Abstract
Despite the great efforts devoted to research on Helicobacter pylori, the prevalence of single-strain infection or H. pylori mixed infection and its implications in the mode of transmission of this bacterium are still controversial. In this study, we explored the usefulness of housekeeping gene amplicon sequencing in the detection of H. pylori microevolution and multiple infections. DNA was extracted from five gastric biopsies from four patients infected with distinct histopathological diagnoses. PCR amplification of six H. pylori-specific housekeeping genes was then assessed on each sample. Optimal results were obtained for the cgt and luxS genes, which were selected for amplicon sequencing. A total of 11,833 cgt and 403 luxS amplicon sequences were obtained, 2042 and 112 of which were unique sequences, respectively. All cgt and luxS sequences were clustered at 97% to 9 and 13 operational taxonomic units (OTUs), respectively. For each sample from a different patient, a single OTU comprised the majority of sequences in both genes, but more than one OTU was detected in all samples. These results suggest that multiple infections with a predominant strain together with other minority strains are the main way by which H. pylori colonizes the human stomach.
1. Introduction
Helicobacter pylori is a Gram-negative bacterium characterized by a high level of intraspecies genetic diversity. This species has been intensely investigated with more than 43,000 research articles cited in PubMed (https://www.ncbi.nlm.nih.gov/pubmed/). One reason for such interest is that H. pylori has evolved with humans for about 100,000 years. Having coexisted with humans for this long period of time, tracing human migrations is still possible today through multilocus sequence analysis (MLSA) of housekeeping genes of H. pylori strains isolated from human populations with distinct geographical origins [1,2].
Humans and some other primates are the unique hosts of H. pylori, which infects gastric mucosa as the primary niche and persists for the lifetime of the host in the absence of treatment. H. pylori infects about half of the world’s population and the infection is mainly acquired in early childhood [3,4]. Infection results in life-long gastric colonization that usually is asymptomatic. However, it can lead to the development of several gastro-intestinal diseases, such as gastric ulcers, mucosa-associated lymphoid tissue (MALT) lymphoma, or chronic inflammation. The latter could potentially progress to multifocal atrophy, intestinal metaplasia, dysplasia, and gastric adenocarcinoma [5,6,7,8].
It has been suggested that the transmission of H. pylori occurs directly via oral–oral, gastro–oral, and fecal–oral routes, although food- and water-borne transmissions could play an important role [7,9]. Another controversial issue related to transmission is the prevalence of H. pylori infection whether by a single strain or by multiple strains, since different methodologies and discordant results have been reported [10,11]. Studies of H. pylori infection at strain level using high-throughput analyses could be key to elucidate the emergence or transmission of antibiotic resistance and the development of gastric diseases.
In addition to the housekeeping genes (atpA, efp, mutY, ppa, trpC, ureI, and yphC) used in MLSA studies to delineate H. pylori/human populations previously described [1], we have recently developed H. pylori specific PCR amplifications of six housekeeping genes (amiA, cgt, cpn60, cpn70, dnaJ, and luxS) related to H. pylori pathogenesis in order to evaluate their usefulness as new virulence markers and for the specific detection of H. pylori, the genetic discrimination at strain level and the detection of multiple infection [11].
The aim of this study was to evaluate the usefulness of amplicon sequencing methodology for the detection of H. pylori microevolution and multiple infections from gastric biopsies of patients with dyspeptic symptoms and different histopathological findings (from atrophy to adenocarcinoma).
2. Results
2.1. Samples and Amplicon Sequences
As the best PCR amplification of six housekeeping genes (amiA, cgt, cpn60, cpn70, dnaJ, and luxS) from DNA biopsies were obtained for the genes cgt and luxS, these genes were selected for amplicon sequencing (see Materials and Methods). The sequencing results of the amplicon libraries (Table 1 and Table 2) yielded a much larger number of cgt sequences (11,833 in total and 2042 unique sequences) than luxS sequences (403 in total and 112 unique sequences). This is probably due to the increased dilution of luxS amplicons in the preparation of the pooling. All sequences from each sample and gene were processed in order to adjust for the length of the sequences (438 nt for cgt sequences and 366 nt for luxS sequences) and to detect duplicate sequences. As a result, a significant number of identical sequences and a low number of unique sequences (11–43%) were obtained (Table 1 and Table 2).

Table 1.
Distribution of cgt amplicon sequences by samples and operational taxonomic units (OTUs).

