Aggregatibacter, a Low Abundance Pathobiont That Influences Biogeography, Microbial Dysbiosis, and Host Defense Capabilities in Periodontitis: The History of a Bug, and Localization of Disease
Abstract
1. Introduction
2. The Disease
2.1. The Dispute
2.2. The Counter Argument
3. The Microbe: Aggregatibacter actinomycetemcomitans
3.1. The Dispute
3.2. The Counter Argument
4. The Evidence Linking the Microbe to the Disease
4.1. A. actinomycetemcomitans as an Amphibiont or Pathobiont
4.2. Molecular Approach
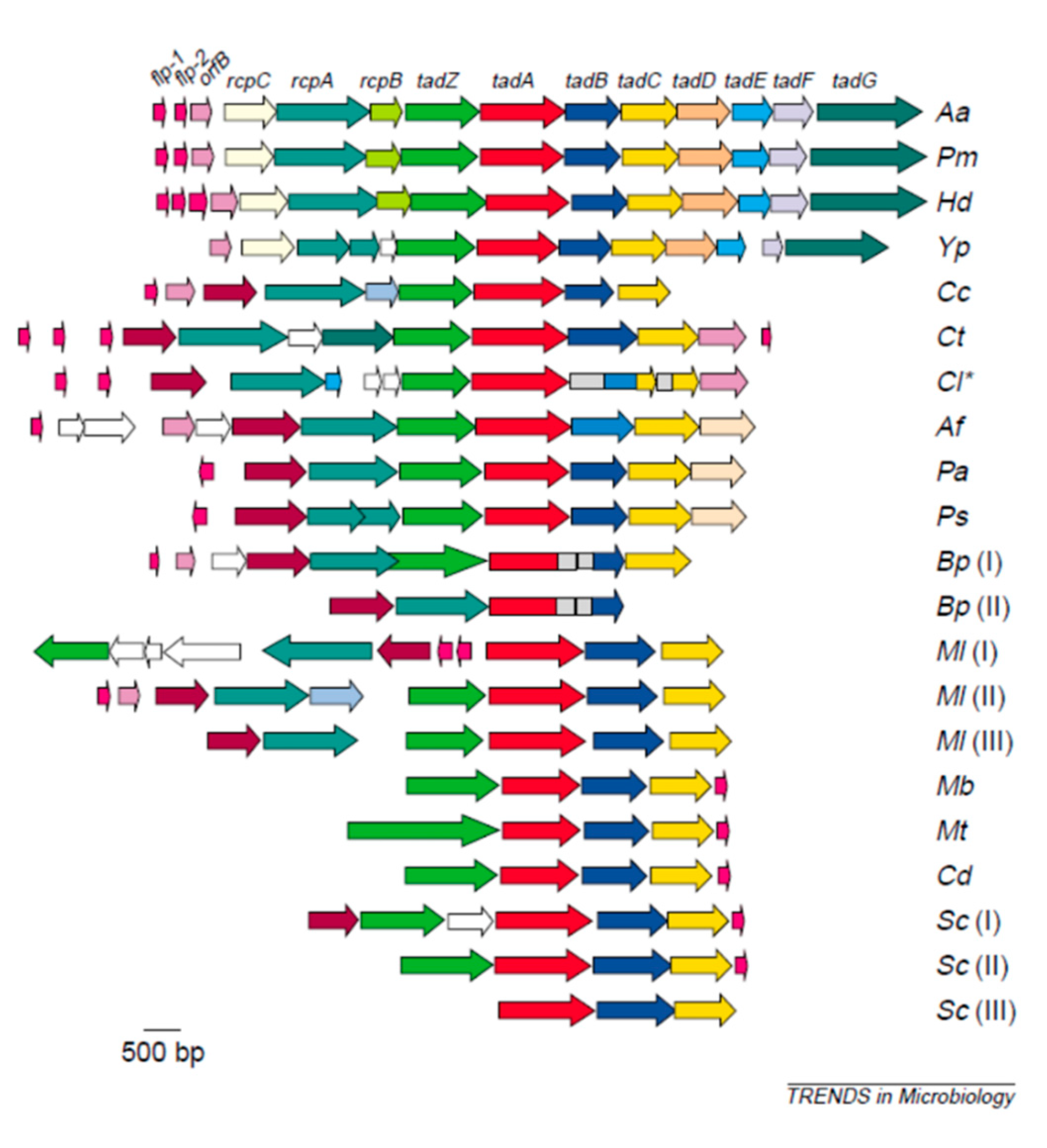
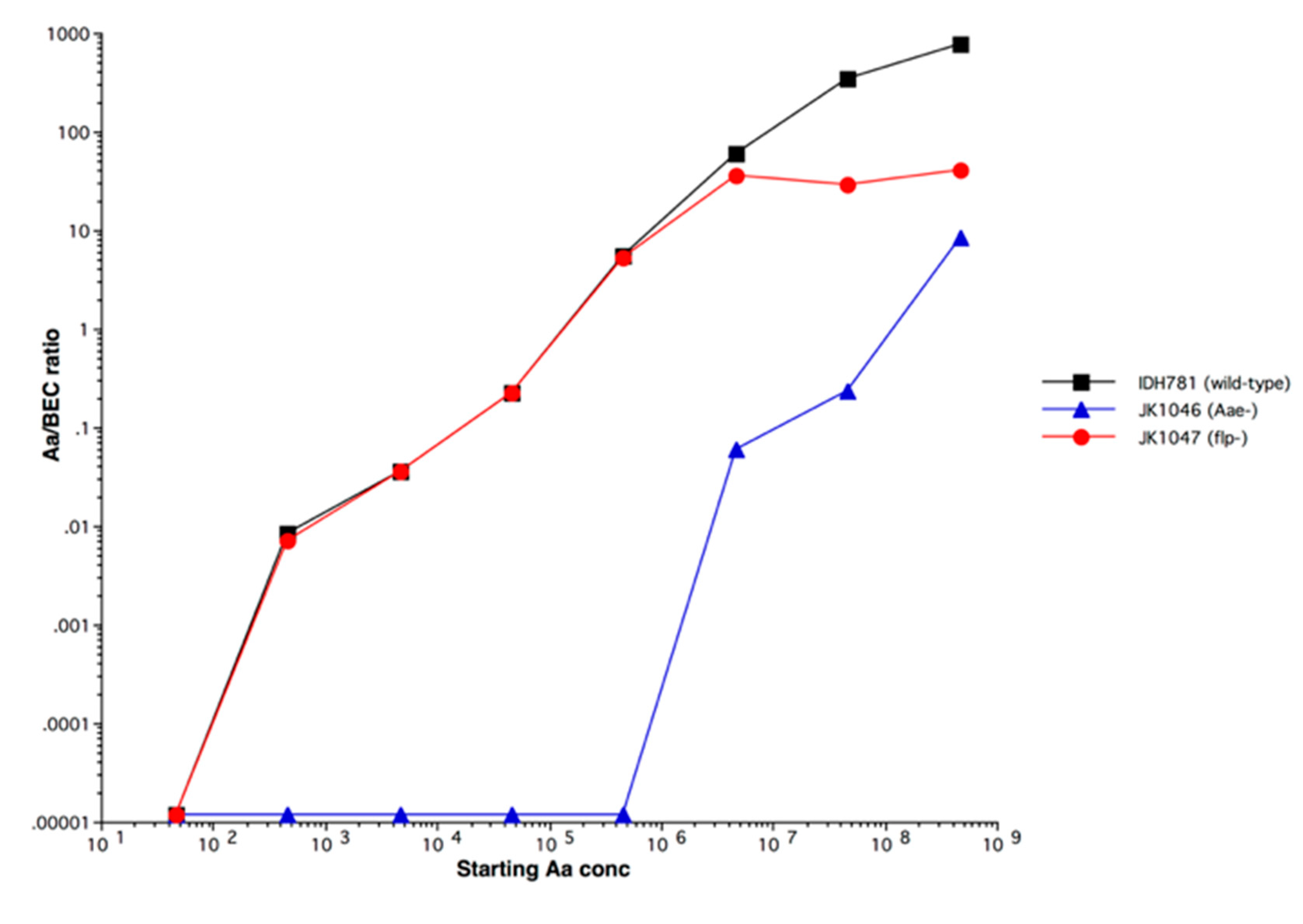
4.2.1. Adherence Genes: Abiotic Adherence
4.2.2. Adherence Genes: Receptor/Adhesin related Adherence
4.2.3. Leukotoxin
4.3. Interventional Studies
4.4. Observational Studies
5. The Defense: Novel Ways of Assessing Periodontitis as an Infectious Disease; Update: “War No More”
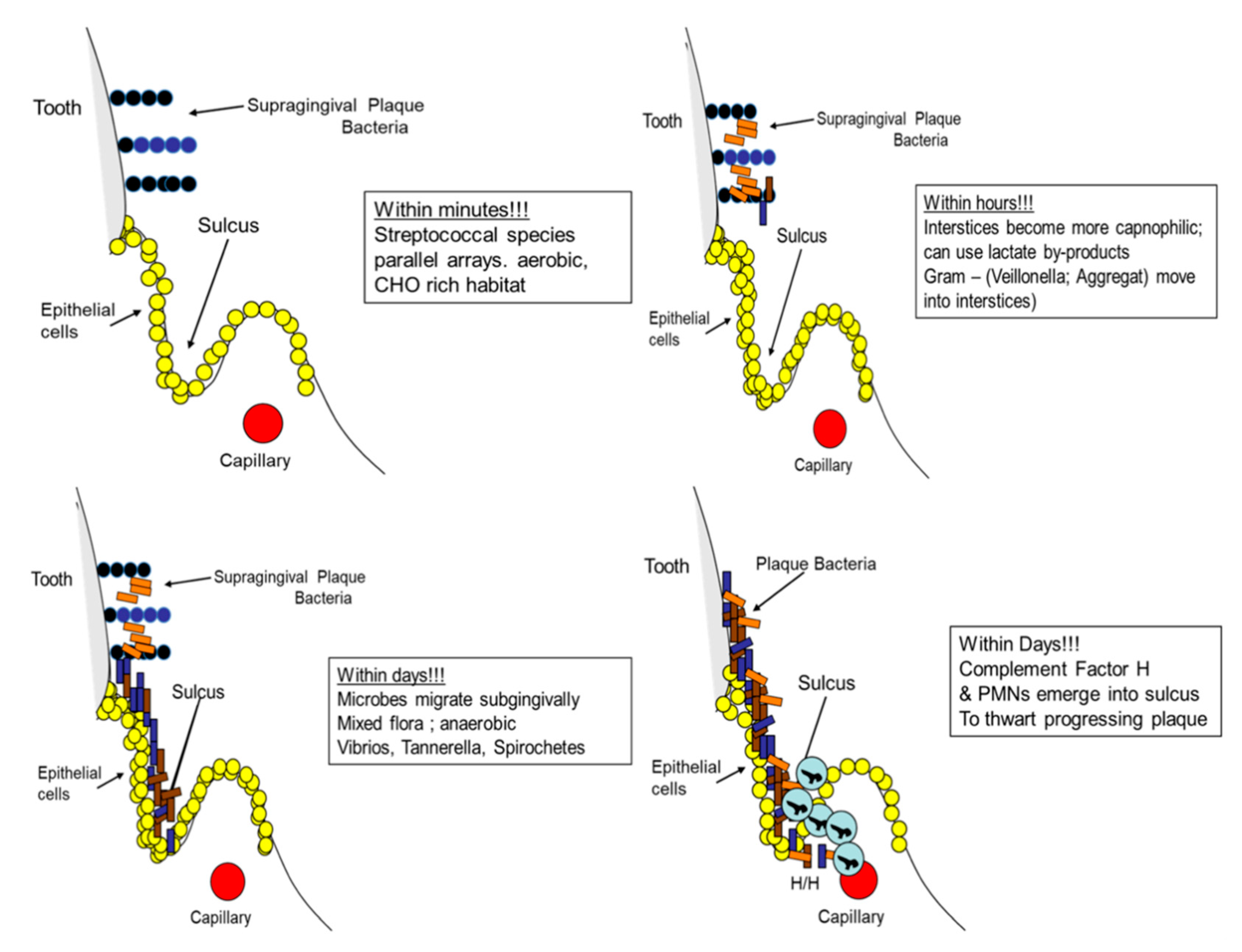
5.1. The Microbial Point of View: Biogeography, Landscape Ecology in the Oral Cavity
5.1.1. Landscapes and Proximal Caries
5.1.2. Landscapes and Proximal Periodontitis
5.2. The Host Point of View: Damage/Response Framework
5.2.1. Localization of Gingivitis and Periodontitis
5.2.2. Toxin localization: The Horseshoe Crab and Periodontitis
6. Concluding Remarks: The Final Verdict, Lesion Localization/Host Survival via Disease Localization.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Haubek, D.; Johansson, A. Pathogenicity of the highly leukotoxiic JP2 clone of Aggregatibacter actinomycetemcomitans and its geographic dissemination and role in aggressive periodontitis. J. Oral Microbiol. 2014, 1–22. [Google Scholar] [CrossRef]
- Belibasakis, G.N.; Maula, T.; Bao, K.; Lindholm, M.; Bostanci, N.; Oscarsson, J.; Ihalin, R.; Johansson, A. Virulence and Pathogenicity of Aggregatibacter actinomycetemcomitans. Pathogens 2019, 222. [Google Scholar] [CrossRef] [PubMed]
- Dahlen, D.; Basic, A.; Bylund, J. Importance of Virulence Factors for the Persisitence of Oral Bacterria in the Inflamed Gingival Crevice and in the Pathogenesis of Periodontal Disease. J. Clin. Microbiol. 2019, 8, 1339. [Google Scholar]
- Fine, D.; Patil, A.; Velusamy, S. Aggregatibacter actinomycetemcomitans (Aa) Under the Radar: Myths and Misunderstandings of Aa and its Role in Aggressive Periodontitis. Front Immunol. 2019, 25. [Google Scholar] [CrossRef]
- Nedergaard, S.; Kobel, C.M.; Nielsen, M.B.; Meller, R.T.; Jensen, A.B.; Norskov-Lauritsen, N. Whole Genome Sequencing of Aggregatibacter actinomycetemcomitans Cultured from Blood Stream Infections Reveals Three Major Phylogenetic Groups Including Novel Lineage Expressing Seerotype a Membrane Polysaccharide. Pathogens 2019, 8, 256. [Google Scholar] [CrossRef]
- Tjokro, N.O.; Kittichotirat, W.; Torittu, A.; Ihalin, R.; Bumgarner, R.E.; Chen, C. Transcriptomic Analysis of Aggregatibacter actinomycetemcomitans Core and Accessory Genes in Different Growth Conditions. Pathogens 2019, 8, 282. [Google Scholar] [CrossRef]
- Klinger, R. Untersuchungen uber enschliche aktinomykose. Cent. Bacteriol. 1912, 62, 191–200. [Google Scholar]
- Fine, D.H.; Cohen, D.W.; Bimstein, E.; Bruckmann, C. A ninety-year history of periodontosis: The legacy of Professor Bernhard Gottlieb. J. Periodontol. 2015, 86, 1–6. [Google Scholar] [CrossRef]
- Slots, J. The predominant cultivable organisms in juvenile periodontitis. Scand. J. Dent. Res. 1976, 84, 1–10. [Google Scholar] [CrossRef]
