Mixed Aetiology of Diarrhoea in Infants Attending Clinics in the North-West Province of South Africa: Potential for Sub-Optimal Treatment
Abstract
1. Introduction
2. Results
2.1. Distribution of Participants According to Age, Mode of Feeding and Presence/Absence of Diarrhoea
2.2. The Overall Distribution of Aetiologic Agents among the Study Population
2.3. Prevalence of Aetiologic Agents in Symptomatic Participants
2.4. Prevalence of Arcobacter and Campylobacter spp. in the Study
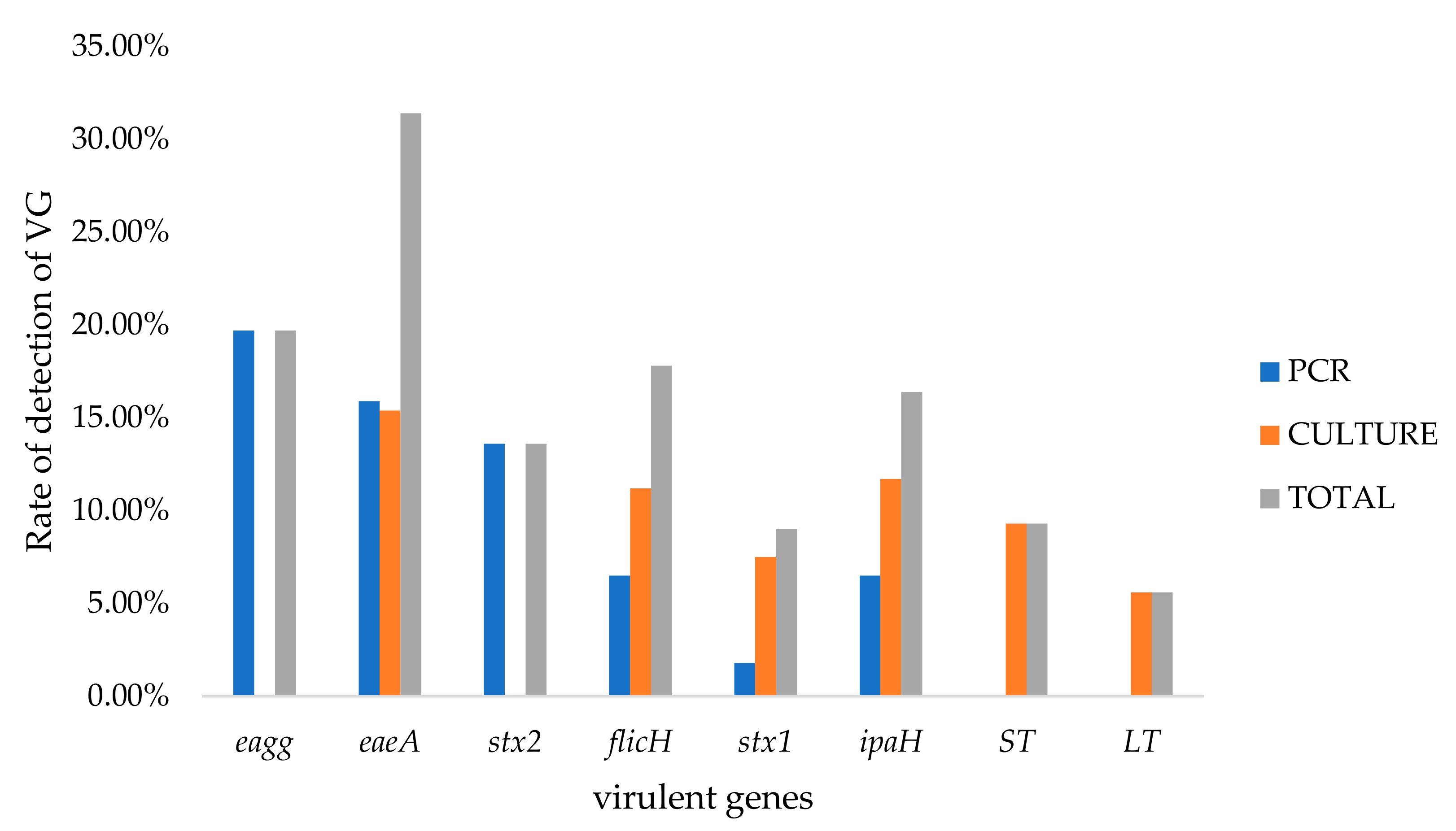
2.5. Prevalence of DEC Virulence-Associated Genes
2.6. Prevalence of Rotavirus and Norovirus
2.7. Prevalence of Mixed Aetiology
3. Discussion
3.1. Occurrence of Bacterial Agents in the Present Study
3.1.1. Occurrence of Campylobacter
3.1.2. Prevalence of Arcobacter
3.1.3. Prevalence of DEC Pathotypes
3.2. Prevalence of Viral Aetiology
3.3. Prevalence of Mixed Aetiology
4. Materials and Methods
4.1. Specimen Collection
4.2. Isolation of Campylobacter and Arcobacter Species
4.3. Preparation of Positive Controls
4.4. Genomic DNA Extraction and PCR Identification of Arcobacter and Campylobacter Species
4.5. Amplification of A. butzleri Virulence Gene
4.6. Isolation of Escherichia coli from Stool Samples
4.7. DNA Extraction and Detection of E. coli Virulence Genes (VGs) from Pure Culture
4.8. Identification of Viral Pathogens
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Rosner, B.M.; Schielke, A.; Didelot, X.; Kops, F.; Breidenbach, J.; Willrich, N.; Gölz, G.; Alter, T.; Stingl, K.; Josenhans, C.; et al. A combined case-control and molecular source attribution study of human Campylobacter infections in Germany, 2011-2014 /692/308/174 /692/499 article. Sci. Rep. 2017, 7, 5139. [Google Scholar] [CrossRef]
- Abajobir, A.A.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abera, S.F.; Aboyans, V.; Adetokunboh, O.; Afshin, A.; Agrawal, A.; Ahmadi, A.; et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar]
- Fischer Walker, C.L.; Rudan, I.; Liu, L.; Nair, H.; Theodoratou, E.; Bhutta, Z.A.; O’Brien, K.L.; Campbell, H.; Black, R.E. Global burden of childhood pneumonia and diarrhoea. Lancet 2013, 381, 1405–1416. [Google Scholar] [CrossRef]
- Becker-Dreps, S.; Bucardo, F.; Vilchez, S.; Zambrana, L.E.; Liu, L.; Weber, D.J.; Peña, R.; Barclay, L.; Vinjé, J.; Hudgens, M.G.; et al. Etiology of childhood diarrhea after rotavirus vaccine introduction: A prospective, population-based study in Nicaragua. Pediatr. Infect. Dis. J. 2014, 33, 1156–1163. [Google Scholar] [CrossRef]
- Platts-Mills, J.A.; Babji, S.; Bodhidatta, L.; Gratz, J.; Haque, R.; Havt, A.; McCormick, B.J.; McGrath, M.; Olortegui, M.P.; Samie, A.; et al. Pathogen-specific burdens of community diarrhoea in developing countries: A multisite birth cohort study (MAL-ED). Lancet Glob. Health 2015, 3, e564–e575. [Google Scholar] [CrossRef]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Steele, A.D.; Duque, J.; Parashar, U.D. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 136–141. [Google Scholar] [CrossRef]
- Steele, A.; Peenze, I.; de Beer, M.; Pager, C.; Yeats, J.; Potgieter, N.; Ramsaroop, U.; Page, N.; Mitchell, J.; Geyer, A.; et al. Anticipating rotavirus vaccines: Epidemiology and surveillance of rotavirus in South Africa. Vaccine 2003, 21, 354–360. [Google Scholar] [CrossRef]
- Groome, M.J.; Page, N.; Cortese, M.M.; Moyes, J.; Zar, H.J.; Kapongo, C.N.; Mulligan, C.; Diedericks, R.; Cohen, C.; Fleming, J.A.; et al. Effectiveness of monovalent human rotavirus vaccine against admission to hospital for acute rotavirus diarrhoea in South African children: A case-control study. Lancet Infect. Dis. 2014, 14, 1096–1104. [Google Scholar] [CrossRef]
- Da Costa, S.T.P.; Fumian, T.M.; De Lima, I.C.G.; Siqueira, J.A.M.; Da Silva, L.D.; Hernández, J.d.M.; De Lucena, M.S.S.; Reymão, T.K.A.; Soares, L.d.S.; Mascarenhas, J.D.P.; et al. High prevalence of norovirus in children with sporadic acute gastroenteritis in Manaus, Amazon region, northern Brazil. Mem. Inst. Oswaldo Cruz 2017, 112, 391–395. [Google Scholar] [CrossRef]
