Abstract
Sulcascaris sulcata (Anisakidae), a pathogenic nematode of sea turtles, may cause ulcerous gastritis with different degrees of severity. Previous studies demonstrated a high prevalence of infection in the Mediterranean loggerhead turtle (Caretta caretta), although no data on the potential intermediate hosts of this nematode has been published thus far from the Mediterranean basin. Here, using molecular analyses, we demonstrated that the cross sections of nematode larvae observed histologically in Mediterranean mussels (Mytilus galloprovincialis) collected from a farm along the Tyrrhenian coast of southern Italy belong to S. sulcata. The BLAST analysis of sequences at the ITS2 region of rDNA and mtDNA cox2 gene loci here obtained from samples of two Mediterranean mussels containing nematode larvae showed 100% homology with those at the same gene loci from the adults of S. sulcata collected from the Mediterranean Sea and deposited in GenBank. To our knowledge, this study is the first to present data on a potential intermediate host of S. sulcata in the Mediterranean basin and to report a nematode parasite from the Mediterranean mussel.
1. Introduction
The anisakid nematode Sulcascaris sulcata is a pathogenic parasite of the esophagus and stomach of sea turtles being able to cause ulcerous gastritis with different degrees of severity predominantly depending on the intensity of infection. S. sulcata infects the loggerhead turtle (Caretta caretta), green turtle (Chelonia mydas), and Kemp’s ridley turtle (Lepidochelys kempii) in the Mediterranean and Caribbean Seas, and the South Atlantic, Western Atlantic, and Western Pacific Oceans [1].
Berry and Cannon [2] demonstrated experimentally that hatchling loggerhead turtles become infected by ingesting scallops infected with fourth stage larvae S. sulcata. Larvae attach at the base of the esophagus where four molts occur about three weeks after infection and mature to adults in at least 5 months. Adult parasites live in the stomach of sea turtles and eggs are shed in the marine environment with host feces. Two molts occur in the egg until the development of the third stage larva, which spontaneously hatches and develops in bivalve and gastropod mollusks to fourth stage larva after 3–4 months. The duration of the natural cycle could be of two years [2].
Recently, we described the pathological changes caused by S. sulcata in two loggerhead populations inhabiting the Tyrrhenian and the Adriatic Seas in the Mediterranean basin [1]; however, no data exists on its intermediate hosts in this geographical area. Published data of intermediate hosts of S. sulcata exists only for Australian and American waters where at least 17 taxa of mollusks have been found infected or have been experimentally infected including six gastropods and 11 bivalves (listed in Table 1).

Table 1.
Marine molluscan hosts for larval forms of Sulcascaris sulcata updated by Lichtenfels et al. [3].
While studying the occurrence of protozoan parasites in the Mediterranean mussel (Mytilus galloprovincialis) along the coast of Campania region of southern Italy, larval forms of nematodes were observed histologically in the tissues of Mediterranean mussels. Herein, using molecular analysis, we report for the first time the occurrence of larvae of S. sulcata in Mediterranean mussels from the Tyrrhenian Sea.
2. Results
A total of five (1.4%) individual Mediterranean mussels collected on February (one mussel), May (one mussel), July (two mussels), and August (one mussel) were histologically positive to one (n = 3), two (n = 1), and three (n = 1) cross sections of nematode larvae, respectively. Cross sections of larvae (n = 4) measured in mean 290.5 μm (range: 178 to 430.9) x 202.5 μm (range: 149.4 to 292.8). Larvae were encysted within the foot of Mediterranean mussels extending to the digestive gland and revealed host inflammatory reaction in all cases. In most cases, larvae appeared to be viable and were surrounded by well-defined hemocytic capsules.
The BLAST analysis of the ITS2 of rDNA and mtDNA cox2 sequences produced here from samples of two Mediterranean mussels containing nematode larvae showed 100% homology with those of adult stages of S. sulcata from the Mediterranean Sea, previously deposited in GenBank (Figure 1 and Figure 2). Sequences obtained in the present study were deposited in GenBank under accession numbers MN736715.1 and MN736716.1 for ITS2 and MN991208 and MN991209 for cox2.