Table 2.
Distribution of luxS amplicon sequences by samples and OTUs.
The distribution of total and unique cgt sequences by sample was as follows (Table 1): 5185 total and 593 (11%) unique sequences from sample B247S; 2422 and 422 (17%) from B373A; 968 and 234 (24%) from B508S; 1414 and 339 (24%) from B508T; and 1844 and 454 (25%) from B601A. In the case of luxS, the distribution of total and unique sequences by sample was as follows (Table 2): 111 total and 34 (31%) unique sequences from sample B247S; 72 and 16 (22%) from B373A; 56 and 18 (32%) from B508S; 115 and 23 (20%) from B508T; and 49 and 21 (43%) from B601A.
2.2. Operational Taxonomic Units (OTUs) Assignment and Distribution
All unique sequences belonging to the same gene were merged in a FASTA file and pairwise distances were calculated to generate a PHYLIP-formatted distance matrix. A cutoff distance of 3% for both genes was defined in order to assign sequences to operational taxonomic units (OTUs). In a previous study [11], it was shown that for the luxS gene this value was the mean distance value between sequences from clones isolated from different patients. The distant value of the cgt gene was calculated to be 1%. However, due to the inherent error rates of Roche 454 platform, it was decided to raise the value to 3%. Therefore, sequences with distance <3% were considered as originating from the same strain. A total of 9 and 13 OTUs were obtained from the cgt (Table 1) and luxS (Table 2) distance matrix, respectively. In all samples, at least two different OTUs were detected. Additionally, most of the total (99% from cgt and 91% from luxS) and unique (98% from cgt and 73% from luxS) sequences clustered in OTUs 01–04. Some sequences from different samples belonged to the same OTU, especially in the case of OTU-03 (containing sequences from samples B508S and B508T) (Table 1 and Table 2).
As seen in the tables, every sample had a dominant OTU. For example, for the gene cgt in the sample B247S, 5184 sequences out of 5185 of the total were in OTU-01. When referring to unique sequences, 592 of 593 are in this OTU. Regarding the gene luxS in the same sample, 98 sequences of a total of 111 were in OTU-01. And referring to unique sequences, 25 of a total of 34 were in this OTU.
2.3. Amplicon Sequence Identification
As cgt and luxS PCRs were specific for H. pylori, all amplicon sequences matched this species. In order to classify the amplicon sequences obtained in this study, two FASTA-formatted sequence files for each gene were generated. One file contained the representative sequence for each OTU (by means of the get.oturep Mothur command) and the other file with reference sequences as detailed below.
For the reference sequences file, cgt and luxS sequences were downloaded from 75 H. pylori complete genomes hosted in GenBank (Supplementary Table S1). These sequences were processed using Mothur (unique.seqs command) in order to avoid duplicate sequences, resulting in 53 cgt and 59 luxS unique sequences. An additional 15 cgt and luxS sequences were also incorporated as reference sequences (Supplementary Table S1). These additional sequences had been previously described in Palau et al. [11]. Furthermore, some of them were obtained from strains isolated from the same biopsies (B247S, B373A, B508S, and B508T) used in this study. All reference sequences were adjusted to the length of 438 nt for cgt and 366 nt for luxS.
Representative OTU sequences from this study were classified using Mothur (classify.seqs command) with the above reference sequences as templates. On the other hand, all sequences (representative OTUs and references) were joined in a single file, one for each gene. Thereafter, the sequence distances were calculated and phylogenetic trees were constructed using MEGA7 [12]. Moreover, representative OTU sequences were also compared using Nucleotide BLAST on the NCBI website.