- Pires, S.M.; Fischer-Walker, C.L.; Lanata, C.F.; Devleesschauwer, B.; Hall, A.J.; Kirk, M.D.; Duarte, A.S.R.; Black, R.E.; Angulo, F.J. Aetiology-specific estimates of the global and regional incidence and mortality of diarrhoeal diseases commonly transmitted through food. PLoS ONE 2015, 10, e0142927. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, S.S.; Green, K.Y.; Korba, B.E. Treatment of norovirus infections: Moving antivirals from the bench to the bedside. Antivir. Res. 2014, 105, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Robilotti, E.; Deresinski, S.; Pinsky, B.A. Norovirus. Clin. Microbiol. Rev. 2015, 28, 134–164. [Google Scholar] [CrossRef] [PubMed]
- Kroneman, A.; Vega, E.; Vennema, H.; Vinjé, J.; White, P.; Hansman, G.; Green, K.; Martella, V.; Katayama, K.; Koopmans, M. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 2013, 158, 2059–2068. [Google Scholar] [CrossRef]
- Delahoy, M.J.; Wodnik, B.; McAliley, L.; Penakalapati, G.; Swarthout, J.; Freeman, M.C.; Levy, K. Pathogens transmitted in animal feces in low- and middle-income countries. Int. J. Hyg. Environ. Health 2018, 221, 661–676. [Google Scholar] [CrossRef]
- Vicente-Martins, S.; Oleastro, M.; Domingues, F.C.; Ferreira, S. Arcobacter spp. at retail food from Portugal: Prevalence, genotyping and antibiotics resistance. Food Control 2018, 85, 107–112. [Google Scholar] [CrossRef]
- Youmans, B.P.; Ajami, N.J.; Jiang, Z.-D.; Campbell, F.; Wadsworth, W.D.; Petrosino, J.F.; DuPont, H.L.; Highlander, S.K. Characterization of the human gut microbiome during travelers’ diarrhea. Gut Microbes 2015, 6, 110–119. [Google Scholar] [CrossRef]
- Kotloff, K.L.; Platts-Mills, J.A.; Nasrin, D.; Roose, A.; Blackwelder, W.C.; Levine, M.M. Global burden of diarrheal diseases among children in developing countries: Incidence, etiology, and insights from new molecular diagnostic techniques. Vaccine 2017, 35, 6783–6789. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Cooper, K.K.; Mandrell, R.E.; Louie, J.W.; Korlach, J.; Clark, T.A.; Parker, C.T.; Huynh, S.; Chain, P.S.; Ahmed, S.; Carter, M.Q.; et al. Comparative genomics of enterohemorrhagic Escherichia coli O145:H28 demonstrates a common evolutionary lineage with Escherichia coli O157:H7. BMC Genom. 2014, 15, 17. [Google Scholar] [CrossRef]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed]
- Keenswijk, W.; Dhont, E.; Raes, A.; Bael, A.; Vande Walle, J. A devastating case of diarrhea-associated hemolytic uremic syndrome associated with extensive cerebral infarction; why we need to do better. Acta Clin. Belg. Int. J. Clin. Lab. Med. 2018, 73, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.C.; Lin, C.H.; Aljuffali, I.A.; Fang, J.Y. Current pathogenic Escherichia coli foodborne outbreak cases and therapy development. Arch. Microbiol. 2017, 199, 811–825. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.; Das, S.; Ramachandran, V.G.; Dar, S.A.; Snehaa, K.; Saha, R.; Shah, D. Spectrum of diarrhoeagenic Escherichia coli in paediatric population suffering from diarrhoea and as commensals in healthy children. Indian J. Med. Microbiol. 2017, 35, 204–210. [Google Scholar] [PubMed]
- Karami, P.; Bazmamoun, H.; Sedighi, I.; Mozaffari Nejad, A.S.; Aslani, M.M.; Alikhani, M.Y. Antibacterial resistance patterns of extended spectrum β-lactamase -producing enteropathogenic Escherichia coli strains isolated from children. Arab J. Gastroenterol. 2017, 18, 206–209. [Google Scholar] [CrossRef]
- Guerra, J.A.; Romero-Herazo, Y.C.; Arzuza, O.; Gómez-Duarte, O.G. Phenotypic and Genotypic Characterization of Enterotoxigenic Escherichia coli Clinical Isolates from Northern Colombia, South America. Biomed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of campylobacter infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef]