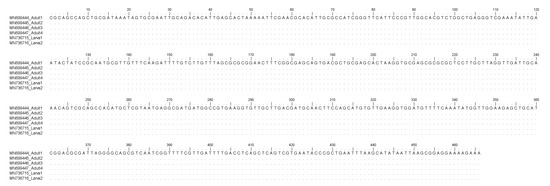
Figure 1.
Nucleotide sequence alignment of the nDNA ITS2 obtained from Sulcascaris sulcata larvae (Accession No. MN736715, MN736716) in comparison with those previously identified as S. sulcata adults (Accession No. MN699444, MN699445, MN699446, MN699447). Dots indicate identity with the consensus sequence.
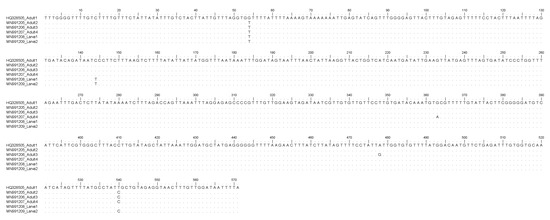
Figure 2.
Nucleotide sequence alignment of the mtDNA cox2 obtained from Sulcascaris sulcata larvae (Accession No. MN991208, MN991209) in comparison with those previously identified as S. sulcata adults (Accession No. HQ328505, MN991205; MN991206; MN991207). Dots indicate identity with the consensus sequence.
3. Discussion
The Mediterranean mussel has been intensively studied for pathogens in the whole Mediterranean as well as the Campanian coastal areas [19,20,21], but to date, our study describes the first finding of a parasite nematode in this bivalve species. According to McElwain et al. [22], parasitic nematodes are uncommon in marine bivalves. In mussels, the only data is by Lauckner [23], reporting in North Atlantic an infection by an anisakid larva thought to be Phocanema (Pseudoterranova) decipiens in a North Atlantic Mytilus edulis.
In the Mediterranean Sea, the occurrence of S. sulcata in loggerhead turtles seems to be limited to its eastern basin and the Tyrrhenian Sea [1,24]. Recently, we observed that all Sulcascaris positive loggerhead turtles from the Tyrrhenian came from the coastal sites located between Castel Volturno and the Naples Gulf (it includes also Monte di Procida), where all Mediterranean mussel farms and wild bivalve beds, registered along the Campania coast are concentrated [1]. According to Berry and Cannon [2], it is plausible to think that the farmed Mediterranean mussels may have been infected by filtering seawater contaminated with S. sulcata eggs and/or larvae laid with feces by an infected loggerhead turtle while feeding on farm ropes of Mediterranean mussels.
Adult individuals of loggerhead turtles are frequently observed feeding on Mediterranean mussel farm ropes along the coast of Campania. Ingestion of large amounts of Mediterranean mussels by loggerhead turtles was confirmed by our unpublished observations at the post-mortem examination of large individuals stranded along the coast of Campania. In those loggerhead turtles, the remains of Mediterranean mussels in the gastrointestinal tract commonly occurred in the presence of S. sulcata individuals within the gastric lumen. This particular feeding habit, rarely observed in other Mediterranean basins [25,26,27], could favor the increase of the incidence of infection with S. sulcata in Mediterranean mussels along the coast of Campania.
