Immune Phenotype and Functionality of Mtb-Specific T-Cells in HIV/TB Co-Infected Patients on Antiretroviral Treatment
Abstract
1. Introduction
2. Results
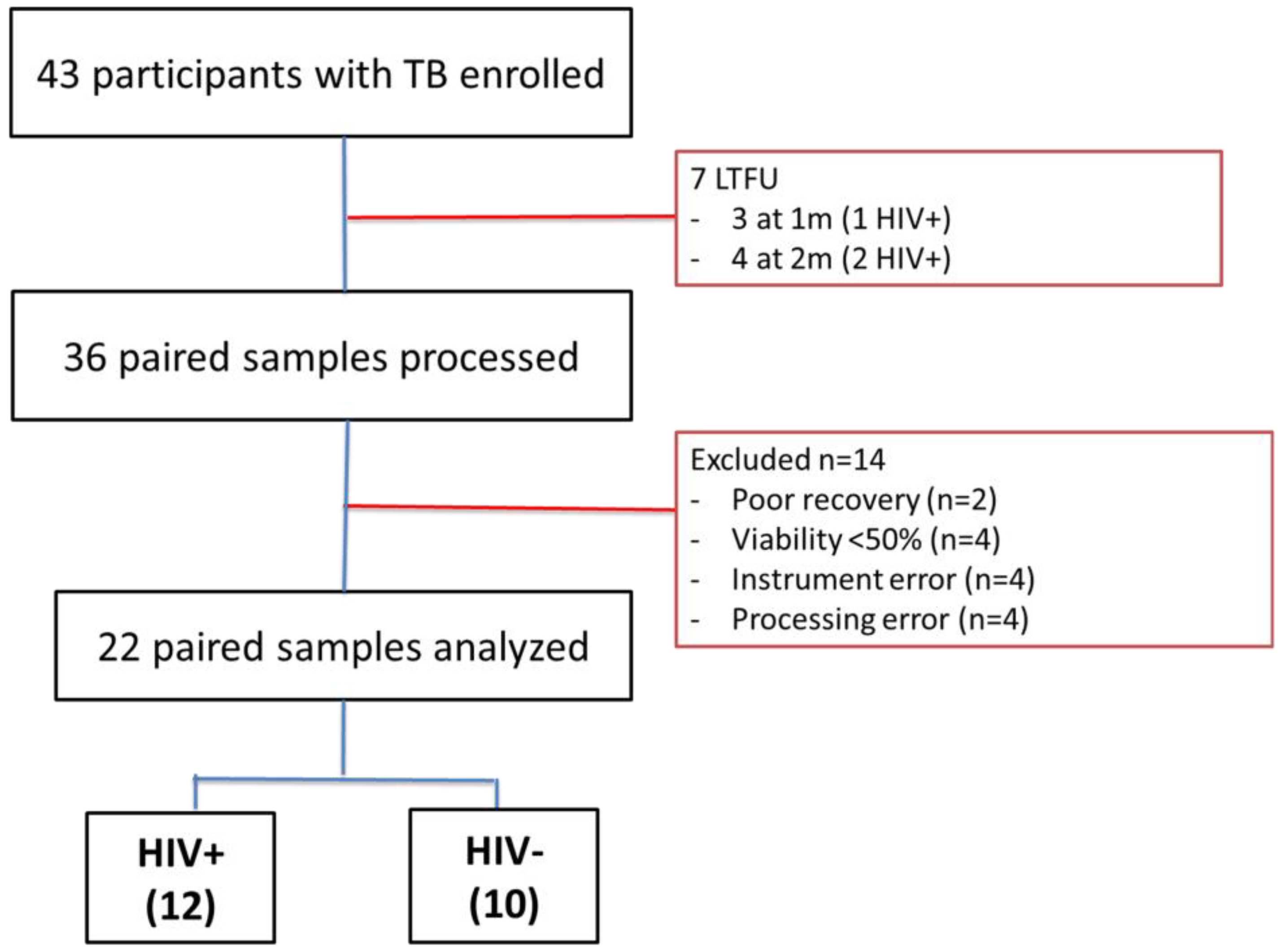
2.1. Cohort Characteristics