- Nilsson, A.; Johansson, C.; Skarp, A.; Kaden, R.; Bertilsson, S.; Rautelin, H. Survival of Campylobacter jejuni and Campylobacter coli water isolates in lake and well water. APMIS 2018, 126, 762–770. [Google Scholar] [CrossRef]
- Heikema, A.P.; Jacobs, B.C.; Horst-Kreft, D.; Huizinga, R.; Kuijf, M.L.; Endtz, H.P.; Samsom, J.N.; van Wamel, W.J.B. Siglec-7 specifically recognizes Campylobacter jejuni strains associated with oculomotor weakness in Guillain-Barr? Syndrome and Miller Fisher syndrome. Clin. Microbiol. Infect. 2013, 19, E106–E112. [Google Scholar] [CrossRef]
- Amour, C.; Gratz, J.; Mduma, E.; Svensen, E.; Rogawski, E.T.; McGrath, M.; Seidman, J.C.; McCormick, B.J.J.; Shrestha, S.; Samie, A.; et al. Epidemiology and Impact of Campylobacter Infection in Children in 8 Low-Resource Settings: Results from the MAL-ED Study. Clin. Infect. Dis. 2016, 63, 1171–1179. [Google Scholar]
- Crushell, E.; Harty, S.; Sharif, F.; Bourke, B. Enteric Campylobacter: Purging Its Secrets? Pediatr. Res. 2004, 55, 3–12. [Google Scholar] [CrossRef]
- Huq, M.; Gonis, G.; Istivan, T. Development and Evaluation of a Multiplex PCR for the Detection of Campylobacter concisus and Other Campylobacter spp. from Gastroenteritis Cases. Open J. Med. Microbiol. 2014, 4, 29–37. [Google Scholar] [CrossRef][Green Version]
- Lastovica, A.J. Emerging Campylobacter spp.: The tip of the iceberg. Clin. Microbiol. Newsl. 2006, 28, 49–56. [Google Scholar] [CrossRef]
- Kimata, K.; Shima, T.; Shimizu, M.; Tanaka, D.; Isobe, J.; Gyobu, Y.; Watahiki, M.; Nagai, Y. Rapid categorization of pathogenic Escherichia coli by multiplex PCR. Microbiol. Immunol. 2005, 49, 485–492. [Google Scholar] [CrossRef]
- Crawford, S.E.; Ramani, S.; Tate, J.E.; Umesh, D.P.; Lennart, S.; Hagbom, M.; Manuel, A.F.; Greenberg, H.B.; O’Ryan, M.; Gagandeep, K.; et al. Rotavirus Infection. Nat. Rev. 2017, 3, 39–41. [Google Scholar]
- Gonzales, L.; Joffre, E.; Rivera, R.; Sjöling, Å.; Svennerholm, A.M.; Iñiguez, V. Prevalence, seasonality and severity of disease caused by pathogenic Escherichia coli in children with diarrhoea in Bolivia. J. Med. Microbiol. 2013, 62, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Bessede, E.; Delcamp, A.; Sifre, E.; Buissonniere, A.; Megraud, F. New Methods for detection of campylobacters in stool samples in comparison to culture. J. Clin. Microbiol. 2011, 49, 941–944. [Google Scholar] [CrossRef] [PubMed]
- Lawson, A.J.; Logan, J.M.J.; O’Neill, G.L.; Desai, M.; Stanley, J. Large-scale survey of Campylobacter species in human gastroenteritis by PCR and PCR-enzyme-linked immunosorbent assay. J. Clin. Microbiol. 1999, 37, 3860–3864. [Google Scholar] [CrossRef]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001940. [Google Scholar]
- Neuzil, K.M.; Kotloff, K.L. Community-acquired diarrhoea in a world with rotavirus vaccine: A glimpse into the future. Lancet Glob. Health 2015, 3, e510–e511. [Google Scholar] [CrossRef][Green Version]
- Taniuchi, M.; Sobuz, S.U.; Begum, S.; Platts-Mills, J.A.; Liu, J.; Yang, Z.; Wang, X.Q.; Petri, W.A.; Haque, R.; Houpt, E.R. Etiology of diarrhea in Bangladeshi infants in the first year of life analyzed using molecular methods. J. Infect. Dis. 2013, 208, 1794–1802. [Google Scholar] [CrossRef] [PubMed]
- Kohler, J.A.; Perkins, A.M.; Bass, W.T. Human milk versus formula after gastroschisis repair: Effects on time to full feeds and time to discharge. J. Perinatol. 2013, 33, 627–630. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Langa, S.; Reviriego, C.; Jimínez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003, 143, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Shobo, C.O.; Bester, L.A.; Baijnath, S.; Somboro, A.M.; Peer, A.K.C.; Essack, S.Y. Antibiotic resistance profiles of campylobacter species in the South Africa private health care sector. J. Infect. Dev. Ctries. 2016, 10, 1214–1221. [Google Scholar] [CrossRef] [PubMed]