Regarding the site of infection, all larval forms of S. sulcata occurred within the foot of the Mediterranean mussels showing an encapsulation-type inflammatory response [28,29]. In Australian and American waters, fourth-stage larvae occurred most commonly within the adductor muscle and gonads of its molluscan hosts (Table 1). The only report of S. sulcata larvae within the foot was in the bivalve Spisula solidissima from the Atlantic coast of United States where larvae were associated with necrosis and hemocytic infiltration [16]. It has been documented that under the influence of environmental changes and the associated degradation of preferred habitats, host switching by parasites may increase [30], and parasite larvae may adapt their tropism changing the intermediate host species [31]. In particular, in the absence of preferred intermediate hosts and rich presence of definitive hosts, other marine mollusks might play a prominent role in the parasite transmission [32], and parasites may adapt in different way to the new hosts. Most of the marine bivalves found as intermediate hosts in Australian and American waters are Pectinidae of commercial importance (Table 1). Contrarily to that observed in other Mediterranean areas (for example the Adriatic Sea), Pectinidae mollusks along the Tyrrhenian coast are rare and do not represent usual food items for loggerhead turtles [33]. The significantly higher values of S. sulcata infection among loggerhead turtles feeding in Adriatic compared to Tyrrhenian have been attributed to the differences of regional habitats supporting higher abundance and diversity of mollusk intermediate hosts [1], and it is plausible to suppose that other intermediate hosts for S. sulcata may occur in other Mediterranean areas where this nematode is known to be widespread.
It is unknown if the adductor muscles of examined Mediterranean mussels were also infected because routinely it has been left on the internal surface of the valves, and it has not been processed by histological examination [34]. Further studies to evaluate the occurrence of nematode larval forms within the adductor muscle are warranted to understand if the prevalence of infection for S. sulcata larvae in Mediterranean mussels from this basin was underestimated and if it was an occasional finding.
Here, the molecular study based on the sequence analysis at the ITS2 region of rDNA and the mtDNA cox2 gene loci, allowed the first molecular identification of the larval nematodes as belonging to S. sulcata in Mediterranean mussels. In particular, sequences analysis of the mtDNA cox2 generally allows the identification of larval stages of anisakid nematodes included in the genera Anisakis, Pseudoterranova, and Contracaecum [35]. Additionally, the same primers used for the anisakid parasites permitted, in recent years, the direct sequencing of several adult nematodes belonging to the species S. sulcata obtained from loggerhead turtles stranded along the Mediterranean coast (Mattiucci, unpublished data). As a consequence, the direct alignment of the mtDNA cox2, here obtained on the two samples with respect to the sequences of adult specimens, have permitted the identification of those ascaridoid larval stages as corresponding to S. sulcata. On the other hand, mtDNA cox2 allowed also the identification of Anisakis pegreffii larvae in human granuloma tissues, surgically removed and formalin-embedded in human cases of anisakiasis [35]. Analogously, the direct sequences analysis of the ITS2 region of rDNA was also previously performed on adults of S. sulcata (Marcer, unpublished data) allowing the identification of its larvae in the same DNA samples. However, in the case of ITS2 region of rDNA, being generally a conservative gene locus in anisakid nematodes [35], no genetic variation was observed between adult and larval specimens of S. sulcata, here examined (Figure 1), whereas the mtDNA cox2 presented, at the intraspecific level, variation at non-diagnostic nucleotide positions (Figure 2); this finding is in accordance with the high polymorphism generally detected at this gene locus at the intraspecific level in several heteroxenous anisakid nematodes, such as those observed in species of the genus Anisakis [35].
This result represents the first report of S. sulcata invading tissues of Mediterranean mussels, and it strongly suggests the Mediterranean mussel as an intermediate host for S. sulcata in the Tyrrhenian Sea. The Mediterranean mussel is the most economically important shellfish produced and consumed in the Western Mediterranean. The Gulf of Naples in southern Italy is among the most important production sites of the Mediterranean mussel in Italy with about 4170 tons per year [36]. Infection of Mediterranean mussels by larval nematodes may cause economic losses due to the prohibition of its marketing. Experimental studies performed on fish, chickens, and cats suggest that infection by S. sulcata may occur only in sea turtles as definitive hosts [2]; however since S. sulcata is a strictly related species to zoonotic anisakid parasites [35,37], the concern regarding public health should not be underestimated.
