Dis. 2019, 69, 1649–1656. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.M.; Caldwell, N.; Barr, I.G. Rapid detection and subtyping of human influenza A viruses and reassortants by pyrosequencing. PLoS ONE 2011, 6, e23400. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, J.; Vemula, S.V.; Lin, C.; Tan, J.; Ragupathy, V.; Wang, X.; Mbondji-Wonje, C.; Ye, Z.; Landry, M.L.; et al. Sensitive Detection and Simultaneous Discrimination of Influenza A and B Viruses in Nasopharyngeal Swabs in a Single Assay Using Next-Generation Sequencing-Based Diagnostics. PLoS ONE 2016, 11, e0163175. [Google Scholar] [CrossRef]
- WHO. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. In CDC Real-Time RT-PCR (rRTPCR) Protocol for Detection and Characterization of Swine Influenza (Version 2009); Centre for Disease Control and Prevention: Atlanta, GA, USA, 2009; Vol. CDC REF. # I-007-05; pp. 1–8. [Google Scholar]
- LeBlanc, J.J.; Li, Y.; Bastien, N.; Forward, K.R.; Davidson, R.J.; Hatchette, T.F. Switching gears for an influenza pandemic: Validation of a duplex reverse transcriptase PCR assay for simultaneous detection and confirmatory identification of pandemic (H1N1) 2009 influenza virus. J. Clin. Microbiol. 2009, 47, 3805–3813. [Google Scholar] [CrossRef]
- Fouchier, R.A.; Bestebroer, T.M.; Herfst, S.; Van Der Kemp, L.; Rimmelzwaan, G.F.; Osterhaus, A.D. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 2000, 38, 4096–4101. [Google Scholar] [CrossRef]