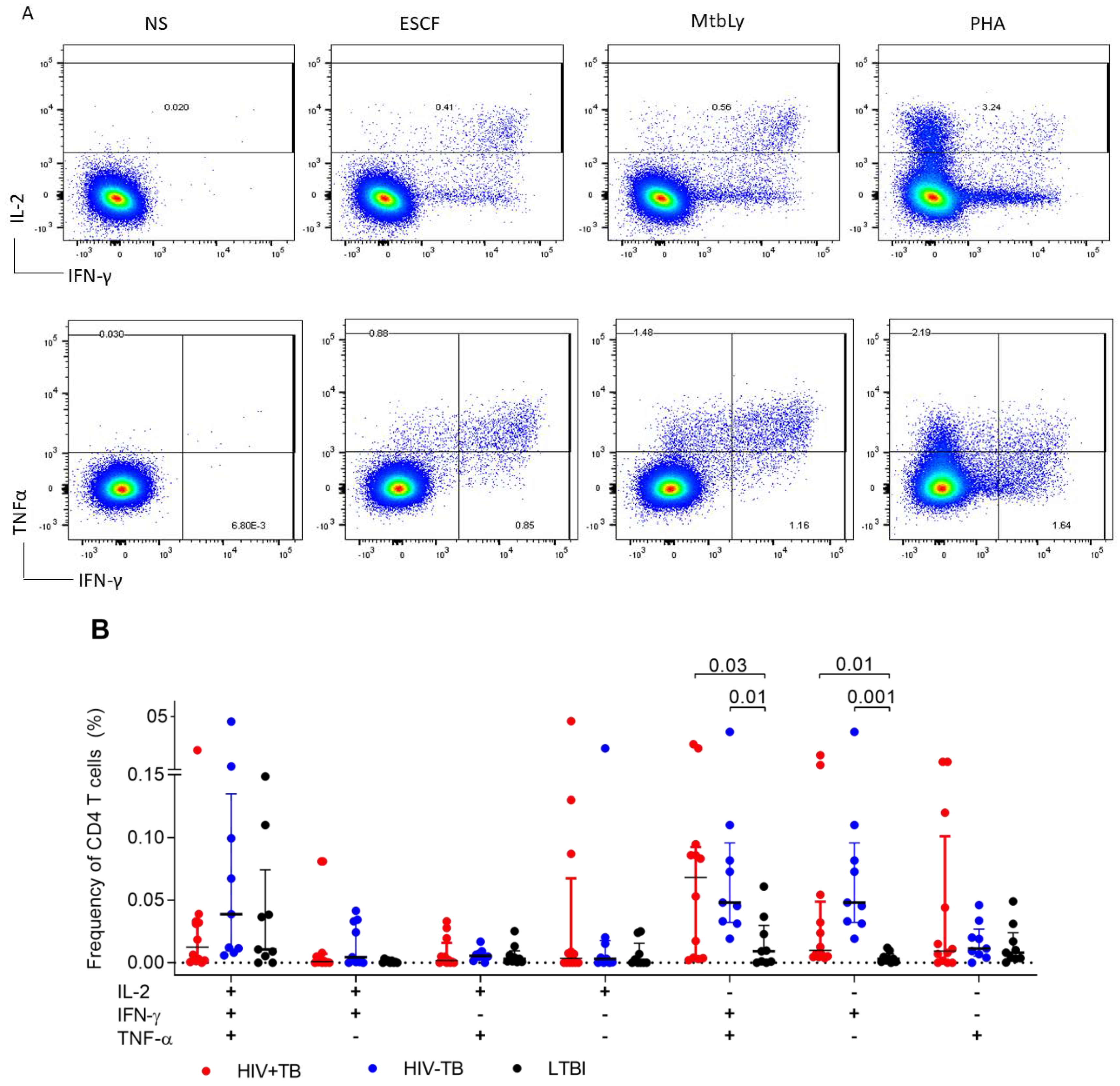
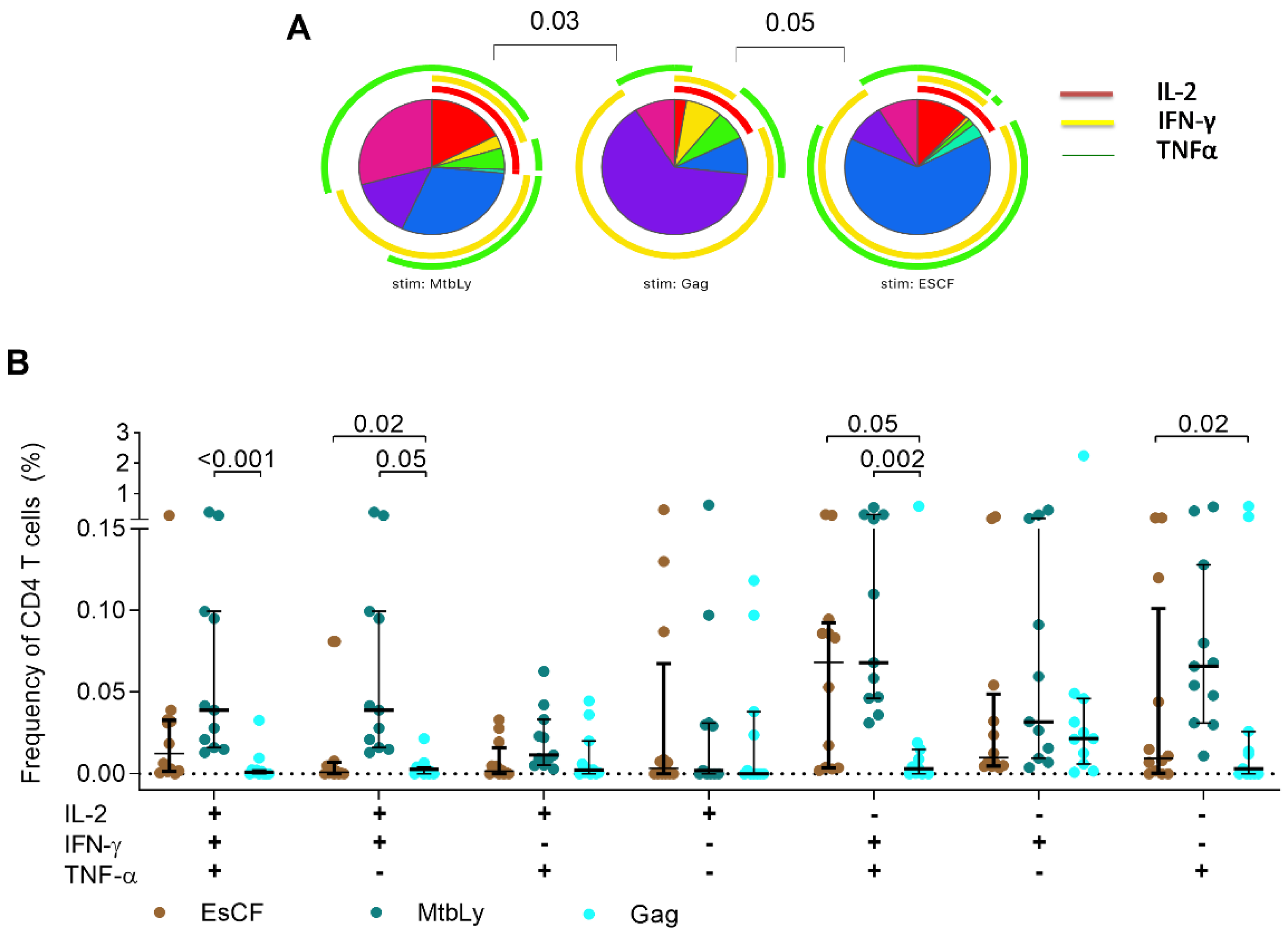
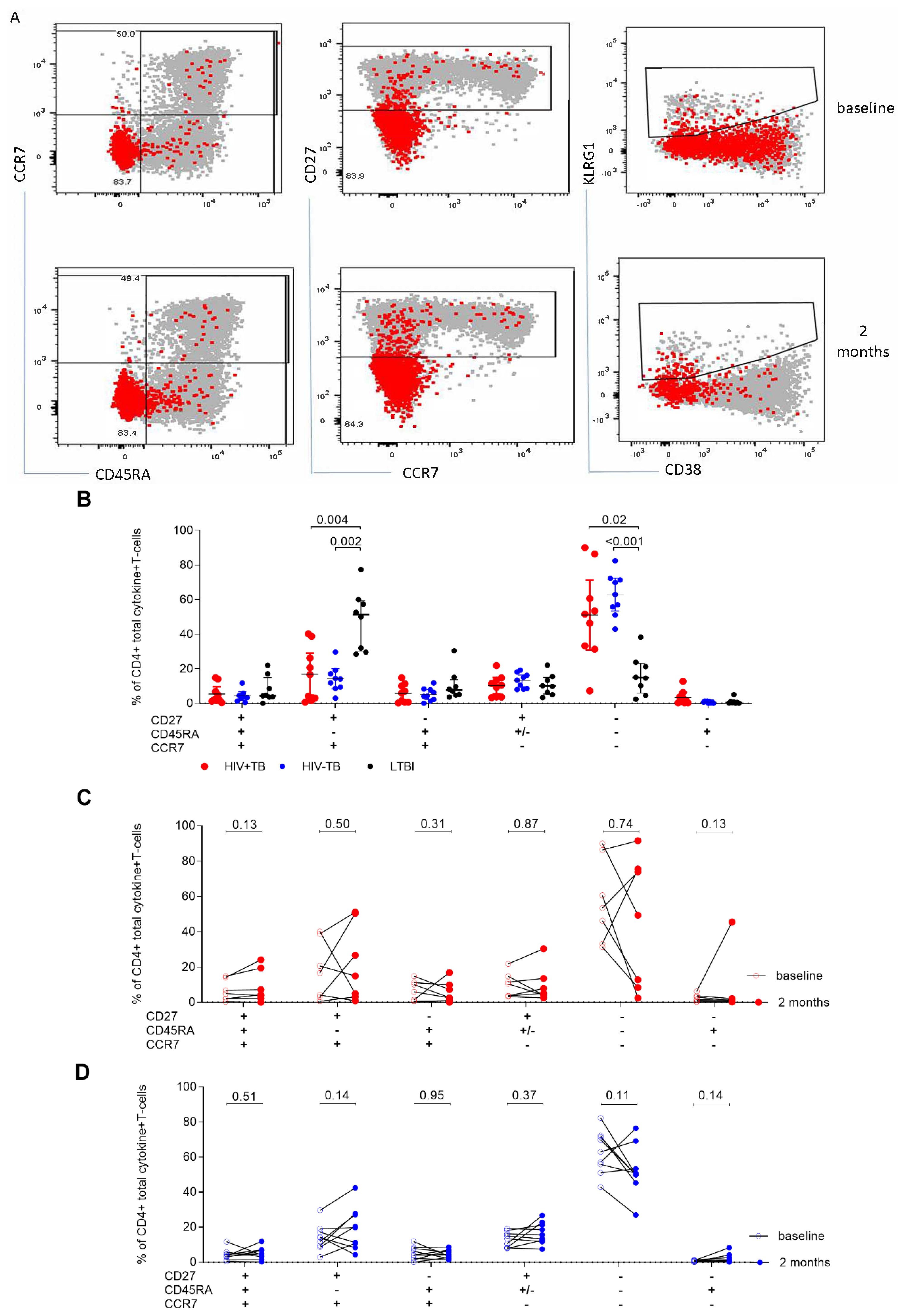
2.2. Single IFN-γ and Dual IFN-γ/TNFα Response Characterizes Active Tuberculosis Regardless of HIV Status