- Samie, A.; Ramalivhana, J.; Igumbor, E.O.; Obi, C.L. Prevalence, haemolytic and haemagglutination activities and antibiotic susceptibility profiles of Campylobacter spp. Isolated from human diarrhoeal stools in Vhembe District, South Africa. J. Health. Popul. Nutr. 2007, 25, 406–413. [Google Scholar]
- Friedman, C.R.; Hoekstra, R.M.; Samuel, M.; Marcus, R.; Bender, J.; Shiferaw, B.; Reddy, S.; Desai Ahuja, S.; Helfrick, D.L.; Hardnett, F.; et al. Risk Factors for Sporadic Campylobacter Infection in the United States: A Case-Control Study in FoodNet Sites. Clin. Infect. Dis. 2004, 38, S285–S296. [Google Scholar] [CrossRef]
- Heikema, A.P.; Islam, Z.; Horst-Kreft, D.; Huizinga, R.; Jacobs, B.C.; Wagenaar, J.A.; Poly, F.; Guerry, P.; van Belkum, A.; Parker, C.T.; et al. Campylobacter jejuni capsular genotypes are related to Guillain–Barré syndrome. Clin. Microbiol. Infect. 2015, 21, e1–e852. [Google Scholar] [CrossRef]
- Nakamura, I.; Omori, N.; Umeda, A.; Ohkusu, K.; Matsumoto, T. First case report of fatal sepsis due to campylobacter upsaliensis. J. Clin. Microbiol. 2015, 53, 713–715. [Google Scholar] [CrossRef][Green Version]
- Ge, B.; Wang, F.; Sjölund-Karlsson, M.; McDermott, P.F. Antimicrobial resistance in Campylobacter: Susceptibility testing methods and resistance trends. J. Microbiol. Methods 2013, 95, 57–67. [Google Scholar] [CrossRef]
- Said, M.M.; El-Mohamady, H.; El-Beih, F.M.; Rockabrand, D.M.; Ismail, T.F.; Monteville, M.R.; Ahmed, S.F.; Klena, J.D.; Salama, M.S. Detection of gyrA mutation among clinical isolates of campylobacter jejuni isolated in Egypt by MAMA PCR. J. Infect. Dev. Ctries 2010, 4, 546–554. [Google Scholar] [CrossRef][Green Version]
- Mukherjee, P.; Ramamurthy, T.; Bhattacharya, M.K.; Rajendran, K.; Mukhopadhyay, A.K. Campylobacter jejuni in hospitalized patients with diarrhea, Kolkata, India. Emerg. Infect. Dis. 2013, 19, 1155–1156. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Pan, W.; Yori, P.P.; Paredes Olortegui, M.; Tilley, D.; Gregory, M.; Oberhelman, R.; Burga, R.; Chavez, C.B.; Kosek, M. Symptomatic and Asymptomatic Campylobacter Infections Associated with Reduced Growth in Peruvian Children. PLoS Negl. Trop. Dis. 2013, 7, e2036. [Google Scholar] [CrossRef] [PubMed]
- Arguello, E.; Otto, C.C.; Mead, P.; Babady, N.E. Bacteremia caused by arcobacter butzleri in an immunocompromised host. J. Clin. Microbiol. 2015, 53, 1448–1451. [Google Scholar] [CrossRef] [PubMed]
- Fernández, H.; Krause, S.; Paz Villanueva, M. Arcobacter butzleri an emerging enteropathogen: Communication of two cases with chronic diarrhea. Braz. J. Microbiol. 2004, 35, 216–218. [Google Scholar] [CrossRef]
- Van den Abeele, A.M.; Vogelaers, D.; Van Hende, J.; Houf, K. Prevalence of Arcobacter species among humans, Belgium, 2008-2013. Emerg. Infect. Dis. 2014, 20, 1731–1734. [Google Scholar] [CrossRef]
- Webb, A.L.; Boras, V.F.; Kruczkiewicz, P.; Selinger, L.B.; Taboada, E.N.; Inglis, G.D. Comparative detection and quantification of Arcobacter butzleri in stools from diarrheic and nondiarrheic people in Southwestern Alberta, Canada. J. Clin. Microbiol. 2016, 54, 1082–1088. [Google Scholar] [CrossRef]
- Douidah, L.; De Zutter, L.; Baré, J.; De Vos, P.; Vandamme, P.; Vandenberg, O.; Van Den Abeele, A.M.; Houf, K. Occurrence of putative virulence genes in Arcobacter species isolated from humans and animals. J. Clin. Microbiol. 2012, 50, 735–741. [Google Scholar] [CrossRef]
- Karadas, G.; Sharbati, S.; Hänel, I.; Messelhäußer, U.; Glocker, E.; Alter, T.; Gölz, G. Presence of virulence genes, adhesion and invasion of Arcobacter butzleri. J. Appl. Microbiol. 2013, 115, 583–590. [Google Scholar] [CrossRef]
- Sekhar, M.S.; Tumati, S.R.; Chinnam, B.K.; Kothapalli, V.S.; Sharif, N.M. Virulence gene profiles of Arcobacter species isolated from animals, foods of animal origin, and humans in Andhra Pradesh, India. Vet. World 2017, 10, 716–720. [Google Scholar] [CrossRef][Green Version]
