Hypermutation as an Evolutionary Mechanism for Achromobacter xylosoxidans in Cystic Fibrosis Lung Infection
Abstract
1. Introduction
2. Results
2.1. Variant Analysis
2.2. Genetic Basis of Hypermutation
2.3. Mobile Genetic Elements
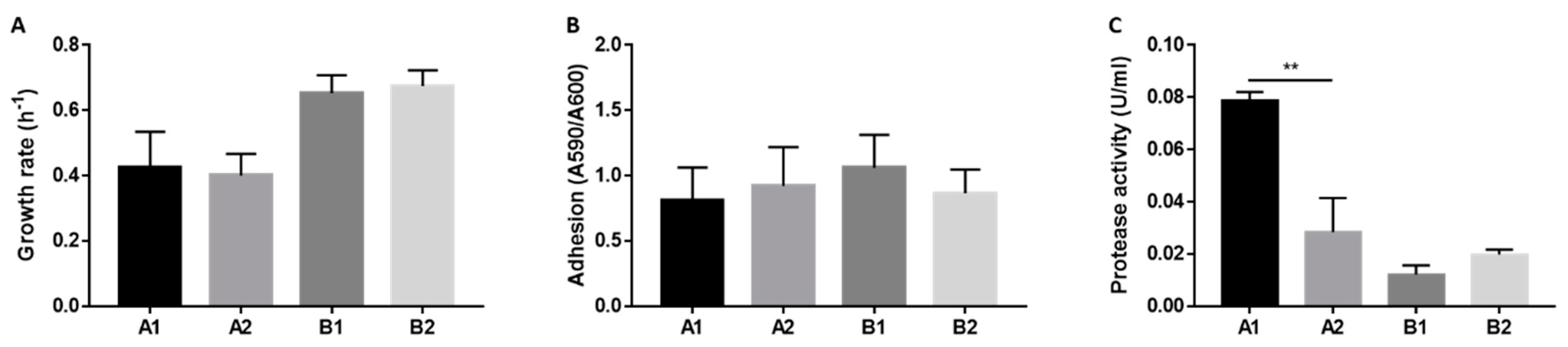
2.4. Phenotypic Features
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. Library Preparation and Whole-Genome Sequencing
4.3. De Novo Assembly
4.4. Variant Analysis
4.5. Mutator Genes Analysis
4.6. Mobilome Analysis
4.7. Growth Curves
4.8. Adhesion Assay
4.9. Protease Activity Measurement
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ciofu, O.; Hansen, C.R.; Hoiby, N. Respiratory bacterial infections in cystic fibrosis. Curr. Opin. Pulm. Med. 2013, 19, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.R.; Pressler, T.; Nielsen, K.G.; Jensen, P.Ø.; Bjarnsholt, T.; Høiby, N. Inflammation in Achromobacter xylosoxidans infected cystic fibrosis patients. J. Cyst. Fibros. 2010, 9, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Ridderberg, W.; Wang, M.; Norskov-Lauritsen, N. Multilocus sequence analysis of isolates of Achromobacter from patients with cystic fibrosis reveals infecting species other than Achromobacter xylosoxidans. J. Clin. Microbiol. 2012, 50, 2688–2694. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.R.; Pressler, T.; Ridderberg, W.; Johansen, H.K.; Skov, M. Achromobacter species in cystic fibrosis: Cross-infection caused by indirect patient-to-patient contact. J. Cyst. Fibros. 2013, 12, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Lambiase, A.; Catania, M.R.; del Pezzo, M.; Rossano, F.; Terlizzi, V.; Sepe, A.; Raia, V. Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Firmida, M.C.; Pereira, R.H.V.; Silva, E.A.S.R.; Marques, E.A.; Lopes, A.J. Clinical impact of Achromobacter xylosoxidans colonization/infection in patients with cystic fibrosis. Braz. J. Med. Biol. Res. 2016, 49, e5097. [Google Scholar] [CrossRef]
- Ridderberg, W.; Nielsen, S.M.; Norskov-Lauritsen, N. Genetic Adaptation of Achromobacter sp. during Persistence in the Lungs of Cystic Fibrosis Patients. PLoS ONE 2015, 10, e0136790. [Google Scholar] [CrossRef]
- Marvig, R.L.; Johansen, H.K.; Molin, S.; Jelsbak, L. Genome analysis of a transmissible lineage of pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet. 2013, 9, e1003741. [Google Scholar] [CrossRef]
- Oliver, A.; Cantón, R.; Campo, P.; Baquero, F.; Blázquez, J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 2000, 288, 1251–1254. [Google Scholar] [CrossRef]
- Jakobsen, T.H.; Hansen, M.A.; Jensen, P.Ø.; Hansen, L.; Riber, L.; Cockburn, A.; Kolpen, M.; Hansen, C.R.; Ridderberg, W.; Eickhardt, S.; et al. Complete genome sequence of the cystic fibrosis pathogen Achromobacter xylosoxidans NH44784-1996 complies with important pathogenic phenotypes. PLoS ONE 2013, 8, e68484. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 10.0; 2020; Available online: http://www.eucast.org/clinical_breakpoints/ (accessed on 15 January 2020).