- Casanova, T.; Rojas, D.; Baeza, C.; Ruiz, A. Immunohistochemistry: A rapid and specific diagnostic tool for influenza virus infection in pigs. J. Dairyveterinary Anim. Res. 2017, 5, 212–214. [Google Scholar] [CrossRef]
- Fuller, T.L.; Gilbert, M.; Martin, V.; Cappelle, J.; Hosseini, P.; Njabo, K.Y.; Abdel Aziz, S.; Xiao, X.; Daszak, P.; Smith, T.B. Predicting hotspots for influenza virus reassortment. Emerg. Infect. Dis. 2013, 19, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Naguib, M.M.; Verhagen, J.H.; Samy, A.; Eriksson, P.; Fife, M.; Lundkvist, A.; Ellstrom, P.; Jarhult, J.D. Avian influenza viruses at the wild-domestic bird interface in Egypt. Infect. Ecol. Epidemiol. 2019, 9, 1575687. [Google Scholar] [CrossRef] [PubMed]
- Abdelwhab, E.M.; Hafez, H.M. An overview of the epidemic of highly pathogenic H5N1 avian influenza virus in Egypt: Epidemiology and control challenges. Epidemiol. Infect. 2011, 139, 647–657. [Google Scholar] [CrossRef]
- Selim, A.A.; Erfan, A.M.; Hagag, N.; Zanaty, A.; Samir, A.H.; Samy, M.; Abdelhalim, A.; Arafa, A.A.; Soliman, M.A.; Shaheen, M.; et al. Highly Pathogenic Avian Influenza Virus (H5N8) Clade 2.3.4.4 Infection in Migratory Birds, Egypt. Emerg. Infect. Dis. 2017, 23, 1048–1051. [Google Scholar] [CrossRef]
- Shope, R.E. The Etiology of Swine Influenza. Science 1931, 73, 214–215. [Google Scholar] [CrossRef]