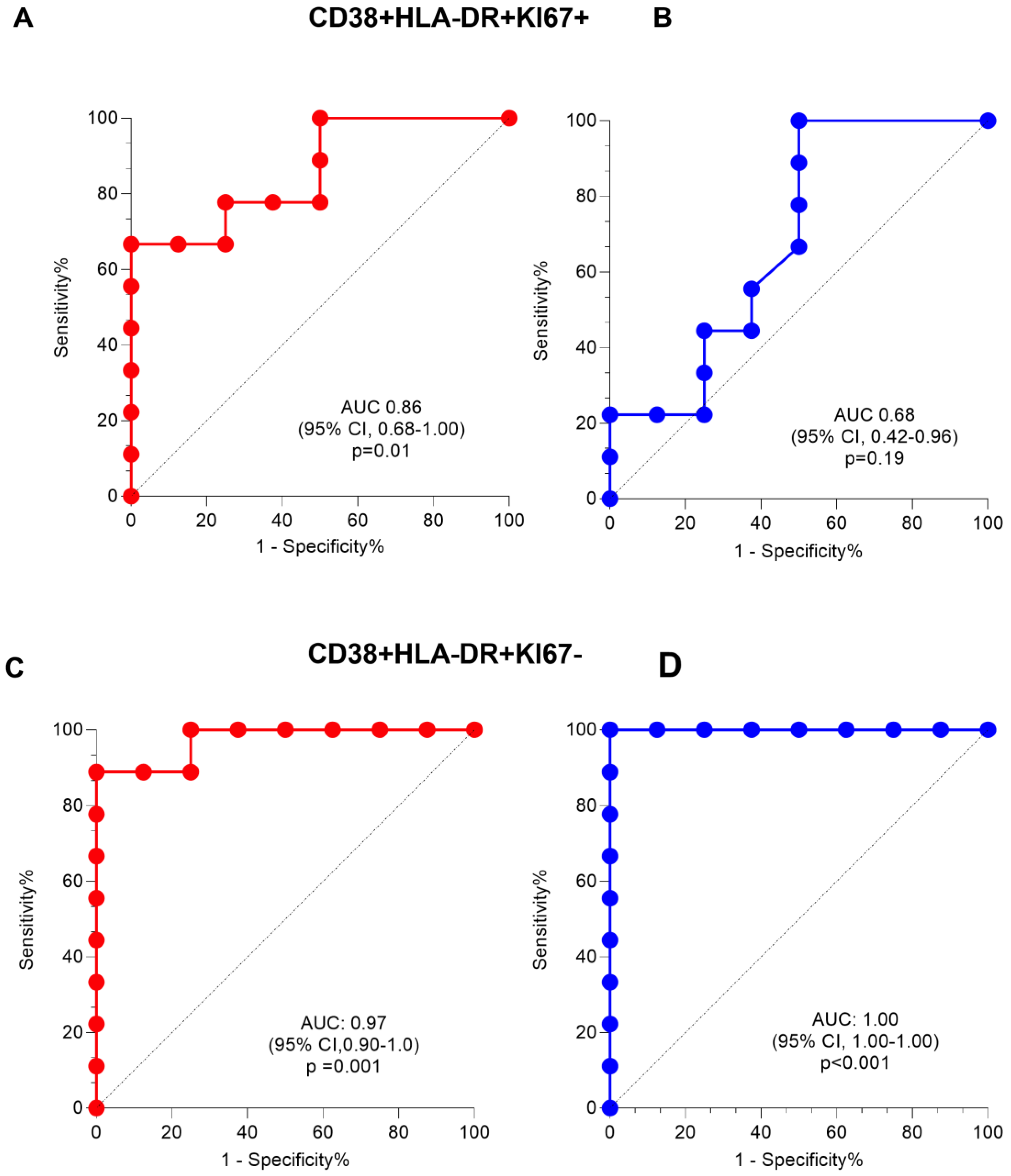
2.3. Frequencies of CD38+HLA-DR+CD4+ T-cells Are Elevated in Active TB Participants Regardless of HIV Status and Decline after Two Months of TB Treatment
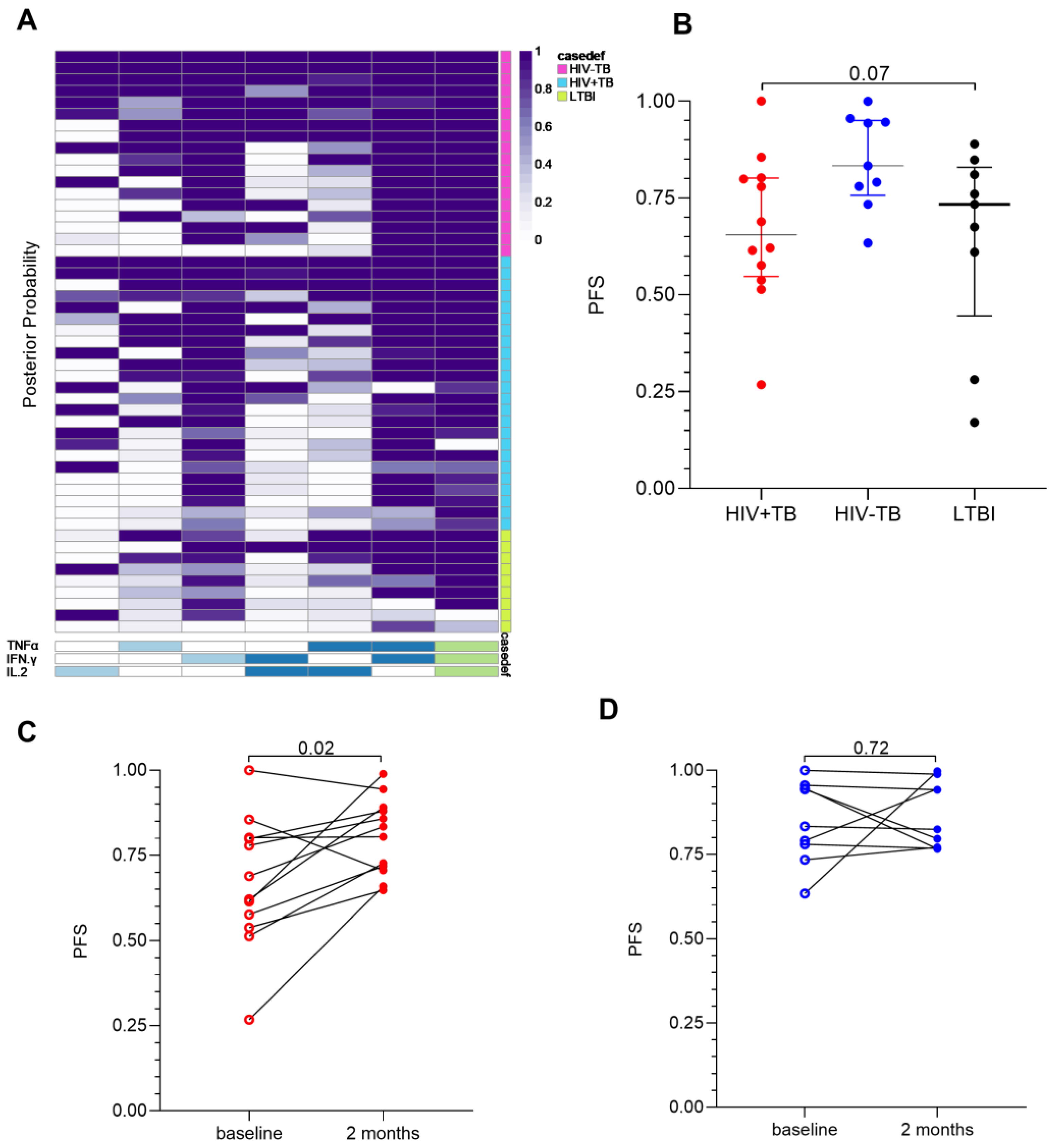
2.4. A High CD27MFI Ratio Discriminates Active TB from Latent TB and Declines after Two Months of TB Treatment
3. Discussion
4. Materials and Methods
4.1. Study Participants
4.2. Laboratory Methods
4.2.1. Cell Activation
4.2.2. Cell Staining
4.3. Statistical Analysis
4.4. Ethical Considerations
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Global TB Report. 2019. Available online: https://www.who.int/tb/publications/global_report/en/ (accessed on 17 October 2019).
