Identification of Divergent Isolates of Banana Mild Mosaic Virus and Development of a New Diagnostic Primer to Improve Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. RNA Extraction
2.3. Crude Extracts Preparation
2.4. Routine Diagnostic Assays
2.5. Immunosorbent Electron Microscopy (ISEM) of Viral Minipreps
2.6. Library Preparation and High-Throughput Sequencing
2.7. Bioinformatics Analysis
2.8. Phylogenetic and Sequence Analysis
2.9. Primer Design
2.10. Gradient PCR
3. Results and Discussion
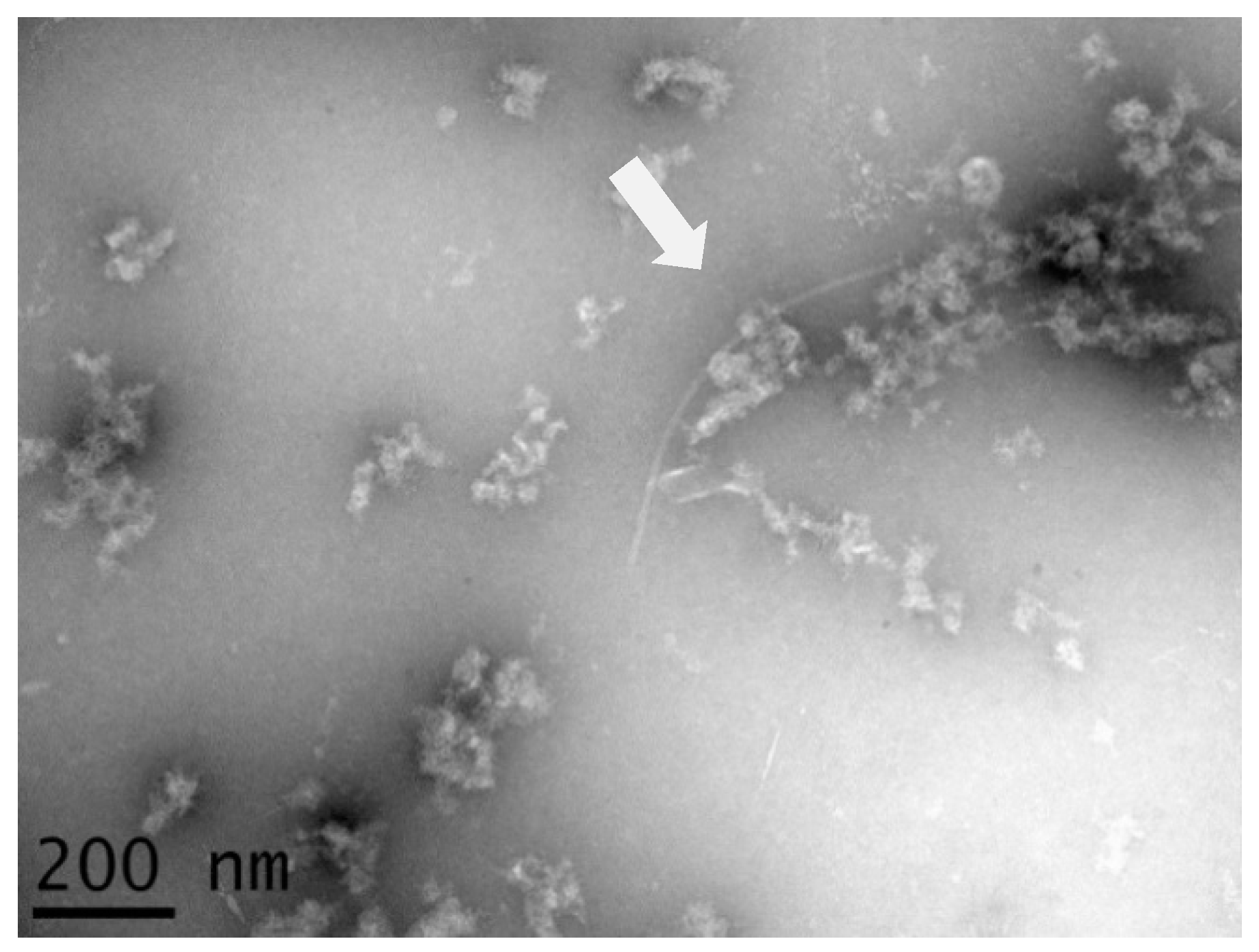
3.1. Immunosorbent Electron Microscopy (ISEM) Analysis
3.2. High-Throughput Sequencing and Bioinformatics Analysis
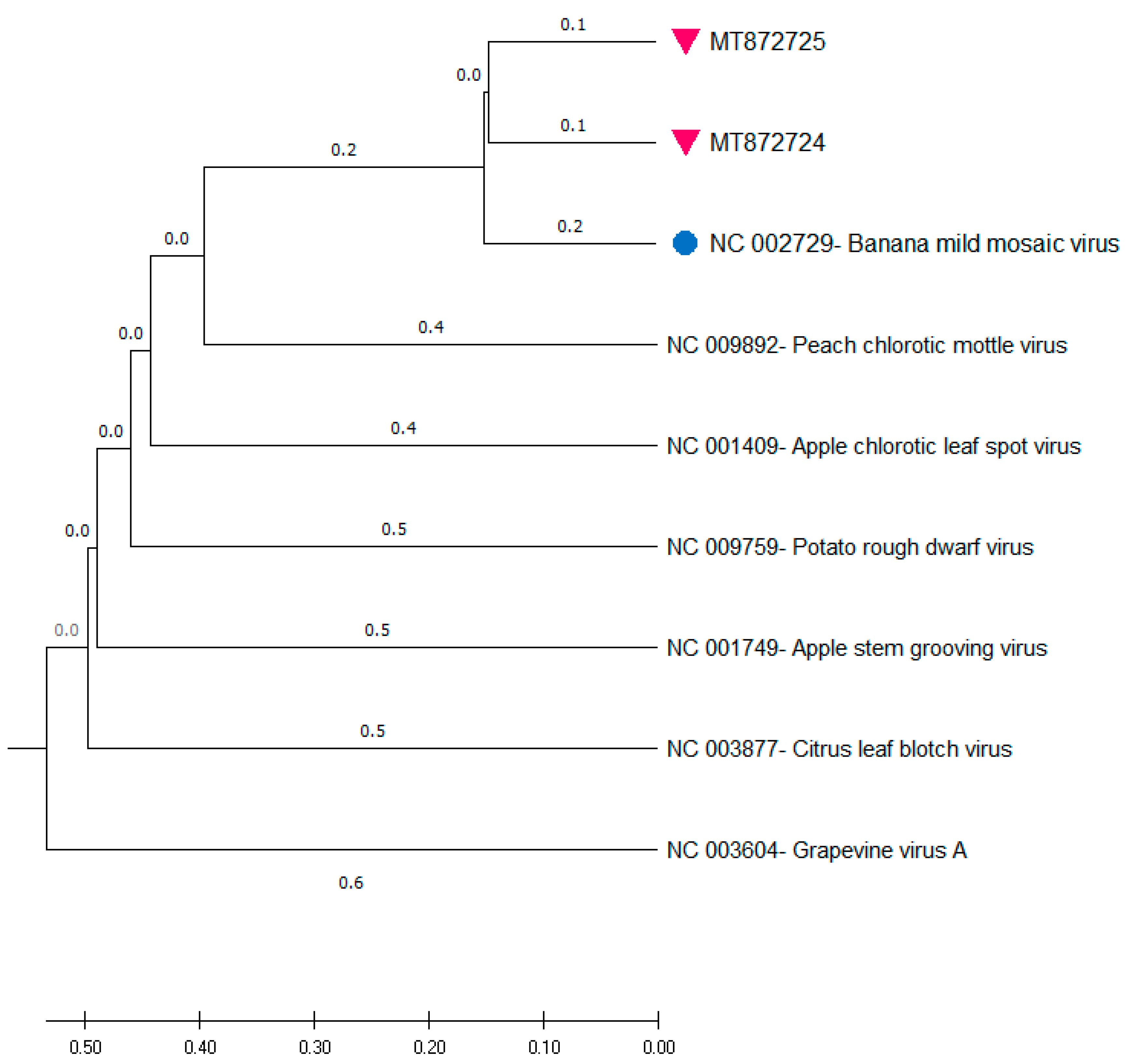
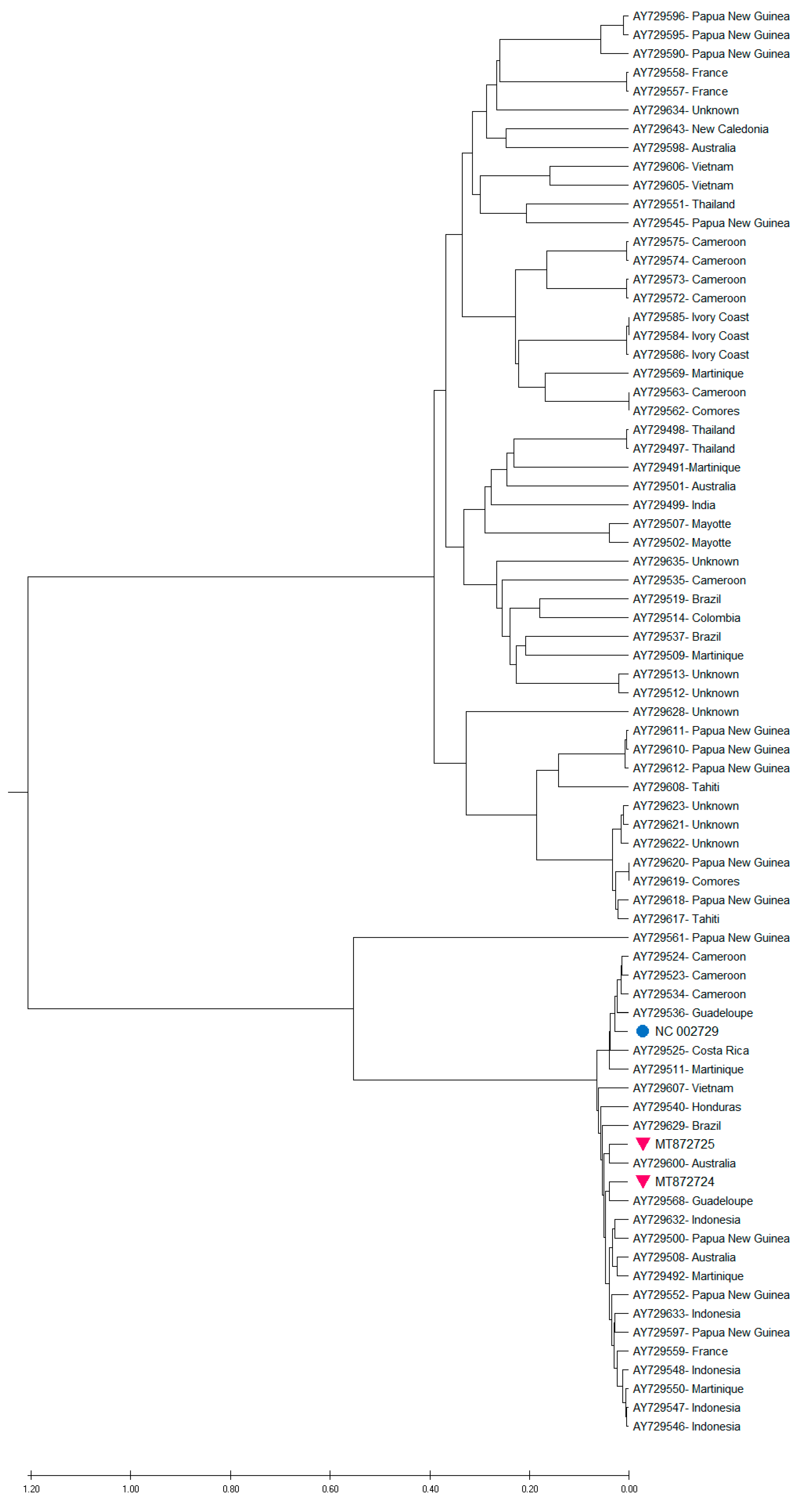
3.3. Phylogenetic Analysis
3.4. Genetic Diversity of the Two Identified Isolates
3.5. Nucleotide Sequence Analysis and Primer Design
3.6. Primer Validation
3.7. Laboratory Validation of the New Primer in Banana Germplasm Collection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kouakou, T.K.; Caroline, D.C.; Therese, A.A.; Ludivine, L.; Olivier, P.; Sebastien, M.; Philippe, L.; Mohamed, H.J. Prevalence of viruses infecting plantain (Musa sp., AAB genome) in the major growing regions in Cote dIvoire. Afr. J. Agric. Res. 2016, 11, 4532–4541. [Google Scholar] [CrossRef]
- Christelová, P.; De Langhe, E.; Hřibová, E.; Čížková, J.; Sardos, J.; Hušáková, M.; Van den houwe, I.; Sutanto, A.; Kepler, A.K.; Swennen, R.; et al. Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers. Conserv. 2017, 26, 801–824. [Google Scholar] [CrossRef]