4. Materials and Methods
4.1. General Data
From February to August 2018, a total of 363 individual Mediterranean mussels (>5 cm in length) were collected from a mussel farm located in Monte di Procida (Naples municipality), under the framework of a control program for protozoan surveillance in the Campania region of southern Italy. The farm was located in the Tyrrhenian Sea about 800 m offshore the Campanian coastline on the sand bottom at a depth of 30 m; the Mediterranean mussels were cultured on ropes suspended from floating buoys at a depth of 5–12 m.
4.2. Histological Procedures
Immediately after collection, the mussels were stored at 4 °C, and within 12 h they were processed by histological examination for pathogen detection [34]. Transverse sections of the mantle, gonad, digestive gland, gills, and foot were obtained in a single slice through the midbody, fixed in buffered 10% formalin, and sectioned at a thickness of 5 mm. Sections were stained with hematoxylin and eosin for histologic evaluation.
4.3. Anisakid DNA Extraction and Molecular Identification
To confirm the identity of larval nematodes observed by histological analysis, we performed the extraction of nematodes’ DNA from microtome slices of five paraffin-embedded infected Mediterranean mussels. The removal of paraffin and extraction of DNA was carried out using NucleoSpin® Tissue Macherey–Nagel kit (GmbH&Co., Germany) according to the manufacturer’s instructions for paraffin embedded tissues.
Amplification of the ITS2 region of rDNA was carried out on the samples using the primers D (5′-GAGTCGATGAAGAACGCAG-3′) and reverse B1 (5′-GAATCCTGGTTAGTTTCTTTTCCT-3′) [38] in a 50 µL reaction, comprising 5 µL DNA, 1.5 mM MgCl2, 0.2 mM dNTPs (MBI Fermentas, Germany), 1X PCR buffer, 0.3 μM each of forward and reverse primer, and 1 U Platinum Taq DNA Polymerase (Invitrogen). Molecular biology grade water was added up to the final volume. The mixture was amplified with an initial activation step at 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, DNA extension at 72 °C for 35 s, and a final extension step of 72 °C for 5 min.
For sequencing the mitochondrial cytochrome C oxidase subunit II (cox2) gene, PCR amplification was performed using the primers 211F (5′-TTTTCTAGTTATATAGATTGRTTTYAT-3′) and 210R (5′-CACCAACTCTTAAAATTATC-3′) [39,40]. PCRs were carried out in a 50 µL volume containing 30 pmol of each primer, MgCl2 2.5 mM (Amersham Pharmacia Biotech. Inc., Piscataway, NJ), 1× PCR buffer (Amersham Pharmacia Biotech. Inc., Piscataway, NJ, USA), DMSO 0.08 mM, dNTPs 0.4 mM (Sigma-Aldrich, St. Louis, MO, USA), 5 U of Taq Polymerase (Amersham Pharmacia Biotech. Inc., Piscataway, NJ, USA), and 10 ng of total DNA. PCR temperature conditions were the following: 94 °C for 3 min (initial denaturation), followed by 34 cycles at 94 °C for 3 min (denaturation), 46 °C for 60 s (annealing), 72 °C for 90 s (extension), followed by post-amplification at 72 °C for 10 min.
The PCR products were resolved in 2% agarose gel (expected fragments length: 500 bp of ITS2 and 629 bp of mtDNA cox2 gene) and sequenced in both directions by Macrogen (Macrogen Europe, The Netherlands). The chromatograms were corrected using the software ChromasPro version 2.4.3 (Technelysium Pty Ltd, Australia). The consensus sequences were assembled with the program SeqMan available in the DNAstar package. The consensus sequences were compared with the non-redundant database available in the GenBank using the software BLASTn [41].