- Yang, Y.; Halloran, M.E.; Sugimoto, J.D.; Longini, I.M., Jr. Detecting human-to-human transmission of avian influenza A (H5N1). Emerg. Infect. Dis. 2007, 13, 1348–1353. [Google Scholar] [CrossRef]
- Ledford, H. Barrier to bird flu transmission found in humans. Nature 2008. [Google Scholar] [CrossRef]
- Gcumisa, S.T.; Oguttu, J.W.; Masafu, M.M. Pig farming in rural South Africa: A case study of uThukela District in KwaZulu-Natal. Indian J. Anim. Res. 2016, 50, 614–620. [Google Scholar] [CrossRef]
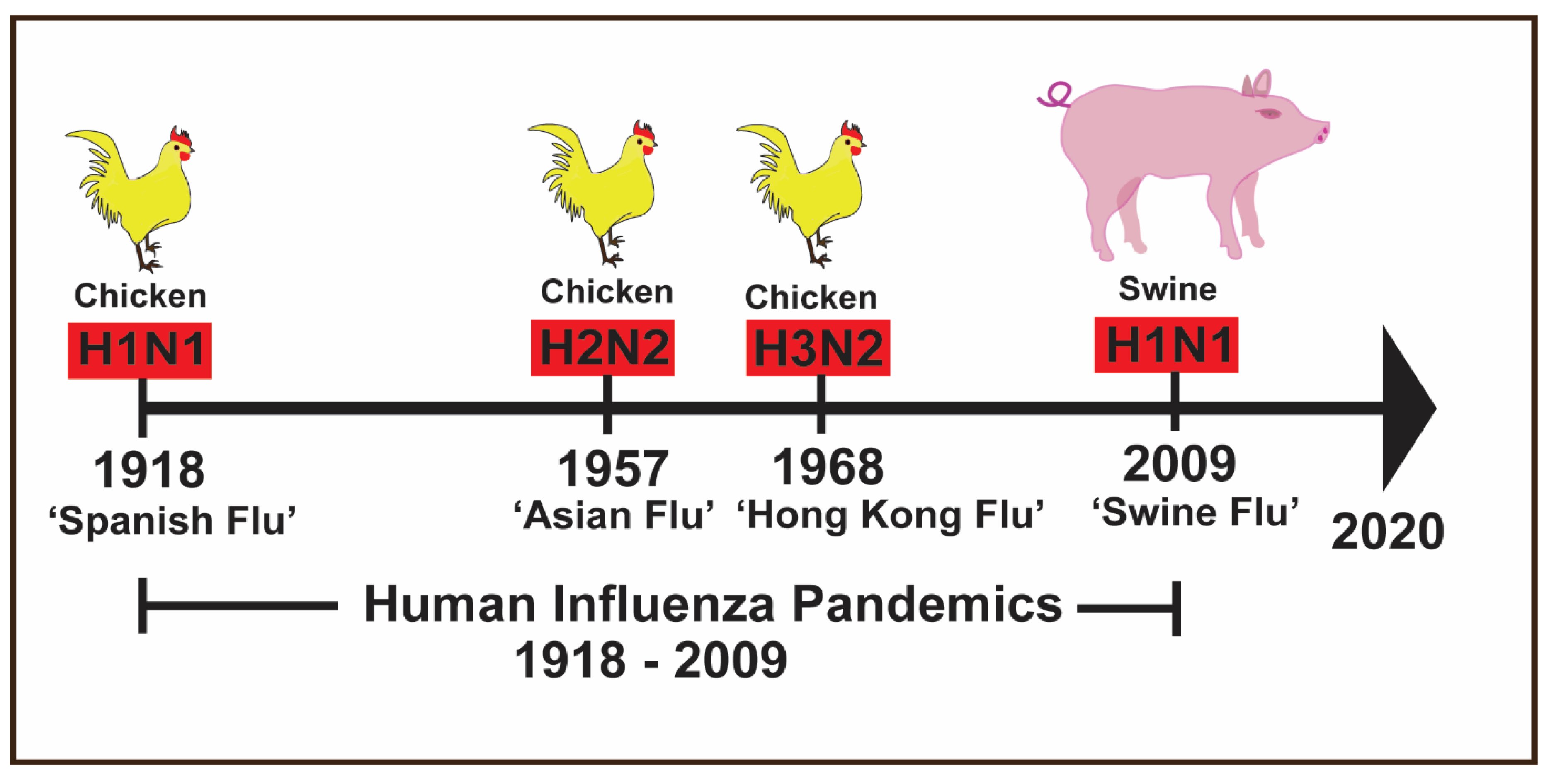
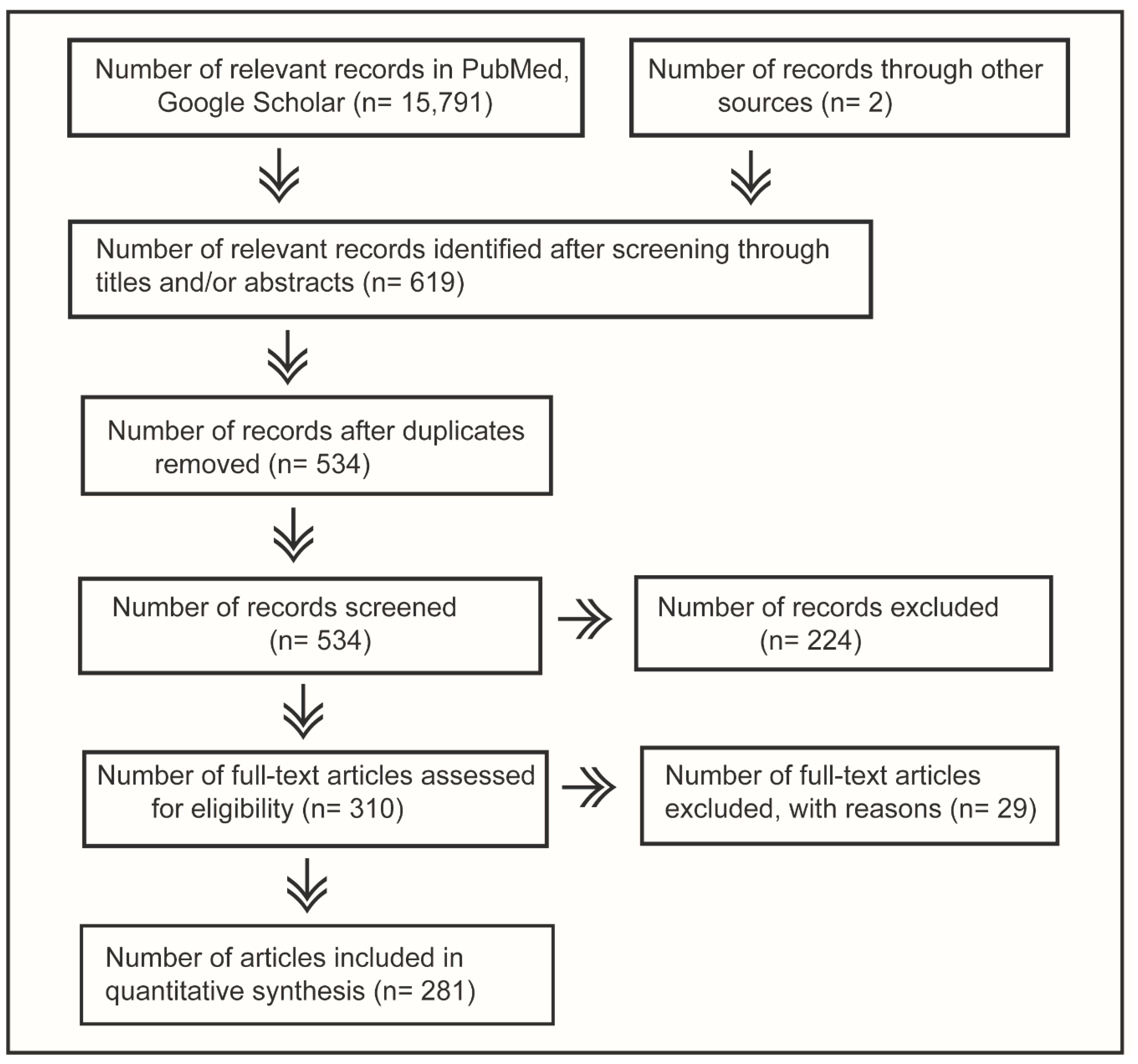
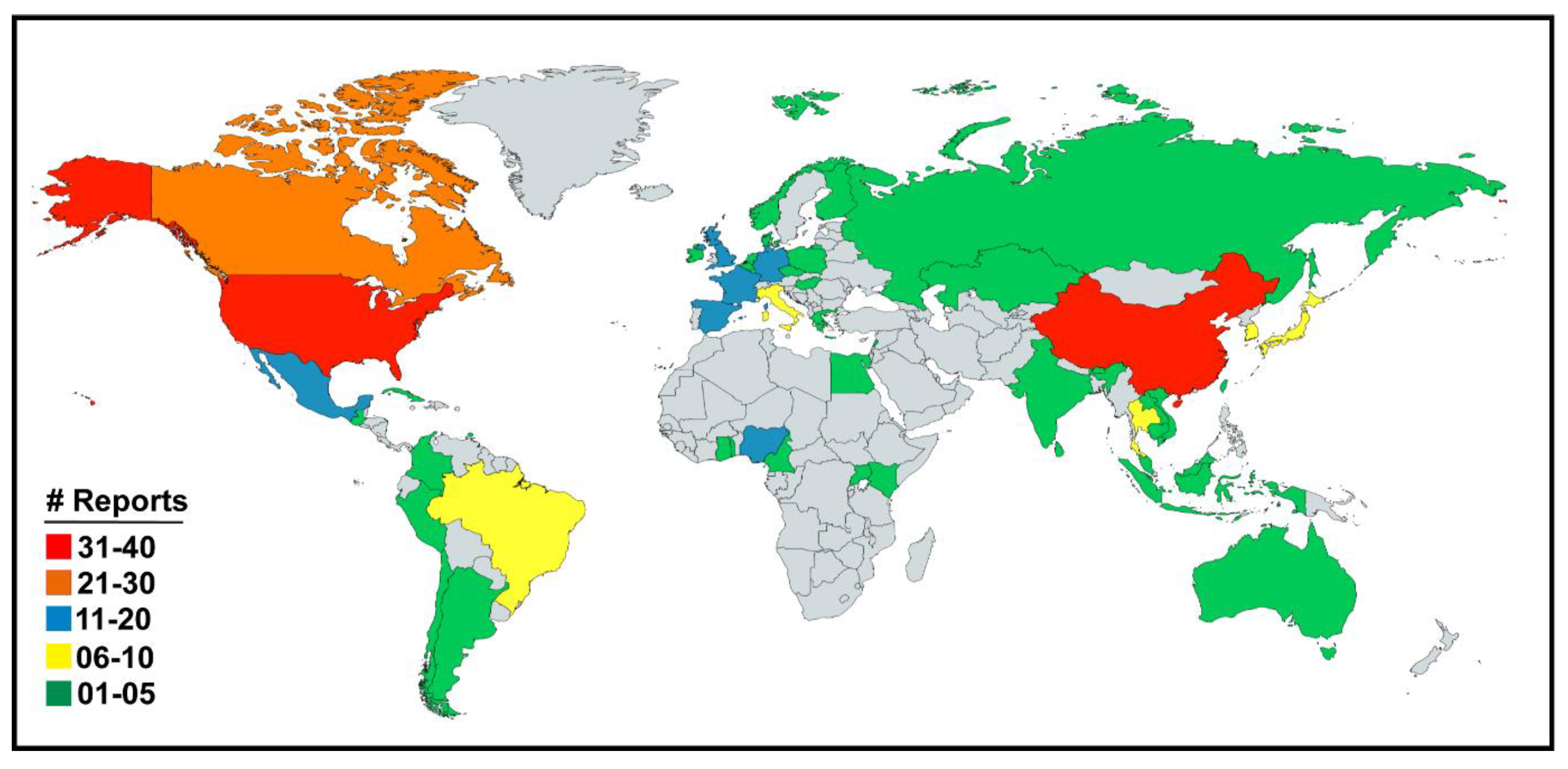
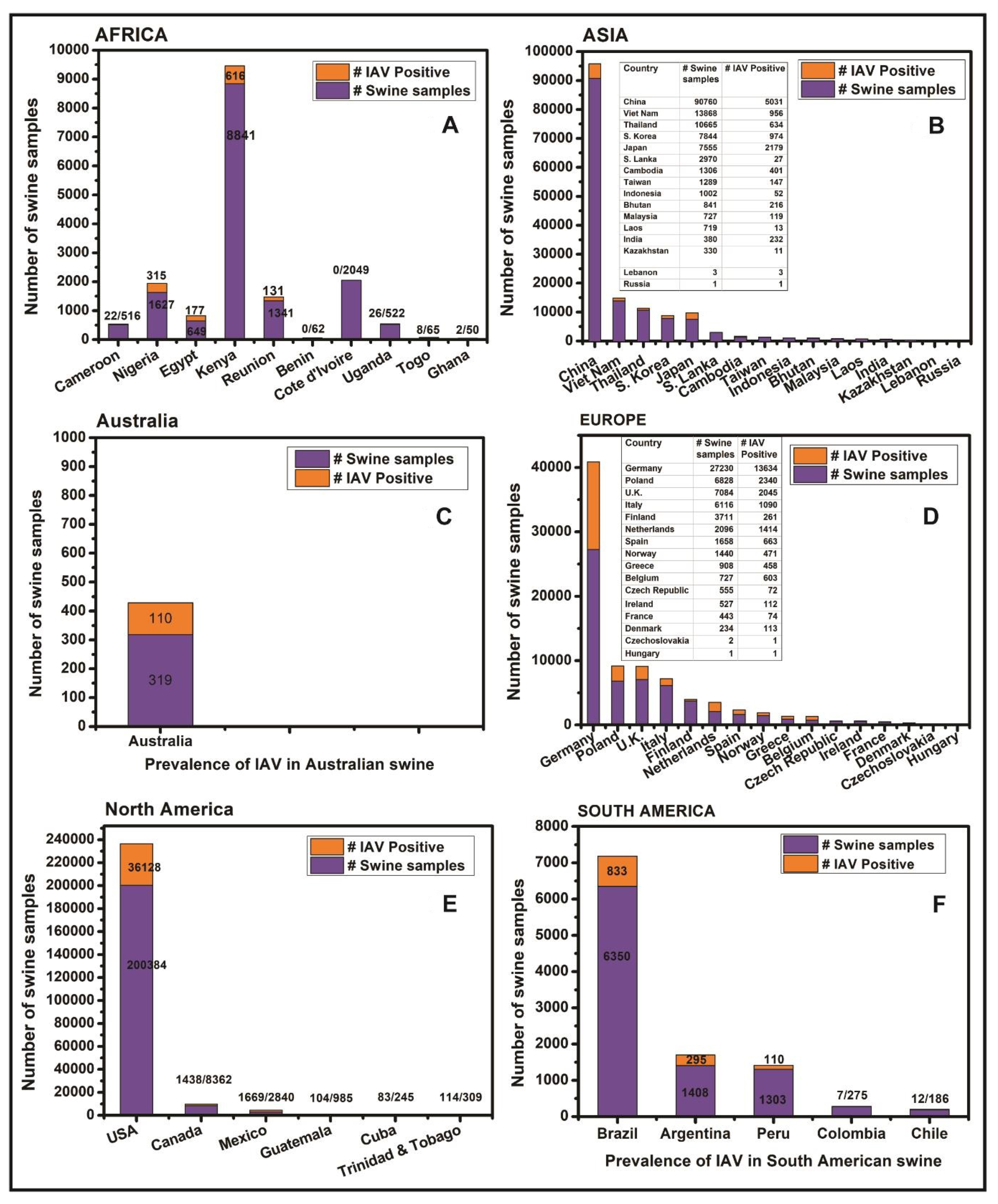
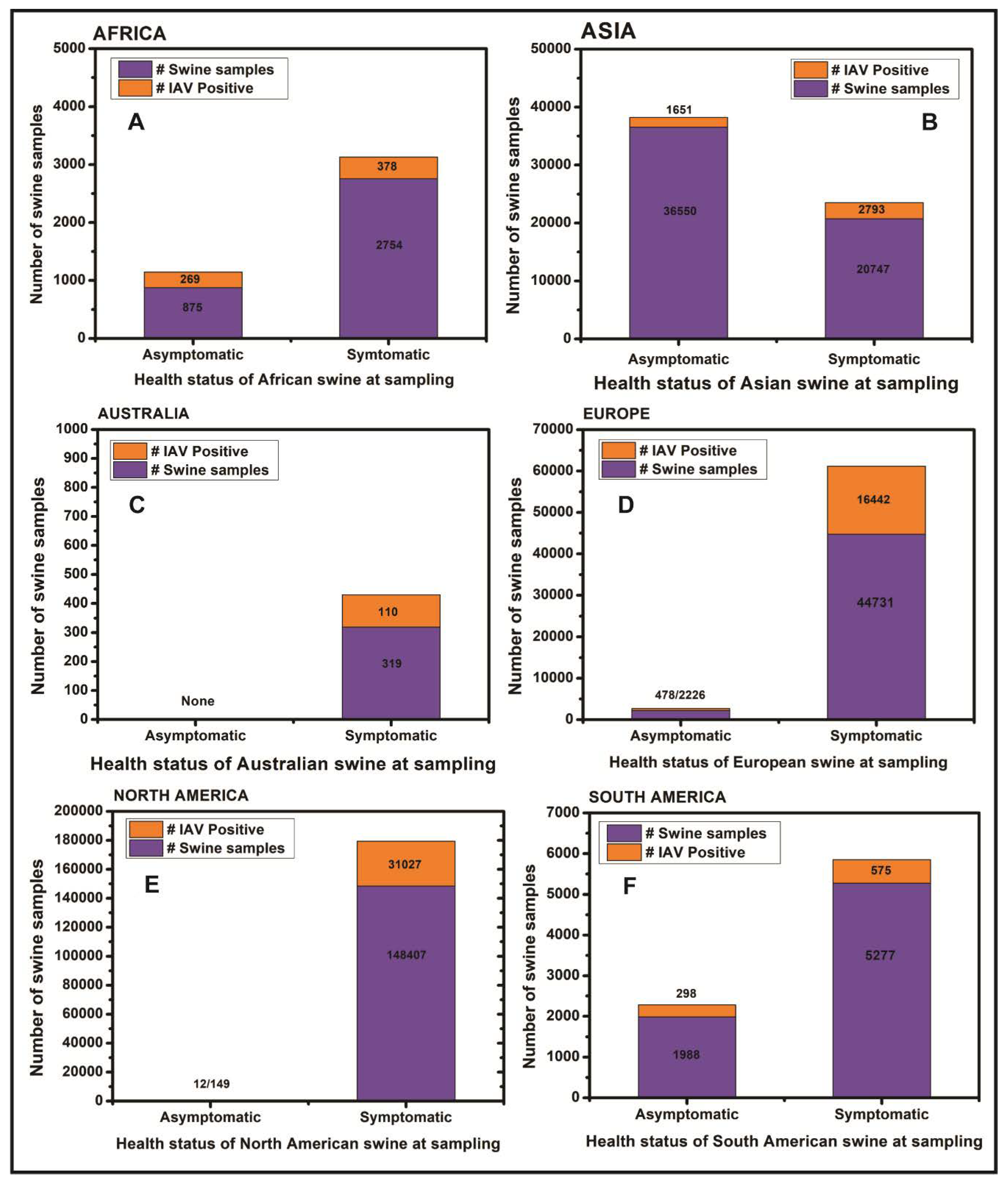
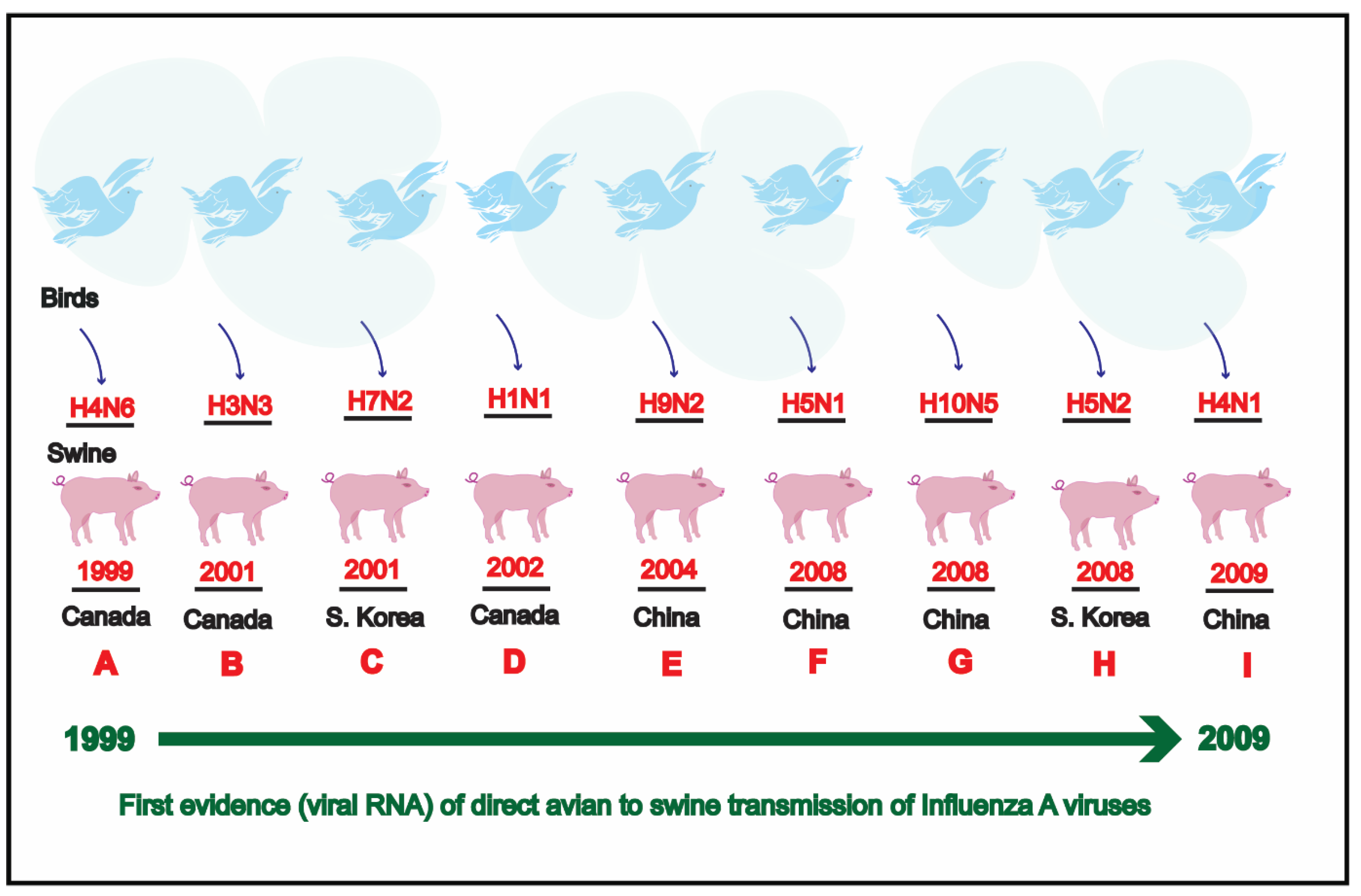
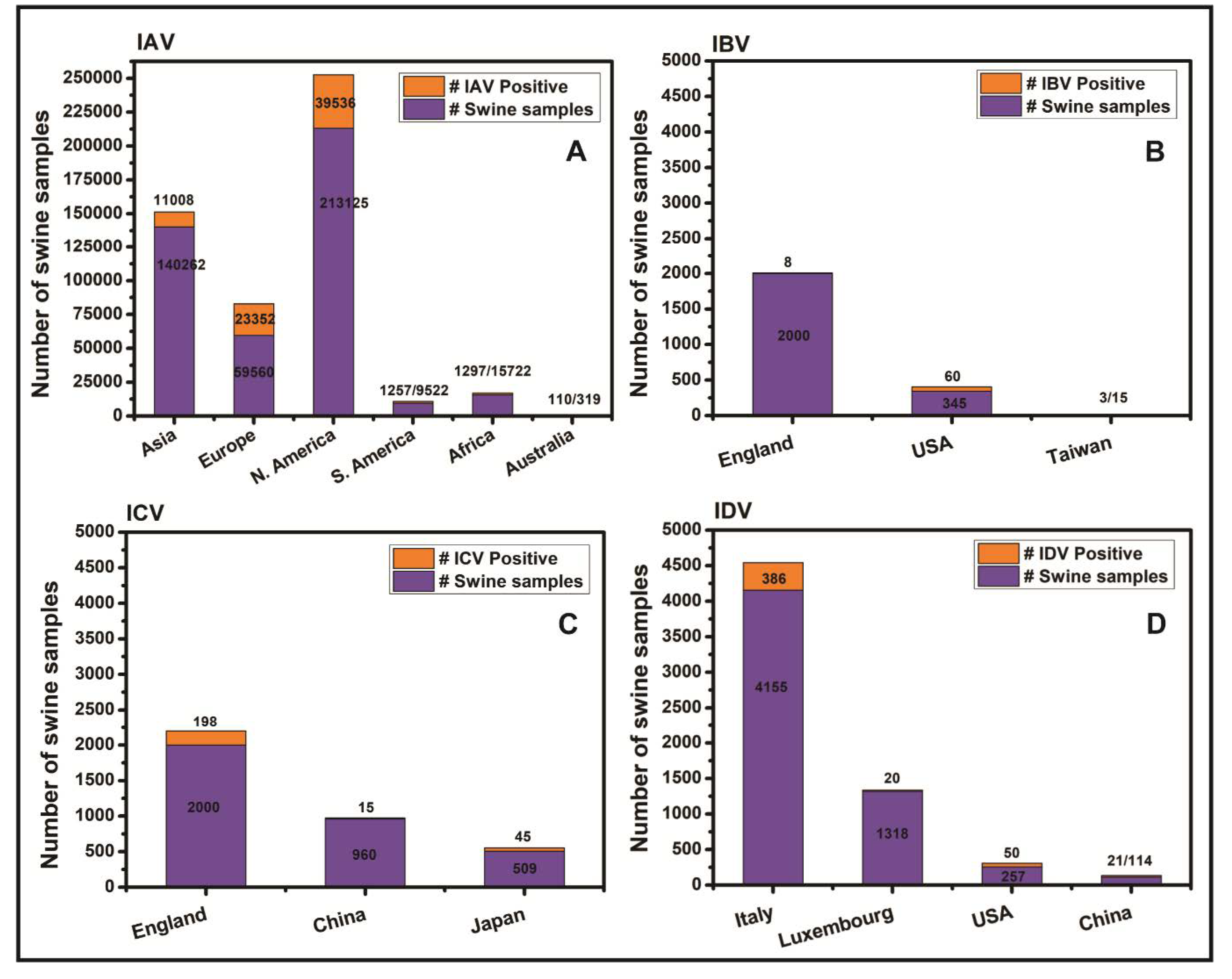
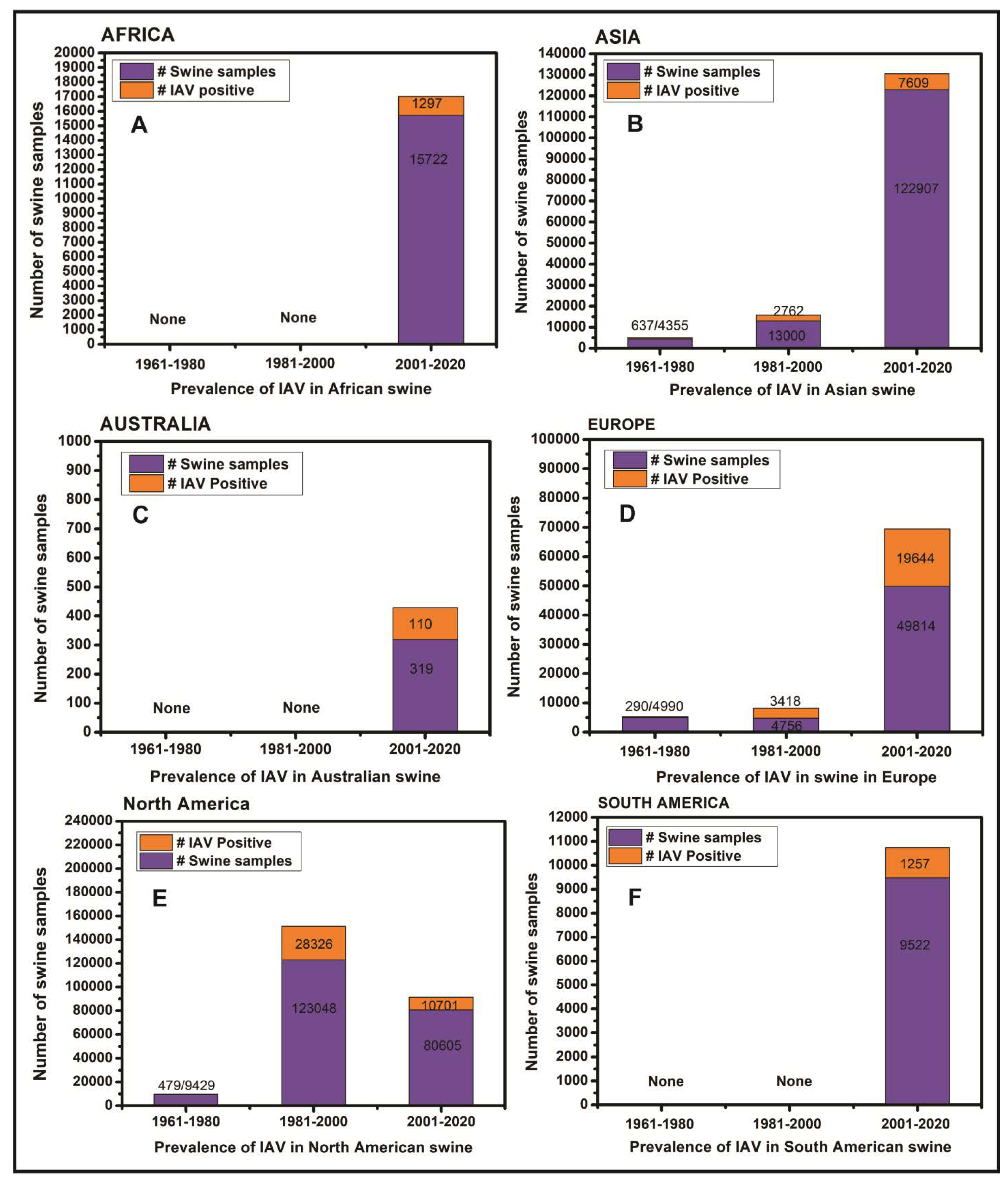
| Continents | Countries | Influenza A Virus (IAV) Subtypes | Other Influenza Virus Types | References |
|---|---|---|---|---|
| Africa | Cameroon | H1N1, A(H1N1)pdm09 | None | [51,52,53] |
| Nigeria | H1N1, H3N2, A(H1N1)pdm09, H5N1 | None | [52,54,55,56,57,58,59] | |
| Ghana | H3N2 | None | [57] | |
| Egypt | H5N1, H5N2, H9N2, A(H1N1)pdm09 | None | [60,61] | |
| Kenya | H1N1, H3N2, A(H1N1)pdm09 | None | [62,63,64] | |
| Benin, Cote d’Ivoire | None | None | [65] | |
| Reunion island | A(H1N1)pdm09 | None | [66] | |
| Togo | A(H1N1)pdm09 | None | [67] | |