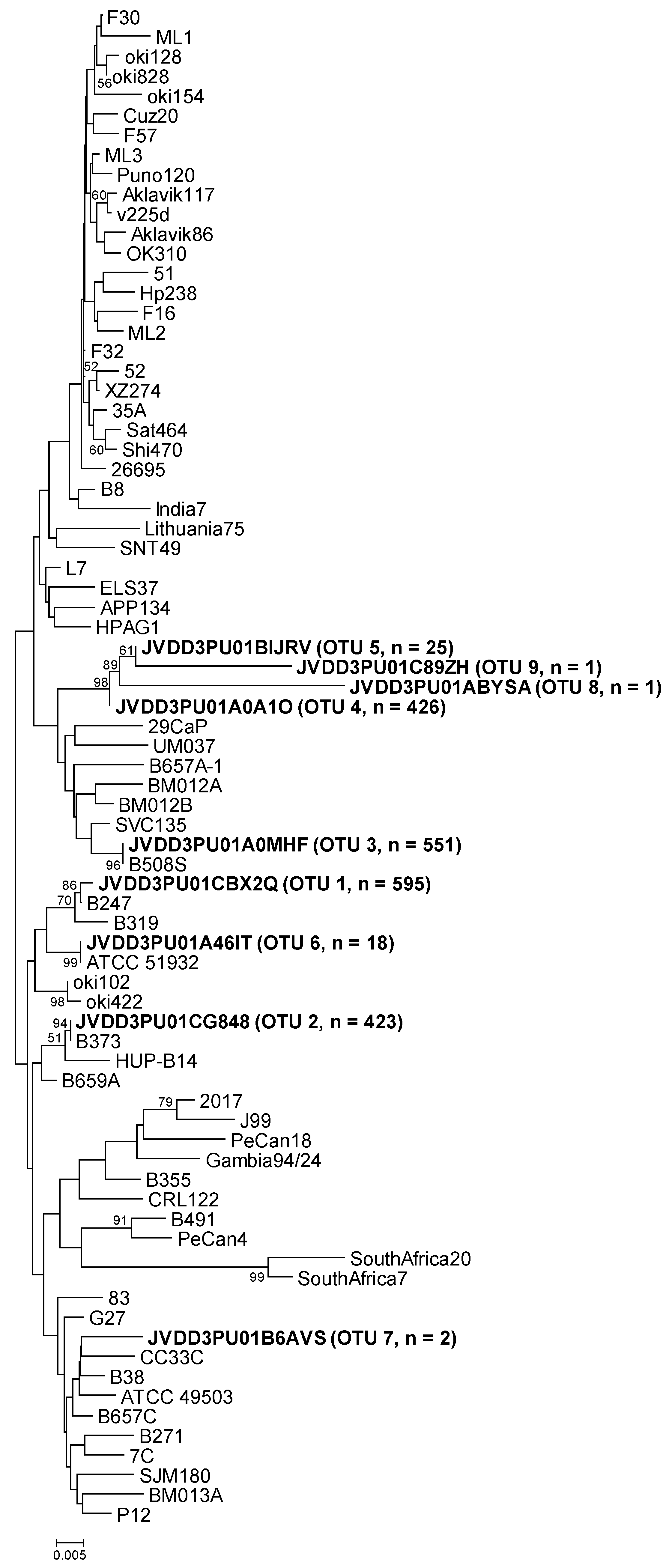
The representative sequence of cgt OTU-01, obtained from sample B247S, showed one nucleotide difference (the first nucleotide of the sequence) with respect to the cgt reference sequence (GenBank accession number KU053362). The reference was obtained from H. pylori strain B247 (Table 3 and Figure 1), which was isolated from replicate biopsy B247S in a previous study [11]. The representative sequence of cgt OTU-02 (from sample B373A) was identical to the cgt reference sequence (GenBank accession number MG950173). In this case, the reference was obtained from H. pylori strain B373, isolated from replicate biopsy B373A in the above-mentioned study. The representative sequence of cgt OTU-03, that comprised most of the sequences from samples B508S and B508T, was identical to the cgt reference sequence that was obtained from H. pylori strain B508S (GenBank accession number KU053367), isolated from replicate biopsy B508S. The representative sequence of the other principal OTU (OTU-04) (from sample B601A) could not be compared to the corresponding strain sequence because no H. pylori strain was isolated from biopsy B601A. The other five cgt OTUs were different from any reference sequence, except in the case of cgt OTU-06. Its sequence was identical to the sequence of H. pylori ATCC 51932 (Table 3 and Figure 1).

Table 3.
Classification of cgt OTUs by Mothur, MEGA7, and BLAST.
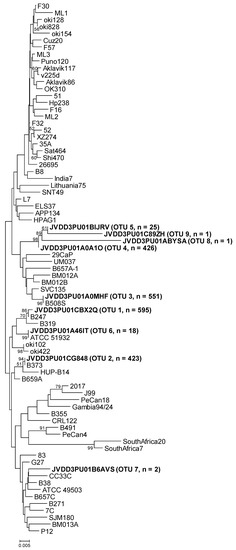
Figure 1.