Author Contributions
Conceptualization: M.S.; molecular analysis: E.M., F.M., M.P., and S.M.; histopathological analysis: B.D.U.; data curation and writing—original draft preparation: M.S.; reviewing and editing: all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Santoro, M.; Marchiori, E.; Iaccarino, D.; Degli Uberti, B.; Cassini, R.; Di Nocera, F.; Cerrone, A.; Galiero, G.; Marcer, F. Epidemiology of Sulcascaris sulcata (Nematoda: Anisakidae) ulcerous gastritis in the Mediterranean loggerhead sea turtle (Caretta caretta). Parasitol. Res. 2019, 118, 1457–1463. [Google Scholar] [CrossRef] [PubMed]
- Berry, G.N.; Cannon, L.R.G. The life history of Sulcascaris sulcata (Nematoda: Ascaridoidea), a parasite of marine molluscs and turtles. Int. J. Parasitol. 1981, 11, 43–54. [Google Scholar] [CrossRef]
- Lichtenfels, J.R.; Sawyer, T.K.; Miller, G.C. New hosts for larval Sulcascaris sp. (Nematoda, Anisakidae) and prevalence in the calico scallop (Argopecten gibbus). Trans. Am. Microsc. Soc. 1980, 99, 448–451. [Google Scholar] [CrossRef]
- Sprent, J.F.A. Ascaridoid nematodes of amphibians and reptiles: Sulcascaris. J. Helminthol. 1977, 51, 379–387. [Google Scholar] [CrossRef]
- Lester, R.J.G.; Blair, D.; Heald, D. Nematodes from scallops and turtles from Shark Bay, Western Australia. Aust. J. Mar. Freshw. Res. 1980, 31, 713–717. [Google Scholar] [CrossRef]
- Hutton, R.F. A second list of parasites from marine and coastal animals of Florida. Trans. Am. Microsc. Soc. 1964, 83, 439–447. [Google Scholar] [CrossRef]
- Cheng, T.C. Marine molluscs as hosts for symbioses with a review of known parasites of commercially important species. Adv. Mar. Biol. 1967, 5. [Google Scholar] [CrossRef]
- Lichtenfels, J.R.; Bier, J.W.; Madden, P.A. Larval anisakid (Sulcascaris) nematodes from Atlantic molluscs with marine turtles as definitive hosts. Trans. Am. Microsc. Soc. 1978, 97, 199–207. [Google Scholar] [CrossRef]
- Barber, B.J.; Blake, N.J.; Moyer, M.A.; Rodrick, G.E. Larval Sulcascaris sulcata from calico scallops, Argopecten gibbus, along the southeast coast of the United States. J. Parasitol. 1987, 73, 476–480. [Google Scholar] [CrossRef]
- Deardorff, T.L. Occurrence of larval Sulcascaris sulcata (Nematoda: Anisakidae) in the calico scallop, Argopecten gibbus, collected along the eastern coast of Florida, with comments on histopathology. Proc. Helminthol. Soc. Wash. 1989, 56, 82–85. [Google Scholar]
- Gutsell, J.S. Natural history of the bay scallop. U.S. Bur. Fish Bull. 1930, 46, 569–632. [Google Scholar]
- Cheng, T.C. Larval nematodes parasitic in shellfish. Mar. Fish Rev. 1978, 40, 39–42. [Google Scholar]
- Cobb, N.A. A nemic parasite of Pecten. J. Parasitol. 1930, 17, 104–105. [Google Scholar] [CrossRef]
- Amato, J.F.R.; Amato, S.B. Sulcascaris sulcata (Nematoda, Anisakinae) infecting sea scallops being exported from southeastern Brazil. Arq. Univ. Fed. Rural Rio Jan. 1982, 5, 61–67. [Google Scholar]
- Cannon, L.R.G. A larval ascaridoid nematode from Queensland scallops. Int. J. Parasitol. 1978, 8, 75–80. [Google Scholar] [CrossRef]
- Murchelano, R.A.; MacLean, S.A. Histopathology Atlas of the Registry of Marine Pathology; National Oceanic and Atmospheric Administration, National Marine Fisheries Service, United States Department of Commerce: Oxford, MD, USA, 1990.