| Uganda | IAV | None | [68] | |
| Asia | China | H1N1, H1N2, H3N2, A(H1N1)pdm09, H5N1, H9N2, H4N1, H4N6, H5N3, H10N5, H4N8, H6N6, H7N9, H3N8 | ICV, IDV | [69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106] |
| Bhutan | H1N1, A(H1N1)pdm09 | None | [107] | |
| Cambodia | H1N1, H3N2, A(H1N1)pdm09 | None | [108,109] | |
| Japan | H1N1, H1N2, H3N2, A(H1N1)pdm09 | ICV | [110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126] | |
| South Korea | H1N1, H1N2, H3N2, A(H1N1)pdm09, H7N2, H5N2, H3N1 | None | [127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143] | |
| Thailand | H1N1, H1N2, H3N2, H3N1, A(H1N1)pdm09 | None | [144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159] | |
| Viet Nam | H1N1, H1N2, H3N2, A(H1N1)pdm09, H5N1 | None | [160,161,162,163] | |
| India | H1N1, H2N2, H3N2, A(H1N1)pdm09 | None | [164,165] | |
| Lebanon | H9N2 | None | [166] | |
| Malaysia | H1N1, H3N2 | None | [167] | |
| Laos | H3N2 | None | [168] | |
| Russia | H1N1 | None | [169] | |
| Taiwan | IAV | IBV | [170,171] | |
| Indonesia | H5N1 | None | [172] | |
| Sri Lanka | H3N2, A(H1N1)pdm09 | None | [173] | |
| Kazakhstan | H1N1, H3N2 | None | [174] | |
| Australia | Australia | H1N1, H1N2, H3N2, A(H1N1)pdm09 | None | [175,176,177,178] |
| Europe | Belgium | H1N1, H1N2, H3N2 | None | [179,180,181] |
| Denmark | H1N1, H1N2, H3N2 | None | [182,183,184] | |