- Feugeas, J.P.; Tourret, J.; Launay, A.; Bouvet, O.; Hoede, C.; Denamur, E.; Tenaillon, O. Links between Transcription, Environmental Adaptation and Gene Variability in Escherichia coli: Correlations between Gene Expression and Gene Variability Reflect Growth Efficiencies. Mol. Biol. Evol. 2016, 33, 2515–2529. [Google Scholar] [CrossRef] [PubMed]
- Elena, S.F.; Lenski, R.E. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat. Rev. Genet. 2003, 4, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Hall, L.M.C.; Henderson-Begg, S.K. Hypermutable bacteria isolated from humans—A critical analysis. Microbiology 2006, 152 Pt 9, 2505–2514. [Google Scholar] [CrossRef]
- Hogardt, M.; Hoboth, C.; Schmoldt, S.; Henke, C.; Bader, L.; Heesemann, J. Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 2007, 195, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Mena, A.; Smith, E.E.; Burns, J.L.; Speert, D.P.; Moskowitz, S.M.; Perez, J.L.; Oliver, A. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J. Bacteriol. 2008, 190, 7910–7917. [Google Scholar] [CrossRef]
- Sundin, G.W.; Weigand, M.R. The microbiology of mutability. FEMS Microbiol. Lett. 2007, 277, 11–20. [Google Scholar] [CrossRef]
- Cooper, V.S.; Lenski, R.E. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 2000, 407, 736–739. [Google Scholar] [CrossRef]
- Notley-McRobb, L.; Seeto, S.; Ferenci, T. Enrichment and elimination of mutY mutators in Escherichia coli populations. Genetics 2002, 162, 1055–1062. [Google Scholar]
- Marvig, R.L.; Sommer, L.M.; Molin, S.; Johansen, H.K. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 2015, 47, 57–64. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Conesa, A.; Garcia-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Oliver, A. Mutators in cystic fibrosis chronic lung infection: Prevalence, mechanisms, and consequences for antimicrobial therapy. Int. J. Med. Microbiol. 2010, 300, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Clausen, P.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Liu, M.; Li, X.; Xie, Y.; Bi, D.; Sun, J.; Li, J.; Tai, C.; Deng, Z.; Ou, H.Y. ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019, 47, D660–D665. [Google Scholar] [CrossRef]
- Sandri, A.; Ortombina, A.; Boschi, F.; Cremonini, E.; Boaretti, M.; Sorio, C.; Melotti, P.; Bergamini, G.; Lleo, M. Inhibition of Pseudomonas aeruginosa secreted virulence factors reduces lung inflammation in CF mice. Virulence 2018, 9, 1008–1018. [Google Scholar] [CrossRef]
| Patient | Isolate | Genome Size (bp) | GC-Content (%) | No. Contigs | N50 | Mean Coverage Depth (x) | No. Coding Sequences | Mapping Reads (%) |
|---|---|---|---|---|---|---|---|---|
| A | A1 | 6913734 | 68.09 | 291 | 78688 | 66 | 6359 | 98.3 |
| A | A2 | 6879357 | 68.08 | 187 | 78799 | 50 | 6339 | 97.87 |
| B | B1 | 6634994 | 67.63 | 178 | 100359 | 49 | 6041 | 98.07 |
| B | B2 | 6628209 | 67.63 | 158 | 93753 | 40 | 6050 | 98.35 |
| Isolate Reads | Longitudinal Isolate de novo Assembly | Mapping Reads vs de novo Assembly (%) | Mapping Reads vs NH44784-1996 (%) |
|---|---|---|---|
| A1 | A2 | 96.49 | 52.37 |
| A2 | A1 | 97.31 | 46.27 |
| B1 | B2 | 98.78 | 83.08 |
| B2 | B1 | 96.73 | 82.98 |
| Analysis | Comparison between Longitudinal Isolates | Comparison with Reference Genome | ||||
|---|---|---|---|---|---|---|