- Lichtenfels, J.R.; Kern, F.G.; Zwerner, D.E.; Bier, J.W.; Madden, P.A. Anisakid nematodes in shellfish of Atlantic continental shelf of North America. Trans. Am. Microsc. Soc. 1976, 95, 265–266. [Google Scholar]
- Payne, W.L.; Gerding, T.A.; Dent, R.G.; Bier, J.W.; Jackson, G.J. Survey of the U.S. Atlantic Coast surf clam, Spisula solidissima, and clam products for anisakine nematodes and hyperparasitic protozoa. J. Parasitol. 1980, 66, 150–153. [Google Scholar] [CrossRef]
- Carella, F.; Aceto, S.; Maiolino, P.; De Vico, G. What is your diagnosis? Pale yellowish digestive gland and watery tissues in Mediterranean mussels. Vet. Clin. Pathol. 2011, 40, 273–274. [Google Scholar] [CrossRef]
- Carella, F.; Aceto, S.; Mangoni, O.; Mollica, M.P.; Cavaliere, G.; Trinchese, G.; Aniello, F.; De Vico, G. Assessment of the health status of mussels Mytilus galloprovincialis along the Campania coastal areas: A multidisciplinary approach. Front. Physiol. 2018, 9, 683. [Google Scholar] [CrossRef]
- Arienzo, M.; Toscanesi, M.; Trifuoggi, M.; Ferrara, L.; Stanislao, C.; Donadio, C.; Grazia, V.; De Vico, G.; Carella, F. Contaminants bioaccumulation and pathological assessment in Mytilus galloprovincialis in coastal waters facing the brownfield site of Bagnoli, Italy. Mar. Pollut. Bull. 2019, 140, 341–352. [Google Scholar] [CrossRef]
- McElwain, A.; Warren, M.B.; Pereira, F.B.; Ksepka, S.P.; Bullard, S.A. Pathobiology and first report of larval nematodes (Ascaridomorpha sp.) infecting freshwater mussels (Villosa nebulosa, Unionidae), including an inventory of nematode infections in freshwater and marine bivalves. Int. J. Parasitol. Parasites Wildl. 2019, 10, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Lauckner, G. Diseases of Mollusca: Bivalvia. In Diseases of Marine Animals. Volume II: Introduction, Bivalvia to Scaphopoda; Kinne, O., Ed.; Biologische Anstalt Helgoland: Hamburg, Germany, 1983; pp. 477–961. [Google Scholar]
- Santoro, M.; Badillo, F.J.; Mattiucci, S.; Nascetti, G.; Bentivegna, F.; Insacco, G.; Travaglini, A.; Paoletti, M.; Kinsella, J.M.; Tomás, J.; et al. Helminth communities of loggerhead turtles (Caretta caretta) from Central and Western Mediterranean Sea: The importance of host’s ontogeny. Parasitol. Int. 2010, 59, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Houghton, J.D.R.; Woolmer, A.; Hays, G.C. Sea turtle diving and foraging behavior around the Greek Island of Kefalonia. J. Mar. Biol. Assoc. UK 2000, 80, 761–762. [Google Scholar] [CrossRef][Green Version]
- Tomas, J.; Aznar, F.J.; Raga, J.A. Feeding ecology of the loggerhead turtle Caretta caretta in the western Mediterranean. J. Zool. Lond. 2001, 255, 525–532. [Google Scholar] [CrossRef]
- Casale, P.; Abbate, G.; Freggi, D.; Conte, N.; Oliverio, M.; Argano, R. Foraging ecology of loggerhead sea turtles Caretta caretta in the Central Mediterranean Sea: Evidence for a relaxed life history model. Mar. Ecol. Prog. Ser. 2008, 372, 265–276. [Google Scholar] [CrossRef]
- De Vico, G.; Carella, F. Morphological features of the inflammatory response in molluscs. Res. Vet. Sci. 2012, 93, 1109–1115. [Google Scholar] [CrossRef]