Neighbor-joining phylogenetic tree obtained from representative OTU sequences and reference sequences of gene cgt. Bar, distance values as calculated by MEGA 7.0. Bootstrap values (> 50%) after 1000 replicates are shown as percentages.
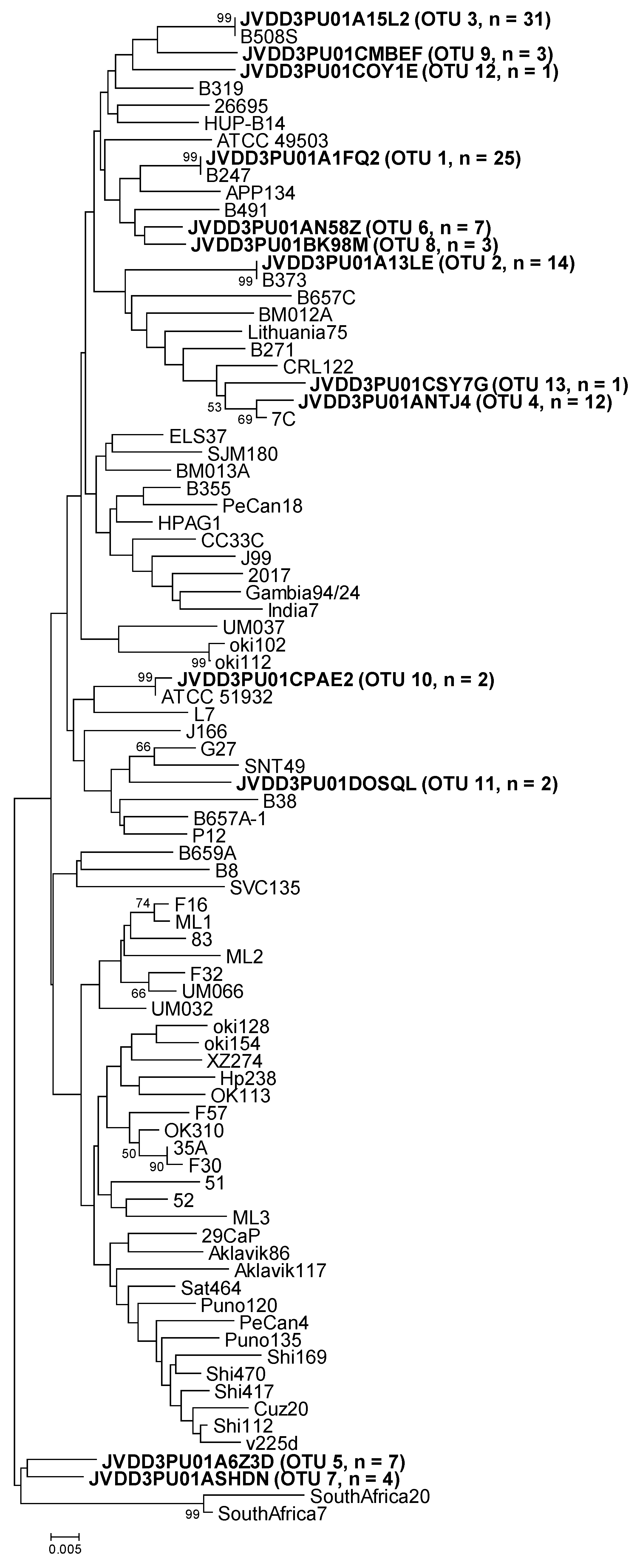
Similarly, the representative sequences of luxS OTU-01, -02, and -03 were identical to the reference sequences (GenBank accession numbers KU053434, MG950172, and KU053439, respectively) that were obtained from the above-mentioned strains, isolated from replicate gastric biopsies of the same patient (B247, B373, and B508, respectively) (Table 4 and Figure 2). The representative sequence of luxS OTU-04 could not be compared either to the corresponding strain sequence because of the lack of H. pylori strain isolation from biopsy B601A. The other nine luxS OTUs were different from any reference sequence (Table 4 and Figure 2).

Table 4.
Classification of luxS OTUs by Mothur, MEGA7, and BLAST.
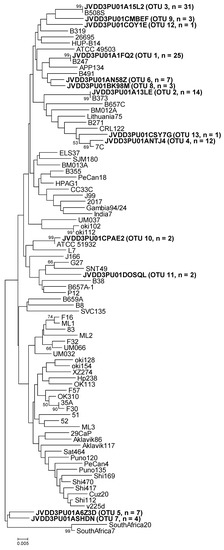
Figure 2.