- Skovgaard, N. Microorganisms in Foods 7: Microbiological Testing in Food Safety Management. Int. J. Food Microbiol. 2003, 89, 291–292. [Google Scholar] [CrossRef]
- Tanih, N.F.; Samie, A.; Nyathi, E.; Barrett, L.; Guerrant, R.; Bessong, P. Prevalence of diarrheagenic Escherichia coli in young children from rural South Africa: The Mal-ED cohort. Int. J. Infect. Dis. 2014, 21, 149. [Google Scholar] [CrossRef][Green Version]
- Rajeshwari, K.; Uppal, B.; Singh, R.; Tiwari, G.; Kumar Mahato, A. Multidrug-resistant enteropathogenic E. coli diarrhea in children. Am. J. Res. Commun. 2015, 3, 27–48. [Google Scholar]
- Nair, G.; Ramamurthy, T.; Bhattacharya, M.; Krishnan, T.; Ganguly, S.; Saha, D.; Rajendran, K.; Manna, B.; Ghosh, M.; Okamoto, K.; et al. Emerging trends in the etiology of enteric pathogens as evidenced from an active surveillance of hospitalized diarrhoeal patients in Kolkata, India. Gut Pathog. 2010, 2, 4. [Google Scholar] [CrossRef]
- Wani, S.A.; Nabi, A.; Fayaz, I.; Ahmad, I.; Nishikawa, Y.; Qureshi, K.; Khan, M.A.; Chowdhary, J. Investigation of diarrhoeic faecal samples for enterotoxigenic, Shiga toxin-producing and typical or atypical enteropathogenic Escherichia coli in Kashmir, India. FEMS Microbiol. Lett. 2006, 261, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Chellapandi, K.; Dutta, T.K.; Sharma, I.; Mandal, S.; Kumar, N.S.; Ralte, L. Prevalence of multi drug resistant enteropathogenic and enteroinvasive Escherichia coli isolated from children with and without diarrhea in Northeast Indian population. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 49. [Google Scholar] [CrossRef] [PubMed]
- Cabal, A.; García-Castillo, M.; Cantón, R.; Gortázar, C.; Domínguez, L.; Álvarez, J. Prevalence of Escherichia coli virulence genes in patients with diarrhea and a subpopulation of healthy volunteers in Madrid, Spain. Front. Microbiol. 2016, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Sumbana, J.; Taviani, E.; Manjate, A.; Paglietti, B.; Santona, A.; Mauro, M. Brief Original Article Genetic determinants of pathogenicity of Escherichia coli isolated from children with acute diarrhea in Maputo, Mozambique. J. Infect. Dev. Ctries 2015, 2, 661–664. [Google Scholar] [CrossRef] [PubMed]
- Hebbelstrup Jensen, B.; Stensvold, C.R.; Struve, C.; Olsen, K.E.P.; Scheutz, F.; Boisen, N.; Röser, D.; Andreassen, B.U.; Nielsen, H.V.; Schønning, K.; et al. Enteroaggregative Escherichia coli in Daycare—A 1-Year Dynamic Cohort Study. Front. Cell. Infect. Microbiol. 2016, 6, 75. [Google Scholar] [CrossRef]
- Lima, I.F.N.; Boisen, N.; Da Silva Quetz, J.; Havt, A.; De Carvalho, E.B.; Soares, A.M.; Lima, N.L.; Mota, R.M.S.; Nataro, J.P.; Guerrant, R.L.; et al. Prevalence of enteroaggregative Escherichia coli and its virulence-related genes in a case-control study among children from north-eastern Brazil. J. Med. Microbiol. 2013, 62, 683–693. [Google Scholar] [CrossRef]
- Bueris, V.; Sircili, M.P.; Taddei, C.R.; dos Santos, M.F.; Franzolin, M.R.; Martinez, M.B.; Ferrer, S.R.; Barreto, M.L.; Trabulsi, L.R. Detection of diarrheagenic Escherichia coli from children with and without diarrhea in Salvador, Bahia, Brazil. Mem. Inst. Oswaldo Cruz 2007, 102, 839–844. [Google Scholar] [CrossRef]
- Meng, C.Y.; Smith, B.L.; Bodhidatta, L.; Richard, S.A.; Vansith, K.; Thy, B.; Srijan, A.; Serichantalergs, O.; Mason, C.J. Etiology of Diarrhea in Young Children and Patterns of Antibiotic Resistance in Cambodia. Pediatr. Infect. Dis. J. 2011, 30, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Paschke, C.; Apelt, N.; Fleischmann, E.; Perona, P.; Walentiny, C.; Löscher, T.; Herbinger, K.H. Controlled study on enteropathogens in travellers returning from the tropics with and without diarrhoea. Clin. Microbiol. Infect. 2011, 17, 1194–1200. [Google Scholar] [CrossRef] [PubMed]