| United Kingdom | H1N1, H1N2, H3N2, A(H1N1)pdm09, H1N7 | IBV, ICV | [185,186,187,188,189,190,191,192] | |
| Finland | H1N1, H1N2, H3N2, A(H1N1)pdm09 | None | [193,194] | |
| France | H1N1, H1N2, H3N2, A(H1N1)pdm09 | None | [195,196,197,198,199] | |
| Germany | H1N1, H1N2, H3N2, A(H1N1)pdm09 | None | [200,201,202,203] | |
| Greece | H1N1, H1N2, H3N2, A(H1N1)pdm09 | None | [204] | |
| Italy | H1N1, H1N2, H3N2, A(H1N1)pdm09 | IDV | [205,206,207,208,209,210,211,212,213,214,215,216,217] | |
| Spain | H1N1, H1N2, H3N2 | None | [218,219,220,221] | |
| Netherlands | H1N1, H1N2, H3N2 | None | [222] | |
| Norway | A(H1N1)pdm09 | None | [223,224,225,226] | |
| Poland | H1N1, H1N2, H3N2, A(H1N1)pdm09 | None | [227,228,229] | |
| Czechoslovakia | H3N2 | None | [230] | |
| Hungary | H1N1 | None | [231] | |
| Czech Republic | H1N1, H1N2, H3N2 | None | [232] | |
| Republic of Ireland | H1N1, H1N2, H3N2 | None | [232] | |
| Luxembourg | None | IDV | [46] | |
| Multiple European Nations | H1N1, H1N2, H3N2 | None | [233] | |
| North America | Canada | H1N1, H1N2, H3N2, A(H1N1)pdm09, H4N6, H3N3 | None | [234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255,256,257,258] |
| USA | H1N1, H1N2, H3N1, H3N2, A(H1N1)pdm09, H4N6, H2N3 | IBV, IDV | [259,260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280,281,282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303] | |
| Mexico | H1N1, H1N2, H3N2, A(H1N1)pdm09, H5N2 | None | [304,305,306,307,308,309] | |