Neighbor-joining phylogenetic tree obtained from representative OTU sequences and reference sequences of gene luxS. Bar, distance values as calculated by MEGA 7.0. Bootstrap values (>50%) after 1000 replicates are shown as percentages.
3. Discussion
Most amplicon sequences were repeated sequences that were largely represented by four OTUs. Representative sequences from these OTUs matched with sequences of strains isolated from replicate gastric biopsies obtained from the same patient (except in the case of sample B601A because of the lack of the corresponding isolate). However, although in the minority, other OTUs were detected in all five gastric biopsies for both genes. These results suggest that H. pylori colonizes the human stomach through different infection events that lead to a gastric multi-infection with a predominant strain together with other minority strains.
Other studies have evaluated the prevalence of mixed H. pylori infections, but with a high disparity of results, even when using similar methodologies. Using randomly amplified polymorphic DNA (RAPD) fingerprinting, Toita et al. [13] reported that all patients were infected with a single H. pylori clone. Contrarily, Sheu et al. [14] found a 23.3% prevalence of mixed infections. Analyzing the cag-PAI status and the s-region or m-region of vacA in H. pylori isolates by PCR, Lai et al. [15] described a prevalence of 28% for mixed infections. Additionally, Kibria et al. [16] studied the prevalence of mixed H. pylori infection using both methods mentioned above (RAPD fingerprinting and multiplex PCR amplification for cagA and vacA alleles). An overall prevalence of 60.2% was obtained. Recently, an event of multiple infection was detected from 52 H. pylori clones isolated from 11 patients by sequencing of six housekeeping genes (amiA, cgt, cpn60, cpn70, dnaJ, and luxS) [11]. However, Raymond et al. [17] detected mixed infection in all six members of a family analyzing 107 clones by multilocus sequence typing (MLST) of two housekeeping genes (hspA and glmM).
To our knowledge, the present work is the first study that uses amplicon sequencing of housekeeping genes to detect H. pylori multi-infections. Amplicon sequencing is a high-throughput technique with greater resolution when compared to other methods (i.e., RAPD fingerprinting, multiplex cagA and vacA PCR, and MLST) used in previous studies. In the present study, multiple H. pylori infections with a predominant strain were detected in all the gastric biopsies analyzed in accordance with results obtained for Raymond et al. [17]. Although further studies are needed to confirm our results, these suggest that mixed H. pylori infections are the main status in the colonization of the human gastric mucosa. In fact, a high frequency of recombination between unrelated strains in mixed colonization is the main cause of the panmictic population structure of H. pylori [18,19,20]. In this study, microevolution by mutation of original strains infecting the gastric mucosa [19,20,21] was also found, since different unique sequences were obtained in each OTU (if consisted of two or more amplicon sequences).
Amplicon sequencing of housekeeping, virulence or antibiotic-resistant genes could be useful in different fields. It could be useful for elucidating the mode of transmission of H. pylori, to perform epidemiological studies. The impact of mixed H. pylori infection in the gastric pathogenesis and the failure of antibiotic treatments also deserves additional investigation.
In conclusion, our study strongly suggests that multiple infections including a predominant strain and multiple minority H. pylori is the predominant pattern of H. pylori infections in humans. The clinical and biological relevance of this finding deserves further study.
4. Materials and Methods
4.1. Gastric Biopsies
Patient recruitment and gastric biopsies obtained by the Digestive Diseases Department of the Hospital Taulí (Sabadell, Catalonia, Spain) were described in previous studies [11,22]. The study was approved by the Ethics Committee in accordance with the Declaration of Helsinki. Biopsies were performed after obtaining the patients’ written informed consent. For the present study, DNA from five gastric biopsies corresponding to four different patients infected with H. pylori were analyzed. Regarding the patients, two suffered from gastric cancer (B247, B508), one from duodenal ulcer (B373), and one was without medical conditions, here labeled as normal stomach (B601). Histological diagnoses from the biopsies were: chronic active gastritis, atrophy, intestinal metaplasia, and adenocarcinoma. Biopsies B508S and B508T were taken from the same patient with gastric adenocarcinoma (B508). Specifically, biopsy B508S was taken from normal gastric tissue and biopsy B508T from the tumoral tissue (Table 5).