- Carella, F.; Feist, S.W.; Bignell, J.P.; De Vico, G. Comparative pathology in bivalves: Aetiological agents and disease processes. J. Invertebr. Pathol. 2015, 131, 107–120. [Google Scholar] [CrossRef]
- Brooks, D.R.; Hoberg, E.P. How will global climate change affect parasite-host assemblages. Trends Parasitol. 2007, 23, 571–574. [Google Scholar] [CrossRef]
- Parker, G.A.; Ball, M.A.; Chubb, J.C. Evolution of complex life cycles in trophically transmitted helminths. I. Host incorporation and trophic ascent. J. Evol. Biol. 2015, 28, 267–291. [Google Scholar] [CrossRef]
- Morley, N.J. Aquatic molluscs as auxiliary hosts for terrestrial nematode parasites: Implications for pathogen transmission in a changing climate. Parasitology 2010, 137, 1041–1056. [Google Scholar] [CrossRef]
- Bentivegna, F.; Ciampa, M.; Mazza, G.; Paglialonga, A.; Travaglini, A. Loggerhead turtle (Caretta caretta) in Tyrrhenian Sea: Trophic role of the Gulf of Naples. In Proceedings of the First Mediterranean Conference on Marine Turtles, Rome, Italy, 24–28 October 2001; pp. 71–75. [Google Scholar]
- OIE. Manual of Diagnostic Tests for Aquatic Animals. Available online: https://www.oie.int/standard-setting/aquatic-manual/access-online/ (accessed on 1 October 2019).
- Mattiucci, S.; Cipriani, P.; Levsen, A.; Paoletti, M.; Nascetti, G. Molecular epidemiology of Anisakis and Anisakiasis: An ecological and evolutionary road map. Adv. Parasitol. 2018, 99, 93–263. [Google Scholar] [CrossRef] [PubMed]
- Prioli, G. La molluschicoltura in Italia. In Estado Actual del Cultivo y Manejo de Moluscos Bivalvos y su Proyección Futura: Factores que Afectan su Sustentabilidad en América Latina; FAO Actas de Pesca y Acuicultura. No. 12.; Lovatelli, A., Farías, A., Uriarte, I., Eds.; FAO: Roma, Italy, 2008; pp. 159–176. [Google Scholar]
- Santoro, M.; Mattiucci, S.; Paoletti, M.; Liotta, A.; Uberti, B.D.; Galiero, G.; Nascetti, G. Molecular identification and pathology of Anisakis pegreffii (Nematoda: Anisakidae) infection in the Mediterranean loggerhead sea turtle (Caretta caretta). Vet. Parasitol. 2010, 174, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; D’Amelio, S.; Paggi, L.; Gasser, R.B. Assessing sequence variation in the internal transcribed spacers of ribosomal DNA within and among members of the Contracaecum osculatum complex (Nematoda: Ascaridoidea: Anisakidae). Parasitol. Res. 2000, 86, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Valentini, A.; Mattiucci, S.; Bondanelli, P.; Webb, S.C.; Mignucci-Giannone, A.; Colom-Llavina, M.M.; Nascetti, G. Genetic relationships among Anisakis species (Nematoda: Anisakidae) inferred from mitochondrial cox2 sequences, and comparison with allozyme data. J. Parasitol. 2006, 92, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Mattiucci, S.; Cipriani, P.; Webb, S.C.; Paoletti, M.; Marcer, F.; Bellisario, B.; Gibson, D.I.; Nascetti, G. Genetic and morphological approaches distinguish the three sibling species of the Anisakis simplex species complex, with a species designation as Anisakis berlandi n. sp. for A. simplex sp. C (Nematoda: Anisakidae). J. Parasitol. 2014, 100, 199–214. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).