- Getahun, H.; Gunneberg, C.; Granich, R.; Nunn, P. HIV infection-associated tuberculosis: The epidemiology and the response. Clin. Infect. Dis. 2010, 3, S201–S207. [Google Scholar] [CrossRef] [PubMed]
- Dye, C.; Williams, B.G. Tuberculosis decline in populations affected by HIV: A retrospective study of 12 countries in the WHO African Region. Bull. World Health Organ. 2019, 97, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Rice, B.; Brown, A.E.; Thomas, H.L.; Zenner, D.; Anderson, L.; Pedrazzoli, D.; Pozniak, A.; Abubakar, I.; Delpech, V.; et al. Does antiretroviral therapy reduce HIV-associated tuberculosis incidence to background rates? A national observational cohort study from England, Wales, and Northern Ireland. Lancet HIV 2015, 2, e243–e251. [Google Scholar] [CrossRef]
- Gupta, A.; Wood, R.; Kaplan, R.; Bekker, L.G.; Lawn, S.D. Tuberculosis incidence rates during 8 years of follow-up of an antiretroviral treatment cohort in South Africa: Comparison with rates in the community. PLoS ONE 2012, 7, e34156. [Google Scholar] [CrossRef] [PubMed]
- Geldmacher, C.; Zumla, A.; Hoelscher, M. Interaction between HIV and Mycobacterium tuberculosis: HIV-1-induced CD4 T-cell depletion and the development of active tuberculosis. Cur. Opin. HIV AIDS 2012, 7, 268–275. [Google Scholar] [CrossRef]
- Geldmacher, C.; Schuetz, A.; Ngwenyama, N.; Casazza, J.P.; Sanga, E.; Saathoff, E.; Boehme, C.; Geis, S.; Maboko, L.; Singh, M.; et al. Early depletion of Mycobacterium tuberculosis-specific T helper 1 cell responses after HIV-1 infection. J. Infect. Dis. 2008, 198, 1590–1598. [Google Scholar] [CrossRef]
- Tonaco, M.M.; Moreira, J.D.; Nunes, F.F.C.; Loures, C.M.G.; Souza, L.R.; Martins, J.M.; Silva, H.R.; Porto, A.H.R.; Toledo, V.; Miranda, S.S.; et al. Evaluation of profile and functionality of memory T cells in pulmonary tuberculosis. Immunol. Lett. 2017, 192, 52–60. [Google Scholar] [CrossRef]
- Riou, C.; Tanko, R.F.; Soares, A.P.; Masson, L.; Werner, L.; Garrett, N.J.; Samsunder, N.; Karim, Q.A.; Karim, S.S.A.; Burgers, W.A. Restoration of CD4+ responses to copathogens in HIV-infected individuals on antiretroviral therapy is dependent on t cell memory phenotype. J. Immunol. 2015, 195, 2273–2281. [Google Scholar] [CrossRef]
- Latorre, I.; Fernandez-Sanmartin, M.A.; Muriel-Moreno, B.; Villar-Hernandez, R.; Vila, S.; Souza-Galvao, M.L.; Stojanovic, Z.; Jimenez-Fuentes, M.A.; Centeno, C.; Ruiz-Manzano, J.; et al. Study of CD27 and CCR4 markers on specific CD4(+) T-Cells as immune tools for active and latent tuberculosis management. Front. Immunol. 2018, 9, 3094. [Google Scholar] [CrossRef]
- Portevin, D.; Moukambi, F.; Clowes, P.; Bauer, A.; Chachage, M.; Ntinginya, N.E.; Mfinanga, E.; Said, K.; Haraka, F.; Rachow, A.; et al. Assessment of the novel t-cell activation marker-tuberculosis assay for diagnosis of active tuberculosis in children: A prospective proof-of-concept study. Lancet Infect. Dis. 2014, 14, 931–938. [Google Scholar] [CrossRef]
- Adekambi, T.; Ibegbu, C.C.; Cagle, S.; Kalokhe, A.S.; Wang, Y.F.; Hu, Y.; Day, C.L.; Ray, S.M.; Rengarajan, J. Biomarkers on patient T cells diagnose active tuberculosis and monitor treatment response. J. Clin. Investig. 2015, 125, 1827–1838. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Oni, T.; Gideon, H.P.; Goliath, R.; Wilkinson, R.J.; Riou, C. Activation Profile of Mycobacterium tuberculosis-Specific CD4(+) T Cells Reflects Disease Activity Irrespective of HIV Status. Am. J. Respir. Crit. Care Med. 2016, 193, 1307–1310. [Google Scholar] [CrossRef] [PubMed]
- Riou, C.; Berkowitz, N.; Goliath, R.; Burgers, W.A.; Wilkinson, R.J. Analysis of the phenotype of Mycobacterium tuberculosis-specific CD4+ T cells to discriminate latent from active tuberculosis in HIV-uninfected and HIV-infected individuals. Front. Immunol. 2017, 8, 968. [Google Scholar] [CrossRef] [PubMed]