Table 5.
Samples included in this study.
4.2. DNA Extraction and PCR Amplification
Extraction and sequential purification of DNA were performed using the MasterPure kit (Epicentre, Illumina). Isolated DNA was quantified with a NanoDrop spectrophotometer (Nano-Drop Technologies, USA).
Using H. pylori specific primers, PCR amplifications of six housekeeping genes (amiA, cgt, cpn60, cpn70, dnaJ, and luxS) were performed as described in a previous work of the authors [11], in which these genes were useful for H. pylori clone’s discrimination. From these genes, cgt and luxS demonstrated the best PCR results (a single, clear band with the corresponding length revealed in agarose gel) and therefore, were selected for amplicon sequencing.
Fusion cgt and luxS primers (Table 6) were designed for unidirectional sequencing following the 454 sequencing system guidelines for amplicon experimental design (454 Life Sciences Corp., Branford, CT, USA). PCR amplifications were carried out with the FastStart High Fidelity PCR System (Roche Diagnostics GmBH, Mannheim, Germany) in a total volume of 50 µL containing 2.5 mM MgCl2, 0.2 mM dNTPs, 2.5 U FastStart polymerase, 40 pmol of each primer (Isogen Life Science, PW De Meern, The Netherlands), and 200 ng DNA. The reaction mixtures were subjected to the following thermal cycling program in a PrimeG Thermal Cycler (Bibby Scientific, Staffordshire, UK): initial denaturation at 95 °C for 5 min; 35 cycles of 95 °C for 1 min, 60 °C (cgt) or 54 °C (luxS) for 1 min, and 72 °C for 1 min; final extension at 72 °C for 5 min. The amplified products were purified using ExoSAP-IT® kit (Affymetrix, Santa Clara, CA, USA).

Table 6.
Fusion primers designed for H. pylori specific PCR and amplicon sequencing.
4.3. Preparation of Libraries
Following the Amplicon Library Preparation Manual (454 Life Sciences Corp., Branford, CT, USA), the amplicons including fusion primers of each sample were sequenced using the 454 GS Junior instrument (Roche, Basel, Switzerland). After PCR purifications, library quantification was done using a QubitTM Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Each amplicon was diluted separately to 1 × 109 molecules/µL in a 1xTE buffer. Diluted amplicons were mixed to prepare the amplicon pool. As the size of cgt and luxS amplicons were different (685 bp for cgt and 499 bp for luxS), mixed volumes were adjusted to the ratio 1.2:1 cgt:luxS. The amplicon pool was diluted to 1 × 107 molecules/µL in molecular biology grade water. Once the libraries were constructed and pooled, emPCR amplification and sequencing were performed in the facilities of the Genomics Unit of Scientific and Technological Centers from the University of Barcelona (CCiTUB).
4.4. Data Processing
Raw data were processed by the GS Run Processor, which resulted in FASTQ and Standard Flowgram Format (SFF) files that were used in subsequent analysis. Amplicon sequences were assigned to each corresponding sample using the multiplex identifiers (MIDs), generating five files for each gene (Table 6). Thereafter, MIDs, sequences <50 bp or containing >5% Ns were removed from FASTQ files [23]. This processing was carried out by the CCiTUB.
Sequences in FASTA files were aligned with cgt or luxS reference sequences [11] using the MEGA7 software [12]. The aim was to check that amplicon sequences corresponded to cgt or luxS sequences. As mentioned above (see Section 2.3), some reference sequences corresponded to H. pylori strains isolated previously from the gastric biopsies B247, B373, and B508; replicate biopsies of the same patient at the same time were used in the present study. Additionally, in this process, the length of the sequences was adjusted. All sequences were deposited in Genbank under the project accession number PRJNA434670.
Sequences were analyzed with Mothur [24] following the subsequent protocol (all commands were run with the default parameters except when explicitly specified): first, the unique.seqs command was used in order to obtain the unique sequences of the sequence file by dereplication. Next, the merge.files command concatenates multiple files, which is useful for merging multiple FASTA-formatted sequence files. The make.group command reads a series of FASTA files and creates a group file, which is used to assign sequences to a specific group. The dist.seqs command then calculates uncorrected pairwise distances between aligned DNA sequences. By default, dist.seqs penalizes terminal gaps. This option was excluded and the default value of countends was changed to False. Afterwards, the cluster command was used to assign sequences to OTUs; the furthest neighbor clustering method was chosen, as it is the most conservative way to select OTUs. The make.shared and the summary.shared commands were used successively to obtain a summary file with the calculated values for each line in the OTU data and for all possible comparisons between the different groups in the group file. The get.oturep command generated a FASTA-formatted sequence file containing only one representative sequence for each OTU. Finally, the classify.seqs command was used to taxonomically classify the sequences.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/2/97/s1, Table S1: Reference sequences used in this study.