| Genome | A | B | A1 | A2 | B1 | B2 |
| Total | 187 | 8 | 162 | 70 | 10 | 42 |
| No. SNPs | 150 | 6 | 150 | 68 | 8 | 39 |
| No. indel | 37 | 2 | 12 | 2 | 2 | 3 |
| No. Synonymous SNPs | 38 | 3 | 87 | 43 | 4 | 24 |
| No. Missense SNPs | 89 | 3 | 53 | 14 | 4 | 10 |
| No. Nonsense SNPs | 5 | 0 | 0 | 0 | 0 | 0 |
| No. Other SNPs | 18 | 0 | 10 | 11 | 0 | 5 |
| Frameshift | 13 | 2 | 8 | 0 | 0 | 2 |
| Disruptive in-frame insertion | 1 | 0 | 0 | 0 | 0 | 0 |
| Disruptive in-frame deletion | 0 | 0 | 0 | 0 | 0 | 1 |
| Stop gain | 5 | 0 | 1 | 0 | 0 | 0 |
| Stop lost | 1 | 0 | 0 | 0 | 0 | 0 |
| Transitions | 128 | 2 | 110 | 43 | 5 | 24 |
| Transversions | 22 | 4 | 40 | 25 | 3 | 15 |
| Transition/transversion ratio | 5.8 | 0.5 | 2.75 | 1.72 | 1.66 | 1.6 |
| Analysis | Comparison between Longitudinal Isolates | Comparison with Reference Genome | ||||||
|---|---|---|---|---|---|---|---|---|
| Functional Category | A | B | Total | A1 | A2 | B1 | B2 | Total |
| Metabolism | 66 | 0 | 66 | 49 | 18 | 3 | 4 | 74 |
| Transcription and translation | 21 | 1 | 22 | 13 | 3 | 1 | 4 | 21 |
| Virulence, disease and defence | 5 | 0 | 5 | 2 | 4 | 0 | 0 | 6 |
| Hypothetical protein | 34 | 2 | 36 | 4 | 1 | 2 | 3 | 10 |
| Transporter | 21 | 3 | 24 | 18 | 6 | 0 | 3 | 27 |
| Iron acquisition and metabolism | 10 | 0 | 10 | 6 | 1 | 0 | 1 | 8 |
| Stress response | 2 | 0 | 2 | 1 | 1 | 0 | 0 | 2 |
| DNA repair | 2 | 0 | 2 | 0 | 3 | 0 | 0 | 3 |
| Antibiotic resistance | 7 | 0 | 7 | 5 | 3 | 0 | 0 | 8 |
| Mobile genetic elements | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other | 3 | 0 | 3 | 4 | 0 | 0 | 0 | 4 |
| Total | 171 | 6 | 177 | 102 | 40 | 6 | 15 | 163 |
| Mobile Elements | A1 | A2 | B1 | B2 |
|---|---|---|---|---|
| PHAGE_Burkho_KS9_NC_013055 | 21 | 21 | - | - |
| PHAGE_Burkho_Bcep176_NC_007497 | 46.7 | 39 | 18.6 | 24.6 |
| PHAGE_Salmon_118970_sal3_NC_031940 | - | 31.6 | - | - |
| PHAGE_Pseudo_YMC11/02/R656_NC_028657 | - | 29.3 | - | - |
| PHAGE_Burkho_BcepMu_NC_005882 | - | - | 40.2 | 40.9 |
| PHAGE_Burkho_KS14_NC_015273 | - | - | 31.7 | - |
| PHAGE_Aeromo_vB_AsaM_56_NC_019527 | - | - | - | - |
| PHAGE_Synech_S_CBS1_NC_016164 | - | - | - | - |
| ICEs | 93 | 227 | - | - |
| IMEs | - | - | 15.6 | 15.6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veschetti, L.; Sandri, A.; Krogh Johansen, H.; Lleò, M.M.; Malerba, G. Hypermutation as an Evolutionary Mechanism for Achromobacter xylosoxidans in Cystic Fibrosis Lung Infection. Pathogens 2020, 9, 72. https://doi.org/10.3390/pathogens9020072
Veschetti L, Sandri A, Krogh Johansen H, Lleò MM, Malerba G. Hypermutation as an Evolutionary Mechanism for Achromobacter xylosoxidans in Cystic Fibrosis Lung Infection. Pathogens. 2020; 9(2):72. https://doi.org/10.3390/pathogens9020072
Chicago/Turabian StyleVeschetti, Laura, Angela Sandri, Helle Krogh Johansen, Maria M. Lleò, and Giovanni Malerba. 2020. "Hypermutation as an Evolutionary Mechanism for Achromobacter xylosoxidans in Cystic Fibrosis Lung Infection" Pathogens 9, no. 2: 72. https://doi.org/10.3390/pathogens9020072
APA StyleVeschetti, L., Sandri, A., Krogh Johansen, H., Lleò, M. M., & Malerba, G. (2020). Hypermutation as an Evolutionary Mechanism for Achromobacter xylosoxidans in Cystic Fibrosis Lung Infection. Pathogens, 9(2), 72. https://doi.org/10.3390/pathogens9020072