- Teycheney, P.Y.; Laboureau, N.; Iskra-Caruana, M.L.; Candresse, T. High genetic variability and evidence for plant-to-plant transfer of Banana mild mosaic virus. J. Gen. Virol. 2005, 86, 3179–3187. [Google Scholar] [CrossRef] [PubMed]
- Gambley, C.F.; Thomas, J.E. Molecular characterisation of Banana mild mosaic virus, a new filamentous virus in Musa spp. Arch. Virol. 2001, 146, 1369–1379. [Google Scholar] [CrossRef]
- Schrader, G.; Unger, J. Plant quarantine as a measure against invasive alien species: The framework of the International Plant Protection Convention and the plant health regulations in the European Union. Biol. Invasions 2003, 5, 357–364. [Google Scholar] [CrossRef]
- Buko, D.H.; Gedebo, A.; Spetz, C. An update of sweet potato viral disease incidence and spread in Ethiopia. Afr. J. Agric. Res. 2020, 16, 1116–1126. [Google Scholar] [CrossRef]
- Le Provost, G.; Iskra-Caruana, M.L.; Acina, I.; Teycheney, P.Y. Improved detection of episomal Banana streak viruses by multiplex immunocapture PCR. J. Virol. Methods 2006, 137, 7–13. [Google Scholar] [CrossRef]
- Thomas, J.E. Technical Guidelines for The Safe Movement of Musa Germplasm; Bioversity International: Rome, Italy, 2015; ISBN 9789292550349. [Google Scholar]
- Zarghani, S.N.; Hily, J.M.; Glasa, M.; Marais, A.; Wetzel, T.; Faure, C.; Vigne, E.; Velt, A.; Lemaire, O.; Boursiquot, J.M.; et al. Grapevine virus T diversity as revealed by full-length genome sequences assembled from high-throughput sequence data. PLoS ONE 2018, 13, e0206010. [Google Scholar] [CrossRef]
- Diaz-lara, A.; Golino, D.; Preece, J.E.; Al, M. Development of RT-PCR degenerate primers to overcome the high genetic diversity of grapevine virus T. J. Virol. Methods 2020, 282, 113883. [Google Scholar] [CrossRef]