| Guatemala | H3N2, A(H1N1)pdm09 | None | [310] | |
| Cuba | H1N1, A(H1N1)pdm09 | None | [311,312] | |
| Trinidad & Tobago | H3N2, A(H1N1)pdm09 | None | [313] | |
| South America | Argentina | H1N1, H1N2, H3N2, A(H1N1)pdm09 | None | [314,315,316,317,318] |
| Brazil | H1N1, H1N2, H3N2, A(H1N1)pdm09 | None | [319,320,321,322,323,324,325,326,327,328,329] | |
| Colombia | A(H1N1)pdm09 | None | [330] | |
| Peru | A(H1N1)pdm09 | None | [331] | |
| Chile | IAV, H1N2 | None | [332,333,334] |
| S. No. | Swine Sample Types | Methods Used for Influenza Virus Detection | Virus Types/Subtypes Detected | References |
|---|---|---|---|---|
| 1. | Nasal swab | RNA extraction, real-time RT-PCR, reverse transcription PCR, multiplex RT-PCR, ligation, partial/whole genome Sanger sequencing, Next-generation sequencing, phylogenetic analysis, virus isolation (MDCK cells/Caco-2 cells/HRT18 cells/SPF chicken eggs), transmission electron microscopy, HI assay, NI assay, ultracentrifugation | H1N1, H1N2, H3N2, H4N6, A(H1N1)pdm09, Reassortant H1N1, Reassortant A(H1N1)pdm09, Triple-reassortant H3N2, H1N7, H5N2, IBV, IDV | [5,37,43,45,46,61,74,88,137,157,171,176,188,191,199,207,215,219,220,221,241,243,248,250,263,289,290,301,306,307,326,334] |
| 2. | Tracheal swab | RNA extraction, real-time RT-PCR, reverse transcription PCR, Sanger sequencing, virus isolation (MDCK cells, SPF chicken eggs) | H5N1, IAV, ICV, H1N1, A(H1N1)pdm09, | [38,59,103,321,331] |