- Newman, M.G.; Socransky, S.S.; Savitt, E.D.; Propas, D.A.; Crawford, A. Studies of the microbiology of periodontosis. J. Periodontol. 1976, 47, 373–379. [Google Scholar] [CrossRef]
- Fine, D.H.; Patil, A.G.; Loos, B.G. Classification and diagnosis of aggressive periodontitis. J. Periodontol. 2018, 89, S103–S119. [Google Scholar] [CrossRef] [PubMed]
- Papapanou, P.N.; Sanz, M.; Budunell, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Peridontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal Disease and Peri-Implant Disease and Conditions. J. Periodontol. 2018, 89, S173–S182. [Google Scholar] [CrossRef] [PubMed]
- Norskov-Lauritsen, N. Classification, Identification, and Clinical Significance of Haemophilu and Aggregatibacter Species with Host Specificity for Humans. Clin. Microbiol. Rev. 2014, 27, 214–240. [Google Scholar] [CrossRef] [PubMed]
- Baer, P.N. The case for periodontosis as a clinical entity. J. Periodontol. 1971, 42, 516–520. [Google Scholar] [CrossRef]
- Offenbacher, S.; Divaris, K.; Barros, S.P.; Moss, K.L.; Marchesan, J.T.; Morelli, T.; Zhang, S.; Kim, S.; Sun, L.; Beck, J.D.; et al. Genome-Wide association study of biologically informed periodontal complex traits offers novel insights into the genetic basis for periodontal disease. Hum. Mol. Genet. 2016, 25, 2113–2129. [Google Scholar] [CrossRef]
- Munz, M.; Richter, G.M.; Loos, B.G.; Jensen, S.; Divaris, K.; Offenbacher, S.; Teumer, A.; Holtfreter, B.; Kocher, T.; Bruckmann, C.; et al. Meta-analysis of genome-wide association studies of aggressive and chronic periodontitis identifies two novel risk loci. Eur. J. Hum. Genet. 2019, 27, 102–113. [Google Scholar] [CrossRef]
- Nibali, L.; Almofareh, S.A.; Bayliss-Chapman, J.; Zhou, Y.; Vieira, A.R.; Divaris, K. Heritability of periodontitis: A systematic review of evidence from animal studies. Arch. Oral Biol. 2020, 109, 1–6. [Google Scholar] [CrossRef]
- Munz, M.; Chen, H.; Jockel-Schneider, Y.; Adam, K.; Hoffman, P.; Berger, K.; Kocher, T.; Meyle, J.; Eickholz, P.; Doerfer, C.; et al. A haplotype block downstream of plasminogen is associated with chronic and aggressive periodontitis. J. Clin. Periodontal. 2017, 44, 962–970. [Google Scholar] [CrossRef]
- Fine, D.H.; Furgang, D.; Schreiner, H.C.; Goncharoff, P.; Charlesworth, J.; Ghazwan, G.; Fitzgerald-Bocarsly, P.; Figurski, D.H. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: Implications for virulence. Microbiology 1999, 145, 1335–1347. [Google Scholar] [CrossRef][Green Version]
- Fine, D.H.; Kaplan, J.B.; Kachlany, S.C.; Schreiner, H.C. How we got attached to Actinobacillus actinomycetemcomitans: A model for infectious diseases. Periodontol. 2000 2006, 42, 114–157. [Google Scholar] [CrossRef]
- Kachlany, S.C.; Fine, D.H.; Figurski, D.H. Secretion of RTX leukotoxin by Actinobacillus actinomycetemcomitans. Infect. Immun. 2000, 68, 6094–6100. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E.; Palmer, R.J., Jr.; Periasamy, S.; Jakubovics, N.S. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 2010, 8, 471–480. [Google Scholar] [CrossRef]
- Fine, D.H.; Furgang, D.; Kaplan, J.; Charlesworth, J.; Figurski, D.H. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary coated hydroxyapatite. Arch. Oral Biol. 1999, 44, 1063–1076. [Google Scholar] [CrossRef]
- Inouye, T.; Ohta, H.; Kokeguchi, S.; Fukui, K.; Kato, K. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol. Lett. 1990, 57, 13–17. [Google Scholar] [CrossRef]