- Da Silva, L.A.; Oliveira, A.S.; Melo, F.L.; Ardisson-Araújo, D.M.P.; Resende, F.V.; Resende, R.O.; Ribeiro, B.M. A new virus found in garlic virus complex is a member of possible novel genus of the family Betaflexiviridae (order Tymovirales). PeerJ 2019, 2019, 1–11. [Google Scholar] [CrossRef]
- Baker, R.; Caffier, D.; Choiseul, J.W.; De Clercq, P.; Gerowitt, B.; Karadjova, O.E.; Lövei, G.; Oude, A.; Makowski, D.; Manceau, C.; et al. Pest risk assessment made by France on Banana bract mosaic virus considered by France as harmful in French overseas departments of French Guiana, Guadeloupe, Martinique and Réunion—Scientific Opinion of the Panel on Plant Health. EFSA J. 2008, 6, 1–21. [Google Scholar] [CrossRef]
- Teycheney, P.Y.; Acina, I.; Lockhart, B.E.L.; Candresse, T. Detection of Banana mild mosaic virus and Banana virus X by polyvalent degenerate oligonucleotide RT-PCR (PDO-RT-PCR). J. Virol. Methods 2007, 142, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Fidan, H.; Koç, G. Occurrence, ecology and phylogeny of banana streak badnavirus (BSV) and cucumber mosaic cucumovirus (CMV) in Musa sp. production areas of the mediterranean coastline of Turkey. Appl. Ecol. Environ. Res. 2019, 17, 5935. [Google Scholar] [CrossRef]
- Foissac, X.; Svanella-Dumas, L.; Gentit, P.; Dulucq, M.J.; Marais, A.; Candresse, T. Polyvalent degenerate oligonucleotides reverse transcription-polymerase chain reaction: A polyvalent detection and characterization tool for trichoviruses, capilloviruses, and foveaviruses. Phytopathology 2005, 95, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Cleary, M. High—Throughput identification and diagnostics of pathogens and pests: Overview and practical recommendations. Mol. Ecol. Resour. 