- Rogawski, E.T.; Guerrant, R.L.; Havt, A.; Lima, I.F.N.; Medeiros, P.H.Q.S.; Seidman, J.C.; McCormick, B.J.J.; Babji, S.; Hariraju, D.; Bodhidatta, L.; et al. Epidemiology of enteroaggregative Escherichia coli infections and associated outcomes in the MAL-ED birth cohort. PLoS Negl. Trop. Dis. 2017, 11, e0005798. [Google Scholar] [CrossRef]
- Contreras, T.J.O. Enteropathogenic, E. coli (EPEC) infection in children. Curr. Opin. Infect. Dis. 2011, 24, 478–483. [Google Scholar]
- Okeke, I.N. Regional Review Diarrheagenic Escherichia coli in sub-Saharan Africa: Status, uncertainties and necessities. J. Infect. Dev. Ctries 2009, 3, 817–842. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.B.; Nataro, J.P.; Bernstein, D.I.; Hawkins, J.; Roberts, N.; Staat, M.A. Prevalence of diarrheagenic Escherichia coli in acute childhood enteritis: A prospective controlled study. J. Pediatr. 2005, 146, 54–61. [Google Scholar] [CrossRef]
- Damian, M.; Tatu-Chiţoiu, D.; Usein, C.-R.; Oprişan, G.; Palade, A.-M.; Dinu, S.; Szmal, C.; Ciontea, S.A.; Ceciu, S.; Condei, M.; et al. Laboratory diagnosis of infectious diarrhoea syndrome; a three years study in two hospitals of infectious diseases. Roum. Arch. Microbiol. Immunol. 2009, 68, 89–94. [Google Scholar] [PubMed]
- Usein, C.R.; Tatu-Chitoiu, D.; Ciontea, S.; Condei, M.; Damian, M. Escherichia coli pathotypes associated with diarrhea in Romanian children younger than 5 years of age. Jpn. J. Infect. Dis. 2009, 62, 289–293. [Google Scholar]
- Ethelberg, S.; Olsen, K.E.P.; Scheutz, F.; Jensen, C.; Schiellerup, P.; Engberg, J.; Petersen, A.M.; Olesen, B.; Gerner-Smidt, P.; Molbak, K. Virulence Factors for Hemolytic Uremic Syndrome, Denmark. In Emerging Infectious Diseases; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2004; Volume 10, pp. 842–847. [Google Scholar]
- Lynn, R.M.; O’Brien, S.J.; Taylor, C.M.; Adak, G.K.; Chart, H.; Cheasty, T.; Coia, J.E.; Gillespie, I.A.; Locking, M.E.; Reilly, W.J.; et al. Childhood hemolytic uremic syndrome, United Kingdom and Ireland. Emerg. Infect. Dis. 2005, 11, 590–596. [Google Scholar] [CrossRef]
- Chokoshvili, O.; Lomashvili, K.; Malakmadze, N.; Geleishvil, M.; Brant, J.; Imnadze, P.; Chitadze, N.; Tevzadze, L.; Chanturia, G.; Tevdoradze, T.; et al. Investigation of an outbreak of bloody diarrhea complicated with hemolytic uremic syndrome. J. Epidemiol. Glob. Health 2014, 4, 249–259. [Google Scholar] [CrossRef]
- Gyles, C.L. Shiga toxin-producing an overview. J. Anim. Sci. 2007, 85, E45. [Google Scholar] [CrossRef] [PubMed]
- Friesema, I.H.M.; Keijzer-Veen, M.G.; Koppejan, M.; Schipper, H.S.; van Griethuysen, A.J.; Heck, M.E.O.C.; van Pelt, W. Hemolytic uremic syndrome associated with Escherichia coli O8:H19 and Shiga toxin 2f gene. Emerg. Infect. Dis. 2015, 21, 168–169. [Google Scholar] [CrossRef] [PubMed]
- Hosseini Nave, H.; Mansouri, S.; Taati Moghadam, M.; Moradi, M. Virulence Gene Profile and Multilocus Variable-Number Tandem-Repeat Analysis (MLVA) of Enteroinvasive Escherichia coli (EIEC) Isolates From Patients with Diarrhea in Kerman, Iran. Jundishapur J. Microbiol. 2016, 9, e33529. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, P.; Kargar, M.; Doosti, A.; Mardaneh, J.; Ghorbani Dalini, S.; Dehyadegari, M.A. Real Time PCR for Characterization of Enteroinvasive E. coli (EIEC) in Children with Diarrhea in Shiraz. Ann. Color. Res. 2015, 2, e22721. [Google Scholar] [CrossRef]
- McLamb, B.L.; Gibson, A.J.; Overman, E.L.; Stahl, C.; Moeser, A.J. Early Weaning Stress in Pigs Impairs Innate Mucosal Immune Responses to Enterotoxigenic E. coli Challenge and Exacerbates Intestinal Injury and Clinical Disease. PLoS ONE 2013, 8, e59838. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.M.; Carvalho-Costa, F.A.; de Mello Volotão, E.; Rose, T.L.; da Silva, M.F.M.; Fialho, A.M.; Assis, R.M.S.; da Silva Ribeiro de Andrade, J.; Sá, A.C.C.; Zeller, M.; et al. Prevalence and genomic characterization of G2P[4] group A rotavirus strains during monovalent vaccine introduction in Brazil. Infect. Genet. Evol. 2014, 28, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Page, N.A.; Seheri, L.M.; Groome, M.J.; Moyes, J.; Walaza, S.; Mphahlele, J.; Kahn, K.; Kapongo, C.N.; Zar, H.J.; Tempia, S.; et al. Temporal association of rotavirus vaccination and genotype circulation in South Africa: Observations from 2002 to 2014. Vaccine 2017, 36, 7231–7237. [Google Scholar] [CrossRef]