| 3. | Nasal wipe | Real-time RT-PCR | H3N2 | [289] |
| 4. | Snout wipe | RNA extraction, real-time RT-PCR, virus isolation | IAV | [286] |
| 5. | Oropharyngeal swab | RNA extraction, real-time RT-PCR, Sanger sequencing | IAV, A(H1N1)pdm09 | [211,276] |
| 6. | Nasopharyngeal swab | RNA extraction, real-time RT-PCR, virus isolation (SPF chicken eggs), Sanger sequencing | H3N2, A(H1N1)pdm09, IBV, ICV | [38,141,211,277] |
| 7. | Oral fluid | RNA extraction, real-time RT-PCR, virus isolation, Sanger sequencing, Next-generation sequencing (MiSeq) | IAV, IDV, H1N1, H1N2, H3N2, A(H1N1)pdm09 | [45,192,227,282,285,287,302,322] |
| 8. | Blood/ serum | IDEXX Ab test, ELISA, HI assay, NI assay, VN assay, MN assay, western blot, virus isolation (MDCK cells) | H1N1, H1N2, H3N2, A(H1N1)pdm09, H5N1, H5N2, H5N3, H9N2, H7N9, H3N1, H4N8, H6N6, H6N2, H7N2, IBV, ICV, IDV | [37,38,46,52,59,70,79,96,97,135,138,153,186,195,200,215,291,306] |
| 9. | Lung/liver/internal organ tissues | RNA extraction, real-time RT-PCR, reverse transcription-PCR, ligation, HI assay, virus isolation (MDCK cells/SPF chicken eggs), Sanger and Next-generation sequencing, hematoxylin-eosin staining, immunohistochemistry, Immunofluorescence | H1N1, H1N2, Reassortant H1N1, H3N2, H2N3, A(H1N1)pdm09, H7N2, IDV | [45,121,127,128,130,201,203,243,245,248,274,314,320,325,327] |