Author Contributions
Conceptualization, M.P. and D.M.-G.; data curation, M.P., A.M.C., G.M.D., M.G.I.L., and D.M.-G.; formal analysis, M.P., A.M.C., G.M.D., M.G.I.L., and D.M.-G.; funding acquisition, X.C. and D.M.-G.; investigation, M.P., M.J.R.-L., S.L., X.C., and D.M.-G.; methodology, M.P., A.M.C., G.M.D., M.J.R.-L., S.L., M.G.I.L., and D.M.-G.; project administration, X.C. and D.M.-G.; resources, M.P., A.M.C., G.M.D., M.J.R.-L., S.L., X.C., M.G.I.L., and D.M.-G.; software, M.P., A.M.C., G.M.D., M.G.I.L., and D.M.-G.; supervision, M.G.I.L. and D.M.-G.; validation, M.P., N.P., and D.M.-G.; visualization, M.P., N.P., A.M.C., G.M.D., M.J.R.-L., S.L., X.C., M.G.I.L., and D.M.-G.; writing—original draft, M.P. and D.M.-G.; writing—review and editing, M.P., N.P., A.M.C., G.M.D., M.J.R.-L., S.L., X.C., M.G.I.L., and D.M.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by philanthropic donations from the FUNDACIÓ LA MARATÓ DE TV3 (Barcelona, Catalonia, Spain), grant number 1007/C/2013.
Acknowledgments
Núria Piqué is a Serra Húnter Fellow. We would like to thank the Genomics Unit of Scientific and Technological Centers from the University of Barcelona (CCiTUB) and Íñigo J. Salanueva from Roche Diagnostics SL (Madrid, Spain) for their technical support. This study was awarded by, and received a prize by Roche Diagnostics SL (award number METAGC14) consisting of a GS Junior run.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Moodley, Y.; Linz, B.; Bond, R.P.; Nieuwoudt, M.; Soodyall, H.; Schlebusch, C.M.; Bernhöft, S.; Hale, J.; Suerbaum, S.; Mugisha, L.; et al. Age of the association between Helicobacter pylori and man. PLoS Pathog. 2012, 8, e1002693. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Shiota, S.; Yamaoka, Y. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. Infect. Genet. Evol. 2012, 12, 203–213. [Google Scholar] [CrossRef]
- Piqué, N.; Palau, M.; Berlanga, M.; Miñana-Galbis, D. Advances in the research of new genetic markers for the detection of Helicobacter pylori infection. In Recent Advances in Pharmaceutical Sciences VI; Transworld Research Network: Kerala, India, 2016; pp. 165–188. [Google Scholar]
- Hashi, K.; Imai, C.; Yahara, K.; Tahmina, K.; Hayashi, T.; Azuma, T.; Miyabe-Nishiwaki, T.; Sato, H.; Matsuoka, M.; Niimi, S.; et al. Evaluating the origin and virulence of a Helicobacter pylori cagA-positive strain isolated from a non-human primate. Sci. Rep. 2018, 8, 15981. [Google Scholar] [CrossRef] [PubMed]
- Correa, P.; Houghton, J. Carcinogenesis of Helicobacter pylori. Gastroenterology 2007, 133, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Buzás, G.M. Benign and malignant gastroduodenal diseases associated with Helicobacter pylori: A narrative review and personal remarks in 2018. Minerva Gastroenterol. Dietol. 2018, 64, 280–296. [Google Scholar] [CrossRef] [PubMed]
- Quaglia, N.C.; Dambrosio, A. Helicobacter pylori: A foodborne pathogen? World J. Gastroenterol. 2018, 24, 3472–3487. [Google Scholar] [CrossRef] [PubMed]
- Kabamba, E.T.; Tuan, V.P.; Yamaoka, Y. Genetic populations and virulence factors of Helicobacter pylori. Infect. Genet. Evol. 2018, 60, 109–116. [Google Scholar] [CrossRef]
- Smith, S.; Fowora, M.; Pellicano, R. Infections with Helicobacter pylori and challenges encountered in Africa. World J. Gastroenterol. 2019, 25, 3183–3195. [Google Scholar] [CrossRef]