- Haase, E.M.; Zmuda, J.L.; Scannapieco, F.A. Identification of rough-colony-specific outer membrane proteins in Actinobacillus actinomycetemcomitans. Infect. Immun. 1999, 67, 2901–2908. [Google Scholar] [CrossRef] [PubMed]
- Planet, P.J.; Kachlany, S.C.; DeSalle, R.; Figurski, D.H. Phylogeny of genes for secretion of NTPases: Identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA 2001, 98, 2503–2508. [Google Scholar] [CrossRef]
- Planet, P.J.; Kachlany, S.C.; Fine, D.H.; DeSalle, R.; Figurski, D.H. The widespread colonization island of Actnobacillus actinomycetemcomitans. Nat. Genet. 2003, 34, 193–198. [Google Scholar] [CrossRef]
- Makino, S.-I.; van Putten, J.P.M.; Meyer, T.F. Phase variation of the opacity outer membrane protein contrals invasion by Neisseria gonorrhea into human epithelial cells. EMBO 1991, 10, 1307–1315. [Google Scholar] [CrossRef]
- Koomey, M.; Gotschlich, E.C.; Robbins, K.; Bergstrom, S.; Swanson, J. Effects of recA on Pilus Anrigenic Variation and Phase Transitions in Neisseria Gonorrhoeae. Genetics 1987, 117, 391–398. [Google Scholar]
- Lenz, J.D.; Dillard, J.P. Pathogenesis of Neisseria gonorrhea and the Host Defense in Ascending Infections of Human Fallopian Tube. Front. Immunol. 2018, 9, 2710. [Google Scholar] [CrossRef]
- Coureuil, M.; Join-Lambert, O.; Lecuyer, H.; Bourdoulous, S.; Marullo, S.; Nassif, X. Mechanism of meningeal invasion by Neisseria meningitides. Virulence 2012, 3, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, C. Mutation analysis of the flp operon in Actinobacillus actinomycetemcomitans. Gene 2005, 351, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lui, A.; Chen, C. Genetic basis for conversion of Rough-to-Smooth Colony Morphology in Actinobacillus actinomycetemcomitans. Infect. Immun. 2005, 73, 3749–3753. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fine, D.H.; Goncharoff, P.; Schreiner, H.C.; Chang, K.M.; Furgang, D.; Figurski, D. Colonization and persistence of rough and smooth colony variants of Actinobacillus actinomycetemcitans in the mouth of rats. Arch. Oral Biol. 2001, 46, 1065–1078. [Google Scholar] [CrossRef]
- Schreiner, H.C.; Sinatra, K.; Kaplan, J.B.; Furgang, D.; Kachlany, S.C.; Planet, P.J.; Perez, B.A.; Figurski, D.H.; Fine, D.H. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc. Natl. Acad. Sci. U S A. 2003, 100, 7295–7300. [Google Scholar] [CrossRef]
- Fine, D.H.; Armitage, G.C.; Genco, R.J.; Griffen, A.L.; Diehl, S.R. Unique etiologic. demographic, and pathologic characteristics of localized aggressive periodontitis support classification as a distinct subcategory of periodontitis. J. Am. Dent. Assoc. 2019, 150, 922–931. [Google Scholar] [CrossRef]
- Kachlany, S.C.; Planet, P.J.; Bhattacharjee, M.K.; Kollia, E.; DeSalle, R.; Fine, D.H.; Figurski, D.H. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J. Bacteriol. 2000, 182, 6169–6176. [Google Scholar] [CrossRef]
- Fine, D.H.; Velliyagonder, K.; Furgang, D.; Kaplan, J.B. The Actinobacillus actinomycetemcomitans autotransporter adhesin Aae exhibits specificity for buccal epithelial cells from humans and old world primates. Infect. Immun. 2005, 73, 1947–1953. [Google Scholar] [CrossRef]
- Danforth, D.R.; Tang-Siegel, G.; Ruiz, T.; Mintz, K.P. A Nonfimbiral Adhesin of Aggregatibacter actinomycetemcomitans Mediates Biofilm Biogenesis. Infect. Immun. 2019, 87, 00704–00718. [Google Scholar]