- Seheri, L.M.; Page, N.A.; Mawela, M.P.B.; Mphahlele, M.J.; Steele, A.D. Rotavirus vaccination within the South African Expanded Programme on Immunisation. Vaccine 2012, 30, C14–C20. [Google Scholar] [CrossRef]
- Mattison, K.; Sebunya, T.K.; Shukla, A.; Noliwe, L.N.; Bidawid, S. Molecular detection and characterization of noroviruses from children in Botswana. J. Med. Virol. 2010, 82, 321–324. [Google Scholar] [CrossRef]
- Abugalia, M.; Cuevas, L.; Kirby, A.; Dove, W.; Nakagomi, O.; Nakagomi, T.; Kara, M.; Gweder, R.; Smeo, M.; Cunliffe, N. Clinical features and molecular epidemiology of rotavirus and norovirus infections in Libyan children. J. Med. Virol. 2011, 83, 1849–1856. [Google Scholar] [CrossRef]
- Mans, J.; Murray, T.Y.; Kiulia, N.M.; Mwenda, J.M.; Musoke, R.N.; Taylor, M.B. Human caliciviruses detected in HIV-seropositive children in Kenya. J. Med. Virol. 2014, 86, 75–81. [Google Scholar] [CrossRef]
- Mans, J.; de Villiers, J.C.; du Plessis, N.M.; Avenant, T.; Taylor, M.B. Emerging norovirus GII.4 2008 variant detected in hospitalised paediatric patients in South Africa. J. Clin. Virol. 2010, 49, 258–264. [Google Scholar] [CrossRef]
- Siebenga, J.J.; Vennema, H.; Renckens, B.; de Bruin, E.; van der Veer, B.; Siezen, R.J.; Koopmans, M. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 2007, 81, 9932–9941. [Google Scholar] [CrossRef] [PubMed]
- Moyo, S.J.; Gro, N.; Matee, M.I.; Kitundu, J.; Myrmel, H.; Mylvaganam, H.; Maselle, S.Y.; Langeland, N. Age specific aetiological agents of diarrhoea in hospitalized children aged less than five years in Dar es Salaam, Tanzania. BMC Pediatr. 2011, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States-Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Nimri, L.F.; Elnasser, Z.; Batchoun, R. Polymicrobial infections in children with diarrhoea in a rural area of Jordan. FEMS Immunol. Med. Microbiol. 2004, 42, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.H.; Saleha, A.A.; Zunita, Z.; Murugaiyah, M.; Aliyu, A.B.; Jafri, N.; Prevalence, Distribution and Antibiotic Resistance of Emergent Arcobacter spp. from Clinically Healthy Cattle and Goats. Transbound. Emerg. Dis. 2013, 60, 9–16. [Google Scholar] [CrossRef][Green Version]
- Best, E.L.; Powell, E.J.; Swift, C.; Grant, K.A.; Frost, J.A. Applicability of a rapid duplex real-time PCR assay for speciation of Campylobacter jejuni and Campylobacter coli directly from culture plates. FEMS Microbiol. Lett. 2003, 229, 237–241. [Google Scholar] [CrossRef]
- Abia, A.L.K.; Ubomba-Jaswa, E.; Momba, M.N.B. Occurrence of diarrhoeagenic Escherichia coli virulence genes in water and bed sediments of a river used by communities in Gauteng, South Africa. Environ. Sci. Pollut. Res. 2016, 23, 15665–15674. [Google Scholar] [CrossRef]
- Chukwu, M.O.; Abia, A.L.K.; Ubomba-Jaswa, E.; Obi, L.; Dewar, J.B. Characterization and Phylogenetic Analysis of Campylobacter Species Isolated from Paediatric Stool and Water Samples in the Northwest Province, South Africa. Int. J. Environ. Res. Public Health 2019, 16, 2205. [Google Scholar] [CrossRef]
- Houf, K.; Tutenel, A.; De Zutter, L.; Van Hoof, J.; Vandamme, P. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii. FEMS Microbiol. Lett. 2000, 193, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Ghunaim, H.; Behnke, J.M.; Aigha, I.; Sharma, A.; Doiphode, S.H.; Deshmukh, A.; Abu-Madi, M.M. Analysis of resistance to antimicrobials and presence of virulence/stress response genes in Campylobacter isolates from patients with severe diarrhoea. PLoS ONE 2015, 10, e0119268. [Google Scholar] [CrossRef] [PubMed]