- Ben Mansour, K.; Fendri, C.; Battikh, H.; Garnier, M.; Zribi, M.; Jlizi, A.; Burucoa, C. Multiple and mixed Helicobacter pylori infections: Comparison of two epidemiological situations in Tunisia and France. Infect. Genet. Evol. 2016, 37, 43–48. [Google Scholar] [CrossRef]
- Palau, M.; Kulmann, M.; Ramírez-Lázaro, M.J.; Lario, S.; Quílez, M.E.; Campo, R.; Piqué, N.; Calvet, X.; Miñana-Galbis, D. Usefulness of housekeeping genes for the diagnosis of Helicobacter pylori infection, strain discrimination and detection of multiple infection. Helicobacter 2016, 21, 481–487. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Toita, N.; Yokota, S.; Fujii, N.; Konno, M. Clonality Analysis of Helicobacter pylori in patients isolated from several biopsy specimens and gastric juice in a Japanese urban population by random amplified polymorphic DNA fingerprinting. Gastroenterol. Res. Pract. 2013, 2013, 721306. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.M.; Sheu, B.S.; Lu, C.C.; Yang, H.B.; Wu, J.J. Mixed infections of Helicobacter pylori: Tissue tropism and histological significance. Clin. Microbiol. Infect. 2009, 15, 253–259. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lai, C.H.; Huang, J.C.; Chiang-Ni, C.; Li, J.-P.; Wu, L.-T.; Wu, H.-S.; Sun, Y.-C.; Lin, M.-L.; Lee, J.-F.; Lin, H.-J. Mixed infections of Helicobacter pylori isolated from patients with gastrointestinal diseases in Taiwan. Gastroenterol. Res. Pract. 2016, 2016, 7521913. [Google Scholar] [CrossRef] [PubMed]
- Kibria, K.M.; Hossain, M.E.; Sultana, J.; Sarker, S.A.; Bardhan, P.K.; Rahman, M.; Nahar, S. The prevalence of mixed Helicobacter pylori infections in symptomatic and asymptomatic subjects in Dhaka, Bangladesh. Helicobacter 2015, 20, 397–404. [Google Scholar] [CrossRef]
- Raymond, J.; Thiberg, J.M.; Chevalier, C.; Kalach, N.; Bergeret, M.; Labigne, A.; Dauga, C. Genetic and transmission analysis of Helicobacter pylori strains within a family. Emerg. Infect. Dis. 2004, 10, 1816–1821. [Google Scholar] [CrossRef]
- Falush, D.; Kraft, C.; Taylor, N.S.; Correa, P.; Fox, J.G.; Achtman, M.; Suerbaum, S. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: Estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. USA 2001, 98, 15056–15061. [Google Scholar] [CrossRef]
- Morelli, G.; Didelot, X.; Kusecek, B.; Schwarz, S.; Bahlawane, C.; Falush, D.; Suerbaum, S.; Achtman, M. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 2010, 6, e1001036. [Google Scholar] [CrossRef]
- Cao, Q.; Didelot, X.; Wu, Z.; Li, Z.; He, L.; Li, Y.; Ni, M.; You, Y.; Lin, X.; Li, Z.; et al. Progressive genomic convergence of two Helicobacter pylori strains during mixed infection of a patient with chronic gastritis. Gut 2015, 64, 554–561. [Google Scholar] [CrossRef]
- Linz, B.; Windsor, H.M.; McGraw, J.J.; Hansen, L.M.; Gajewski, J.P.; Tomsho, L.P.; Hake, C.M.; Solnick, J.V.; Schuster, S.C.; Marshall, B.J. A mutation burst during the acute phase of Helicobacter pylori infection in humans and rhesus macaques. Nat. Commun. 2014, 5, 4165. [Google Scholar] [CrossRef]
- Lario, S.; Ramírez-Lázaro, M.J.; Sanjuan-Herráez, D.; Brunet-Vega, A.; Pericay, C.; Gombau, L.; Junquera, F.; Quintás, G.; Calvet, X. Plasma sample based analysis of gastric cancer progression using targeted metabolomics. Sci. Rep. 2017, 7, 17774. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomics datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).