- Schreiner, H.; Li, Y.; Cline, J.; Tsiagbe, V.K.; Fine, D.H. A comparison of Aggregatibacter actinomycetemcomitans (Aa) virulence traits in a rat model for periodontal disease. PLoS ONE. 2013, 8, e69382. [Google Scholar] [CrossRef]
- Yue, G.; Kaplan, J.B.; Furgang, D.; Mansfield, K.G.; Fine, D.H. A second Aggregatbacter actinomycetemcomitans autotransporter adhesin that exhibits specificity for buccal epithelial cells of humans and Old World Primates. Infect. Immun. 2007, 75, 4440–4448. [Google Scholar] [CrossRef]
- Fine, D.H.; Markowitz, K.; Furgang, D.; Velliyagounder, K. Aggregatibacter actinomycetemcomitans as an early colonizer of oral tissues: Epithelium as a reservoir? J. Clin. Microbiol. 2010, 48, 4464–4473. [Google Scholar] [CrossRef]
- Baehni, P.; Tsai, C.C.; Taichman, N.S.; McArthur, W.P. Interaction of inflammatory cells and oral microorganisms. V. Electron microscopic and biochemical study on the mechanisms of release of lysosomal constituents from human polymorphonuclear leukocytes exposed to dental plaque. J. Periodontal. Res. 1978, 13, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.C.; McArthur, W.P.; Baehni, P.C.; Hammond, B.F.; Taichman, N.S. Extraction and partial characterization of a leukotoxin from a plaque-derived Gram-negative microorganism. Infect. Immun. 1979, 25, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Lally, E.T.; Golub, E.E.; Kieba, I.R.; Taichman, N.S.; Rosenblum, J.; Rosenblum, J.C.; Gibson, C.W.; Demuth, D.R. Analysis of the Actinobacillus actinomycetemcomitans leukotoxin gene. J. Biol. Chem. 1989, 264, 15451–15456. [Google Scholar] [PubMed]
- Koldrubetz, D.; Dailey, T.; Ebersole, J. Cloning and expression of the leukotoxin gene from Actinobacillus actinomycetemcomitans. Infect. Immun. 1989, 57, 1465–1469. [Google Scholar] [CrossRef]
- Haubek, D.; Ennibi, O.K.; Poulsen, K.; Benzarti, N.; Baelum, V. The highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans and progression of periodontal attachment loss. J. Dent. Res. 2004, 83, 767–770. [Google Scholar] [CrossRef] [PubMed]
- Haubek, D.; Ennibi, O.K.; Poulsen, K.; Vaeth, M.; Poulsen, S.; Kilian, M. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: A prospective longitudinal cohort study. Lancet 2008, 371, 237–242. [Google Scholar] [CrossRef]
- Kachlany, S.C. Aggregatibacter actinomycetemcomitans leukotoxin from threat to therapy. J. Dent. Res. 2010, 89, 561–570. [Google Scholar] [CrossRef]
- Johansson, A.; Claesson, R.; Aberg, C.H.; Haubek, D.; Lindholm, M.; Jasim, S.; Oscarsson, J. Genetic Profiling of Aggragatibacter actinomycetemcomitans Serotype B Isolated from Periodontitis Patients Living in Sweden. Pathogens 2019, 8, 153. [Google Scholar] [CrossRef]
- Sampathkumar, V.; Velusamy, S.K.; Godboley, D.; Fine, D.H. Increased leukotoxin production: Characterization of 100 base pairs within the 530 base pair leukotoxin promoter region of Aggregatibacter actinomycetemcomitans. Sci. Rep. 2017, 7, 1887. [Google Scholar] [CrossRef] [PubMed]
- Graves, D.T.; Fine, D.; Teng, Y.T.; Van Dyke, T.E.; Hajishengallis, G. The use of rodent models to investigate host-bacterial interactions related to periodontal diseases. J. Clin. Periodontol. 2008, 35, 89–105. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, H.C.; Markowitz, K.; Miryalkar, M.; Moore, D.; Diehl, S.; Fine, D.H. Aggregatibacter actinomycetemcomitans-Induced Bone Loss and Antibody Response in Three Rat Strains. J. Periodontol. 2011, 82, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Fine, D.H.; Karched, M.; Furgang, D.; Sampathkumar, V.; Velusamy, S.; Godboley, D. Colonization and Persistence of Labeled and Foreign Strains of Inoculated into the Mouths of Rhesus Monkeys. J. Oral Biol. (Northborough) 2015, 2. [Google Scholar] [CrossRef]