- Musvosvi, M.; Duffy, D.; Filander, E.; Africa, H.; Mabwe, S.; Jaxa, L.; Bilek, N.; Llibre, A.; Rouilly, V.; Hatherill, M.; et al. T-cell biomarkers for diagnosis of tuberculosis: Candidate evaluation by a simple whole blood assay for clinical translation. Eur. Respir. J. Off. J. Eur. Soc. Clin. Respir. Physiol. 2018, 51, 1800153. [Google Scholar] [CrossRef] [PubMed]
- Chun, T.-W.; Justement, J.S.; Sanford, C.; Hallahan, C.W.; Planta, M.A.; Loutfy, M.; Kottilil, S.; Moir, S.; Kovacs, C.; Fauci, A.S. Relationship between the frequency of HIV-specific CD8+T cells and the level of CD38+CD8+T cells in untreated HIV-infected individuals. Proc. Nat. Acad. Sci. USA 2004, 101, 2464–2469. [Google Scholar] [CrossRef]
- Fletcher, H.A.; Snowden, M.A.; Landry, B.; Rida, W.; Satti, I.; Harris, S.A.; Matsumiya, M.; Tanner, R.; O’Shea, M.K.; Dheenadhayalan, V.; et al. T-cell activation is an immune correlate of risk in BCG vaccinated infants. Nat. Commun. 2016, 7, 11290. [Google Scholar] [CrossRef]
- Tanko, R.F.; Soares, A.P.; Masson, L.; Garrett, N.J.; Samsunder, N.; Karim, Q.A.; Karim, S.S.A.; Riou, C.; Burgers, W.A. Residual T cell activation and skewed CD8+ T cell memory differentiation despite antiretroviral therapy-induced HIV suppression. Clin. Immunol. 2018, 195, 127–138. [Google Scholar] [CrossRef]
- Lin, L.; Finak, G.; Ushey, K.; Seshadri, C.; Hawn, T.R.; Frahm, N.; Scriba, T.J.; Mahomed, H.; Hanekom, W.; Bart, P.-A.; et al. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nat. Biotechnol. 2015, 33, 610–616. [Google Scholar] [CrossRef]
- Schuetz, A.; Haule, A.; Reither, K.; Ngwenyama, N.; Rachow, A.; Meyerhans, A.; Maboko, L.; Koup, R.A.; Hoelscher, M.; Geldmacher, C. Monitoring CD27 expression to evaluate Mycobacterium tuberculosis activity in HIV-1 infected individuals in vivo. PLoS ONE 2011, 6, e27284. [Google Scholar] [CrossRef]
- Riou, C.; Bunjun, R.; Muller, T.L.; Kiravu, A.; Ginbot, Z.; Oni, T.; Goliath, R.; Wilkinson, R.J.; Burgers, W.A. Selective reduction of IFN-gamma single positive mycobacteria-specific CD4+ T cells in HIV-1 infected individuals with latent tuberculosis infection. Tuberculosis 2016, 101, 25–30. [Google Scholar] [CrossRef]
- Sutherland, R.; Yang, H.; Scriba, T.J.; Ondondo, B.; Robinson, N.; Conlon, C.; Suttill, A.; McShane, H.; Fidler, S.; McMichael, A.; et al. Impaired IFN-gamma-secreting capacity in mycobacterial antigen-specific CD4 T cells during chronic HIV-1 infection despite long-term HAART. AIDS 2006, 20, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Shi, R.; Liu, X.; Yuan, X.; Zheng, S.; Zhang, G.; Wang, W.; Wang, J.; England, K.; Via, L.E.; et al. Interferon-gamma response to the treatment of active pulmonary and extra-pulmonary tuberculosis. Int. J. Tuberc. Lung Dis 2017, 21, 1145–1149. [Google Scholar] [CrossRef] [PubMed]
- Harari, A.; Rozot, V.; Enders, F.B.; Perreau, M.; Stalder, J.M.; Nicod, L.P.; Cavassini, M.; Calandra, T.; Blanchet, C.L.; Jaton, K.; et al. Dominant TNF-alpha+ Mycobacterium tuberculosis-specific CD4+ T cell responses discriminate between latent infection and active disease. Nat. Med. 2011, 17, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Chiacchio, T.; Petruccioli, E.; Vanini, V.; Cuzzi, G.; Pinnetti, C.; Sampaolesi, A.; Antinori, A.; Girardi, E.; Goletti, D. Polyfunctional T-cells and effector memory phenotype are associated with active TB in HIV-infected patients. J. Infect. 2014, 69, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Geldmacher, C.; Koup, R.A. Pathogen-specific T cell depletion and reactivation of opportunistic pathogens in HIV infection. Trends Immunol. 2012, 33, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Chiacchio, T.; Petruccioli, E.; Vanini, V.; Cuzzi, G.; La Manna, M.P.; Orlando, V.; Pinnetti, C.; Sampaolesi, A.; Antinori, A.; Caccamo, N.; et al. Impact of antiretroviral and tuberculosis therapies on CD4+ and CD8+ HIV/M. tuberculosis-specific T-cell in co-infected subjects. Immunol. Lett. 2018, 198, 33–43. [Google Scholar] [CrossRef]