| 10. | Lung homogenate | RNA extraction, real-time RT-PCR, multiplex RT-PCR, single step RT-PCR, virus isolation (MDCK cells/Caco-2 cells/SPF chicken eggs), Sanger sequencing, membrane enzyme immunoassay, HI assay | H1N1, H1N2, H3N2, Reassortant H1N2, A(H1N1)pdm09 | [130,213,218,292] |
| 11. | Fecal slurry | RNA extraction, qRT-PCR | IAV | [88] |
| 12. | Rectal swab | Nucleic acid extraction, reverse transcription, metagenomic sequencing | IAV | [300] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chauhan, R.P.; Gordon, M.L. A Systematic Review Analyzing the Prevalence and Circulation of Influenza Viruses in Swine Population Worldwide. Pathogens 2020, 9, 355. https://doi.org/10.3390/pathogens9050355
Chauhan RP, Gordon ML. A Systematic Review Analyzing the Prevalence and Circulation of Influenza Viruses in Swine Population Worldwide. Pathogens. 2020; 9(5):355. https://doi.org/10.3390/pathogens9050355
Chicago/Turabian StyleChauhan, Ravendra P., and Michelle L. Gordon. 2020. "A Systematic Review Analyzing the Prevalence and Circulation of Influenza Viruses in Swine Population Worldwide" Pathogens 9, no. 5: 355. https://doi.org/10.3390/pathogens9050355
APA StyleChauhan, R. P., & Gordon, M. L. (2020). A Systematic Review Analyzing the Prevalence and Circulation of Influenza Viruses in Swine Population Worldwide. Pathogens, 9(5), 355. https://doi.org/10.3390/pathogens9050355