- Velusamy, S.K.; Sampathkumar, V.; Godboley, D.; Fine, D.H. Profound Effects of Aggregatibacter actinomycetemcomitans Leukotoxin Mutation on Adherence Properties Are Clarified in in vitro Experiments. PLoS ONE 2016, 11, e0151361. [Google Scholar] [CrossRef]
- Narayanan, A.M.; Ramsey, M.M.; Stacy, A.; Whiteley, M. Defining Genetic Fitness Determinants and Creating Genomic Resources for an Oral Pathogen. Appl. Environ. Microbiol. 2017, 83, e00797-17. [Google Scholar] [CrossRef]
- Loe, H.; Brown, L.J. Early onset periodontitis in the United States of America. J. Periodontol. 1991, 62, 608–616. [Google Scholar] [CrossRef]
- Fine, D.H.; Markowitz, K.; Furgang, D.; Fairlie, K.; Ferrandiz, J.; Nasri, C.; McKiernan, M.; Gunsolley, J. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: Longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 2007, 45, 3859–3869. [Google Scholar] [CrossRef]
- Fine, D.H.; Markowitz, K.; Fairlie, K.; Tischio-Bereski, D.; Ferrendiz, J.; Furgang, D.; Paster, B.J.; Dewhirst, F.E. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J. Clin. Microbiol. 2013, 51, 2850–2861. [Google Scholar] [CrossRef]
- Fine, D.H.; Markowitz, K.; Fairlie, K.; Tischio-Bereski, D.; Ferrandiz, J.; Godboley, D.; Furgang, D.; Gunsolley, J.; Best, A. Macrophage inflammatory protein-1alpha shows predictive value as a risk marker for subjects and sites vulnerable to bone loss in a longitudinal model of aggressive periodontitis. PLoS ONE 2014, 9, e98541. [Google Scholar] [CrossRef]
- Al-Sabbagh, M.; Alladah, A.; Lin, Y.; Keyscio, R.J.; Thomas, M.V.; Ebersole, J.L.; Miller, C.S. Bone remodelling-associated biomarker MIP-1alpha distinguishes periodontal disease from health. J. Periodontol. Res. 2012, 47, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Fine, D.H. Dr. Theodor Rosebury: Grandfather of Modern Oral Microbiology. J. Dent. Res. 2006, 85, 990–995. [Google Scholar] [CrossRef] [PubMed]
- Falkow, S. Is persistent bactertial infection good for your health? Cell 2006, 124, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Pirofski, L.; Casadevall, A. The Damage-Response Framework as a Tool for the Physician-Scientist to Understand the Pathogenesis of Infectious Diseases. J. Infect. Dis. 2018, 218, S7–S11. [Google Scholar] [CrossRef] [PubMed]
- Rook, G.A.W.; Lowry, C.A.; Raison, C.L. Microbial Old Friends immunoregulation and stress resilience. Evol. Med. Public Health 2013, 2013, 46–64. [Google Scholar] [CrossRef] [PubMed]
- Levin, S.A.; Paine, R.T. Disturbance, patch formation, and community struture. PNAS 1974, 71, 2744–2747. [Google Scholar] [CrossRef]
- Mark Welch, J.L.; Dewhirst, F.E.; Borisy, G.G. Biogeographty of the Oral Microbiome: The Site-Specific Hyothesis. Ann. Rev. Microbiol. 2019, 73, 335–358. [Google Scholar] [CrossRef]
- Picket, S.T.; Cadenasso, M.L. Landscape ecolotgy: Spatial heterogeneity in ecological systems. Science 1995, 269, 331–334. [Google Scholar] [CrossRef]