- Abia, A.L.K.; Ubomba-Jaswa, E.; Momba, M.N.B. High prevalence of multiple-antibiotic-resistant (MAR) Escherichia coli in river bed sediments of the Apies River, South Africa. Environ. Monit. Assess. 2015, 187, 652. [Google Scholar] [CrossRef] [PubMed]
- Chukwu, M.O.; Abia, A.L.K.; Ubomba-Jaswa, E.; Obi, L.C.; Dewar, J.B. Antibiotic Resistance Profile and Clonality of E. coli Isolated from Water and Paediatric Stool Samples in the North-West, Province South Africa. J. Pure Appl. Microbiol. 2019, 13, 517–530. [Google Scholar] [CrossRef]
- Abia, A.L.K.; Schaefer, L.; Ubomba-Jaswa, E.; Le Roux, W. Abundance of pathogenic escherichia coli virulence-associated genes in well and borehole water used for domestic purposes in a peri-urban community of South Africa. Int. J. Environ. Res. Public Health 2017, 14, 320. [Google Scholar] [CrossRef] [PubMed]
- Onori, M.; Coltella, L.; Mancinelli, L.; Argentieri, M.; Menichella, D.; Villani, A.; Grandin, A.; Valentini, D.; Raponi, M.; Russo, C. Evaluation of a multiplex PCR assay for simultaneous detection of bacterial and viral enteropathogens in stool samples of paediatric patients. Diagn. Microbiol. Infect. Dis. 2014, 79, 149–154. [Google Scholar] [CrossRef]
| Age (Months) | Total | Gender | Exclusive Breastfeeding | Mixed Feeding | Diarrhoea | Non-Diarrhoea | Bloody Diarrhoea | Vomiting | Fever | |
|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | |||||||||
| 0–12 | 414 | 206 | 208 | 184 | 230 | 266 | 148 | 61 | 122 | 167 |
| 13–24 | 81 | 48 | 33 | 0 | 81 | 75 | 6 | 19 | 32 | 37 |
| 25–36 | 9 | 3 | 6 | 0 | 9 | 8 | 1 | 1 | 4 | 4 |
| 37–48 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| Sub total | 505 | 257 | 248 | 184 | 321 | 350 | 155 | 82 | 159 | 209 |
| Aetiologic agent (n) | Diarrhoea (%) | Fever (%) | Vomiting (%) | Bloody Diarrhoea (%) |
|---|---|---|---|---|
| Campylobacter (274) | 152 (55.5) | 150 (54.7) | 109 (39.7) | 32 (11.6) |
| E. coli (235) | 151 (64.2) | 99 (42) | 95 (40.4) | 36 (15.3) |
| Rotavirus (118) | 68 (57.6) | 47 (39.8) | 35 (29.6) | 0 |
| Norovirus (101) | 77 (76.2) | 55 (54.4) | 40 (39.6) | 0 |
| Arcobacter (80) | 56 (70) | 46 (57.7) | 37 (64.2) | 20 (25) |
| Parameters | Stx1 | flicH | eaeA | eagg | ipaH | Stx2 | ST | LT |
|---|---|---|---|---|---|---|---|---|
| Exclusive breastfeeding | 2 | 4 | 7 | 6 | 3 | 9 | 6 | 0 |
| mix-feeding | 2 | 10 | 27 | 36 | 6 | 20 | 14 | 5 |
| Diarrhoea | 15 | 20 | 22 | 32 | 24 | 22 | 17 | 6 |
| Non-diarrhoea | 2 | 8 | 11 | 10 | 3 | 7 | 3 | 3 |
| Bloody diarrhoea | 6 | 6 | 8 | 1 | 6 | 3 | 4 | 2 |
| Fever | 13 | 13 | 13 | 19 | 18 | 12 | 11 | 6 |
| No-fever | 0 | 11 | 20 | 23 | 4 | 17 | 9 | 3 |
| Vomiting | 12 | 14 | 20 | 15 | 17 | 9 | 11 | 6 |
| No-vomiting | 0 | 10 | 23 | 27 | 5 | 20 | 9 | 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chukwu, M.O.; Abia, A.L.K.; Ubomba-Jaswa, E.; Dewar, J.B.; Obi, C.L. Mixed Aetiology of Diarrhoea in Infants Attending Clinics in the North-West Province of South Africa: Potential for Sub-Optimal Treatment. Pathogens 2020, 9, 198. https://doi.org/10.3390/pathogens9030198
Chukwu MO, Abia ALK, Ubomba-Jaswa E, Dewar JB, Obi CL. Mixed Aetiology of Diarrhoea in Infants Attending Clinics in the North-West Province of South Africa: Potential for Sub-Optimal Treatment. Pathogens. 2020; 9(3):198. https://doi.org/10.3390/pathogens9030198
Chicago/Turabian StyleChukwu, Martina O., Akebe Luther King Abia, Eunice Ubomba-Jaswa, John Barr Dewar, and C.L. Obi. 2020. "Mixed Aetiology of Diarrhoea in Infants Attending Clinics in the North-West Province of South Africa: Potential for Sub-Optimal Treatment" Pathogens 9, no. 3: 198. https://doi.org/10.3390/pathogens9030198
APA StyleChukwu, M. O., Abia, A. L. K., Ubomba-Jaswa, E., Dewar, J. B., & Obi, C. L. (2020). Mixed Aetiology of Diarrhoea in Infants Attending Clinics in the North-West Province of South Africa: Potential for Sub-Optimal Treatment. Pathogens, 9(3), 198. https://doi.org/10.3390/pathogens9030198