- Ahmed, M.I.M.; Ntinginya, N.E.; Kibiki, G.; Mtafya, B.A.; Semvua, H.; Mpagama, S.; Mtabho, C.; Saathoff, E.; Held, K.; Loose, R.; et al. Phenotypic Changes on Mycobacterium Tuberculosis-Specific CD4 T Cells as Surrogate Markers for Tuberculosis Treatment Efficacy. Front. Immunol. 2018, 9, 2247. [Google Scholar] [CrossRef]
- Saharia, K.K.; Petrovas, C.; Ferrando-Martinez, S.; Leal, M.; Luque, R.; Ive, P.; Luetkemeyer, A.; Havlir, D.; Koup, R.A. Tuberculosis therapy modifies the cytokine profile, maturation state, and expression of inhibitory molecules on Mycobacterium tuberculosis-Specific CD4+ T-Cells. PLoS ONE 2016, 11, e0158262. [Google Scholar] [CrossRef]
- Nikitina, I.Y.; Kondratuk, N.A.; Kosmiadi, G.A.; Amansahedov, R.B.; Vasilyeva, I.A.; Ganusov, V.V.; Lyadova, I.V. Mtb-Specific CD27low CD4 T Cells as markers of lung tissue destruction during pulmonary tuberculosis in humans. PLoS ONE 2012, 7, e43733. [Google Scholar] [CrossRef]
- Burgers, W.A.; Riou, C.; Mlotshwa, M.; Maenetje, P.; De Assis Rosa, D.; Brenchley, J.; Mlisana, K.; Douek, D.C.; Koup, R.; Roederer, M.; et al. Association of HIV-specific and total CD8+ T memory phenotypes in subtype C HIV-1 infection with viral set point. J. Immunol 2009, 182, 4751–4761. [Google Scholar] [CrossRef]
- Lee, J.A.; Spidlen, J.; Boyce, K.; Cai, J.; Crosbie, N.; Dalphin, M.; Furlong, J.; Gasparetto, M.; Goldberg, M.; Goralczyk, E.M.; et al. MIFlowCyt: The minimum information about a flow cytometry experiment. Cytom. Part A J. Int. Soc. Anal. Cytol. 2008, 73, 926–930. [Google Scholar] [CrossRef] [PubMed]
- Roederer, M.; Nozzi, J.L.; Nason, M.C. SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytom. Part A J. Int. Soc. Anal. Cytol. 2011, 79, 167–174. [Google Scholar] [CrossRef] [PubMed]


| Characteristic | HIV+TB (n = 12) | HIV−TB (n = 9) | LTBI (n = 9) |
|---|---|---|---|
| Age, median years (Q1, Q3) | 39 (33, 48) | 28 (25, 40) | 31 (28, 36) |
| Female (%) | 7 (58) | 2 (22) | 4 (44) |
| Previous TB diagnosis | 2 (17) | 0 | 0 |
| Cough duration (%) | N/A | ||
| <1 m | 8 (67) | 0 | |
| 1–2 | 3 (25) | 4 (44) | |
| 2–3 | 1 (8) | 3 (33) | |
| >3 m | 0 | 2 (22) | |
| CD4 T-cell count, median (Q1, Q3) | 288 (172, 357) | N/A | N/A |
| CD4:CD8 ratio, median (Q1, Q3) | 0.77 (0.4, 1.12) | N/A | N/A |
| Viral load, copies/mL 1 median (Q1, Q3) | 317,212 (76, 327,725) | N/A | N/A |
| ART duration, median years (Q1, Q3) | 1.3 (0.2, 6) | N/A | N/A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mupfumi, L.; Mpande, C.A.M.; Reid, T.; Moyo, S.; Shin, S.S.; Zetola, N.; Mogashoa, T.; Musonda, R.M.; Kasvosve, I.; Scriba, T.J.; et al. Immune Phenotype and Functionality of Mtb-Specific T-Cells in HIV/TB Co-Infected Patients on Antiretroviral Treatment. Pathogens 2020, 9, 180. https://doi.org/10.3390/pathogens9030180
Mupfumi L, Mpande CAM, Reid T, Moyo S, Shin SS, Zetola N, Mogashoa T, Musonda RM, Kasvosve I, Scriba TJ, et al. Immune Phenotype and Functionality of Mtb-Specific T-Cells in HIV/TB Co-Infected Patients on Antiretroviral Treatment. Pathogens. 2020; 9(3):180. https://doi.org/10.3390/pathogens9030180
Chicago/Turabian StyleMupfumi, Lucy, Cheleka A.M. Mpande, Tim Reid, Sikhulile Moyo, Sanghyuk S. Shin, Nicola Zetola, Tuelo Mogashoa, Rosemary M. Musonda, Ishmael Kasvosve, Thomas J. Scriba, and et al. 2020. "Immune Phenotype and Functionality of Mtb-Specific T-Cells in HIV/TB Co-Infected Patients on Antiretroviral Treatment" Pathogens 9, no. 3: 180. https://doi.org/10.3390/pathogens9030180
APA StyleMupfumi, L., Mpande, C. A. M., Reid, T., Moyo, S., Shin, S. S., Zetola, N., Mogashoa, T., Musonda, R. M., Kasvosve, I., Scriba, T. J., Nemes, E., & Gaseitsiwe, S. (2020). Immune Phenotype and Functionality of Mtb-Specific T-Cells in HIV/TB Co-Infected Patients on Antiretroviral Treatment. Pathogens, 9(3), 180. https://doi.org/10.3390/pathogens9030180