- Proctor, D.M.; Shelef, K.M.; Gonzalez, A.; Davis, C.L.; Dethlefsen, L.; Burns, A.R.; Loomer, P.M.; Armitage, G.C.; Ryder, M.I.; Millman, M.E.; et al. Microbial biogeography and ecology of the mouth and implications for periodontal diseases. Periodontol. 2000 2019, 82, 26–41. [Google Scholar] [CrossRef]
- Schroeder, H.E.; Listgarten, M.A. The gingival tissues: The architecture of periodontal protection. Periodontol. 2000 1997, 13, 91–120. [Google Scholar] [CrossRef]
- Scannapieco, F. Saliva-bacterial interactions in oral microbial ecology. Crit. Rev. Oral Biol. Med. 1994, 5, 203–248. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.A. Landscape Mosaics and Ecological Theory; Springer Netherland: London, UK, 1995. [Google Scholar]
- Proctor, D.M.; Relman, D.A. The Landscape Ecology and Microbiota of the Ear, Nose and Throught. Cell Host Microbe 2017, 21, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Loe, H.; Theilade, E.; Jensen, S.B. Experimental Gingivitis in Man. J. Periodontol. 1965, 36, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Ritz, H.L. Microbial population shifts in developing human plaque. Arch. Oral Biol. 1967, 12, 1561–1568. [Google Scholar] [CrossRef]
- Kolenbrander, P.E.; Palmer, R.J., Jr.; Rickard, A.H.; Jakubovics, N.S.; Chalmers, N.I.; Diaz, P.I. Bacterial interactions and successions during plaque development. Periodontol. 2000 2006, 42, 47–79. [Google Scholar] [CrossRef]
- Von der Fehr, F.R.; Loe, H.; Theilade, E. Experimental caries in man. Caries Res. 1970, 4, 131–148. [Google Scholar] [CrossRef]
- Marsh, P.D. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 1994, 8, 263–271. [Google Scholar] [CrossRef]
- Marsh, P.D.; Zaura, E. Dental biofilm: Ecological interactions in health and disease. J. Clin. Periodontol. 2017, 44, S12–S22. [Google Scholar] [CrossRef]
- Van der Hoeven, J.S.; Toorop, A.I.; Mikx, F.H.M. Symbiotic Relationship of Veillonella alcalescens and Streptococcus mutans in Dental Plaque in Gnotobiotic Rats. Caries Res. 1978, 12, 142–147. [Google Scholar] [CrossRef]
- Ramsey, M.M.; Rumbaugh, K.P.; Whiteley, M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011, 7, e1002012. [Google Scholar] [CrossRef]
- Mandel, I.D. The functions of saliva. J. Dent. Res. 1987, 66, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Pirofski, L.-A. Host-Pathogen Interactions: Redefining the Basic Concepts of Virulence and Pathogenicity. Infect. Immun. 1999, 67, 3703–3713. [Google Scholar] [CrossRef] [PubMed]
- Jakubovics, N.S.; Kolenbrander, P.E. The road to ruin: The formation of disease-associated oral biofilms. Oral Dis. 2010, 16, 729–739. [Google Scholar] [CrossRef]
- Moore, L.V.; Moore, W.E.; Cato, E.P.; Smibert, R.M.; Burmeister, J.A.; Best, A.M.; Ranney, R.R. Bacteriology of human gingivitis. J. Dent. Res. 1987, 66, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Moore, W.E.; Moore, L.H.; Ranney, R.R.; Smibert, R.M.; Burmeister, J.A.; Schenkein, H.A. The microflora of periodontal sites showing active destructive progression. J. Clin. Periodontol. 1991, 18, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Moore, W.E.C.; Moore, L.V.H. The bacteria of periodontal disease. Periodontol. 2000 1994, 5, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Listgarten, M.A. Ultrastructure of the dento-gingival junction after gingivectomy. J. Periodontal Res. 1972, 7, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, H.E.; Listgarten, M.A. Fine structure of the developing epithelial attachment to teeth. Monogr. Dev. Biol. 1971, 2, 33–46. [Google Scholar]
- Listgarten, M.A. The structure of dental plaque. Periodontol. 2000 1994, 5, 52–65. [Google Scholar] [CrossRef]
- Page, R.C.; Schroeder, H.E. Pathogenesis of inflammatory periodontal disease. A summary of current work. Lab. Invest. 1976, 34, 235–249. [Google Scholar]
- Fine, D.H.; Furgang, D.; McKiernan, M.; Tereski-Bischio, D.; Ricci-Nittel, D.; Zhang, P.; Araugo, M.W.B. An investigation of the effect of an essential oil mouthrinse on induced bacteremia: A pilot study. J. Clin. Periodontol. 2010, 37, 840–847. [Google Scholar] [CrossRef] [PubMed]
- Griffen, A.L.; Beall, C.J.; Campbell, J.H.; Firestone, N.D.; Kumar, P.S.; Yang, Z.K.; Podar, M.; Leys, E.J. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME 2012, 6, 1176–1185. [Google Scholar] [CrossRef]
- Fine, D.H.; Korik, I.; Furgang, D.; Myers, R.; Olshan, A.; Barnett, M.L.; VIncent, J. Assessing Pre-Procedural Subgingival Irrigation and Rinsing with an Antiseptic Mouthrinse to Reduce Bacteremia. JADA 1996, 127, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasan, P.K.; Tischio-Bereski, D.; Fine, D.H. Reduction in bacteremia after brushing with a triclosan/copolymer dentifrice-A randomized clinical study. J. Clin. Periodontol. 2017, 44, 1020–1028. [Google Scholar] [CrossRef] [PubMed]
- Figuero, E.; Sanchez-Beltran, M.; Cuesta-Frechoso, S.; Tejerina, J.M.; del Castro, J.A.; Gutierrez, J.M.; Herrera, D.; Sanz, M. Detection of Periodontal Bacteria in Atheromatous Plaque by Nested Polymerase Chain Reaction. J. Periodontol. 2011, 82, 1469–1477. [Google Scholar] [CrossRef]
- Lee, S.A.; Liu, F.; Riordan, S.; Lee, C.S.; Zhang, L.G. Global Investigations of Fusobacterium nucleatum in Human Colorectal Cancer. Front. Oncol. 2019, 9, 566. [Google Scholar] [CrossRef]
- Roblin, X.; Neut, C.; Darfeuille-Michaud, A.; Colombel, J.F. Local appendiceal dysbiosis: The missing link between the appendix and ulcerative colitis? Gut 2012, 61, 635–636. [Google Scholar] [CrossRef]
- Offenbacher, S.; Beck, J.D. Changing Paradigms in the Oral Disease-Systemic Disease Relationship. J. Periodontol. 2014, 85, 761–763. [Google Scholar] [CrossRef]
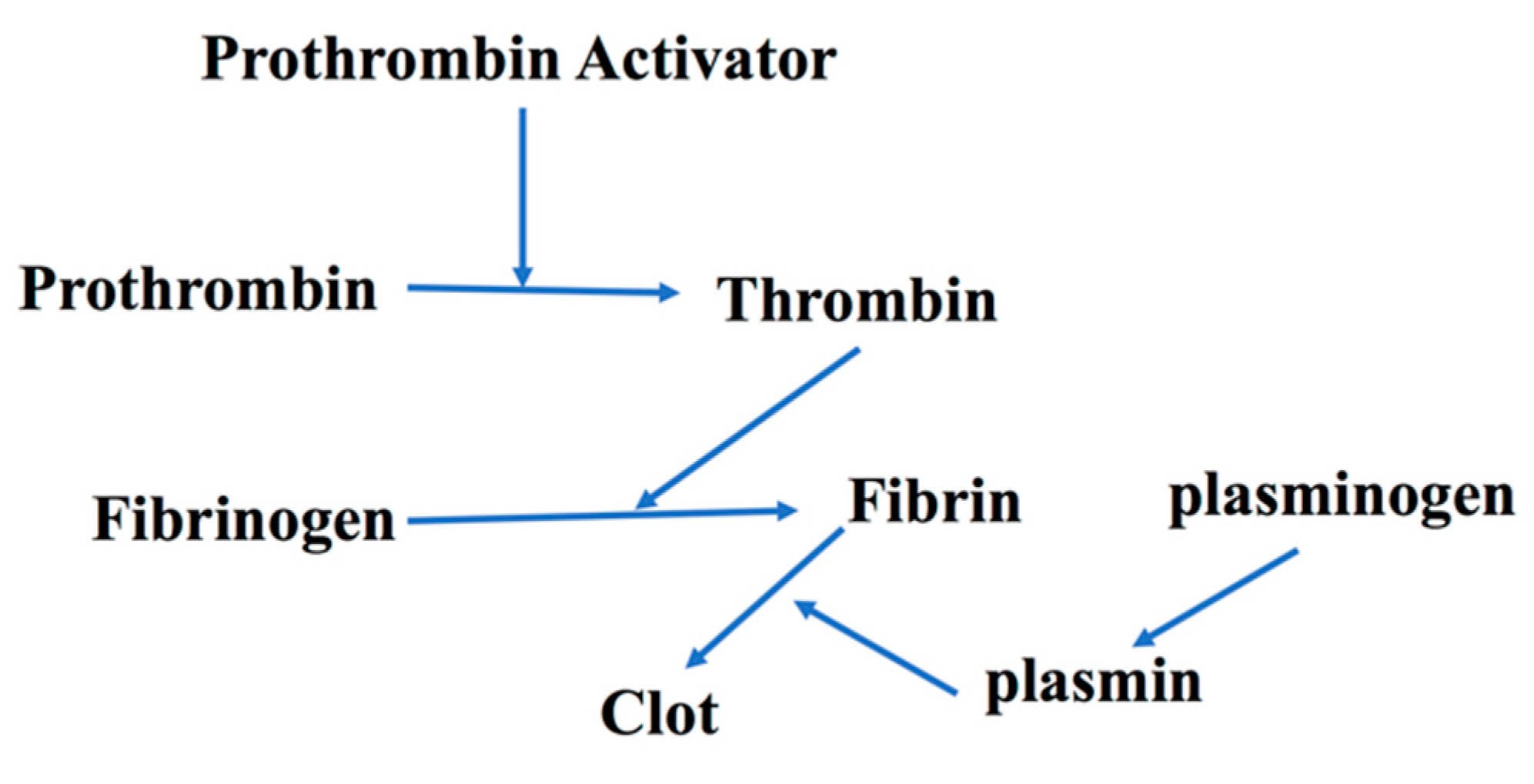
- IIwanaga, S. Biochemical principle of Limulus test for detecting bacterial endotoxins. Proc. Jpn. Acad. Ser 2007, 83, 110–119. [Google Scholar] [CrossRef]
- Levin, J.; Bang, F.B. Clottable protein in Limulus: Its localization and kinetics of its coagulation by endotoxin. Thromb. Diath. Haemorrh. 1968, 19, 186–194. [Google Scholar] [CrossRef]
- Chalin, A.B.; Opal, J.M.; Opal, S.M. Whatever happened to the Schwartzman phenomenon? Innate Immun. 2018, 24, 466–479. [Google Scholar]
- Young, N.S.; Levin, J.; Prendergast, R.A. An Invertebrate Coagulation System Activated by Endotoxin: Evidence for Enzymatic Mediation. J. Clin. Investig. 1972, 51, 1790–1797. [Google Scholar] [CrossRef]
- Kweider, M.; Lowe, G.D.O.; Murray, G.D.; Kinane, D.F.; McGowan, D.A. Dental Disease, Fibringogen and White Cell Count; Links with Myocardial Infaction? Scot. Med. J. 1993, 38, 73–74. [Google Scholar] [CrossRef] [PubMed]
- Stashenko, P.; Teles, R. Periapical Inflammatory Responses and Their Modulation. Crit. Rev. Oral Biol. Med. 1998, 9, 498–521. [Google Scholar] [CrossRef] [PubMed]
- Graves, D.T.; Chen, C.-P.; Douville, C.; Jiang, Y. Interleukin-1 Receptor Signalling Rather than That of Tumor Necrosis Factor Is Critical in Protecting the Host from the Severe Consquences of a Polymicrobial Anaerobic Infection. Infect. Immun. 2000, 68, 4746–4751. [Google Scholar] [CrossRef] [PubMed]
- Kuchler, E.C.; Mazzi-Chaves, J.F.; Antunes, L.S.; Baratto-Filho, F.; Sousa-Neto, M.D. Current treens of genetics in apical periodontitis research. Braz. Oral Res. 2018, 32, 126–132. [Google Scholar] [CrossRef]
- Garlet, G.P.; Cardoso, C.R.; Silva, T.A.; Ferreira, B.R.; Avila-Campos, M.J.; Cunha, F.Q.; Silva, J.S. Cytokine pattern determines the progression of experimental periodontal disease induced by Actinobacillus actinomycetemcomitans through the modulation of MMPs, RANKL, and their physiological inhibitors. Oral Microbiol. Immunol. 2006, 21, 12–20. [Google Scholar] [CrossRef]
- Armitage, G.C. Periodontal diseases: Diagnosis. Ann. Periodontol. 1996, 1, 37–215. [Google Scholar] [CrossRef]
- Ebersole, J.L.; Dawson, D.; Emecan-Huja, P.; Nagarajan, R.; Howard, K.; Grady, M.E.; Thompson, K.; Peyyala, R.; AL-Attar, A.; Lethbridge, K.; et al. The periodontal war: Microbes and immunity. Periodontol. 2000 2017, 75, 52–115. [Google Scholar] [CrossRef]
- Van der Velden, U. Diagnosis of periodontitis. J. Clin. Periodontol. 2000, 27, 960–961. [Google Scholar]
- Armitage, G.C. Comparison of the microbiological features of chronic and aggressive periodontitis. Periodontol. 2000 2010, 53, 70–88. [Google Scholar] [CrossRef] [PubMed]
- Herrera, D.; Sanz, M.; Jepsen, S.; Needleman, I.; Roldan, S. A systematic review on the effect of systemic antimicrobials as an adjunct to scaling and root planing in periodontitis patients. J. Clin. Periodontol. 2002, 29, 136–159. [Google Scholar] [CrossRef] [PubMed]


| Gene Symbol | Gene Name | SNP Region | Reference |
|---|---|---|---|
| PTGR1 | Prostaglandin reductase | rs758534 | [15] |
| DGKD | Diacylglycerol kinase delta 130 kDa | rs10980953 | [15,16,17] |
| INPPSF | Inositol polyphosphate phosphatase 1F | rs17680667 | [15,16,17] |
| HTR1D | 5-hydoxytryptamine (serotonin) receptor 1D, G protein coupled | rs16828047 | [15,16,17] |
| LUZP1 | Lucine zipper protein1 | rs94266589 | [15,17] |
| PLG | Plasminogen * | rs4252120 | [18] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fine, D.H.; Schreiner, H.; Velusamy, S.K. Aggregatibacter, a Low Abundance Pathobiont That Influences Biogeography, Microbial Dysbiosis, and Host Defense Capabilities in Periodontitis: The History of a Bug, and Localization of Disease. Pathogens 2020, 9, 179. https://doi.org/10.3390/pathogens9030179
Fine DH, Schreiner H, Velusamy SK. Aggregatibacter, a Low Abundance Pathobiont That Influences Biogeography, Microbial Dysbiosis, and Host Defense Capabilities in Periodontitis: The History of a Bug, and Localization of Disease. Pathogens. 2020; 9(3):179. https://doi.org/10.3390/pathogens9030179
Chicago/Turabian StyleFine, Daniel H., Helen Schreiner, and Senthil Kumar Velusamy. 2020. "Aggregatibacter, a Low Abundance Pathobiont That Influences Biogeography, Microbial Dysbiosis, and Host Defense Capabilities in Periodontitis: The History of a Bug, and Localization of Disease" Pathogens 9, no. 3: 179. https://doi.org/10.3390/pathogens9030179
APA StyleFine, D. H., Schreiner, H., & Velusamy, S. K. (2020). Aggregatibacter, a Low Abundance Pathobiont That Influences Biogeography, Microbial Dysbiosis, and Host Defense Capabilities in Periodontitis: The History of a Bug, and Localization of Disease. Pathogens, 9(3), 179. https://doi.org/10.3390/pathogens9030179