2019, 47–76. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.C.; Hafner, G.J.; Revill, P.A.; Dale, J.L.; Harding, R.M. Sequence diversity of South Pacific isolates of Taro bacilliform virus and the development of a PCR-based diagnostic test. Arch. Virol. 2003, 148, 1957–1968. [Google Scholar] [CrossRef]
- Olmos, A.; Boonham, N.; Candresse, T.; Gentit, P.; Giovani, B.; Kutnjak, D.; Liefting, L.; Maree, H.J.; Minafra, A.; Moreira, A.; et al. High-throughput sequencing technologies for plant pest diagnosis: Challenges and opportunities. EPPO Bull. 2018, 48, 219–224. [Google Scholar] [CrossRef]
- Massart, S.; Candresse, T.; Gil, J.; Lacomme, C.; Predajna, L.; Ravnikar, M.; Reynard, J.S.; Rumbou, A.; Saldarelli, P.; Škoric, D.; et al. A framework for the evaluation of biosecurity, commercial, regulatory, and scientific impacts of plant viruses and viroids identified by NGS technologies. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Villamor, D.E.V.; Mekuria, T.A.; Pillai, S.S.; Eastwell, K.C. High-Throughput Sequencing Identifies Novel Viruses in Nectarine: Insights to the Etiology of Stem-Pitting Disease. Phytopathology 2016, 106, 519–527. [Google Scholar] [CrossRef]
- Massart, S.; Olmos, A.; Jijakli, H.; Candresse, T. Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 2014, 188, 90–96. [Google Scholar] [CrossRef]
- Jones, S.; Baizan-Edge, A.; MacFarlane, S.; Torrance, L. Viral diagnostics in plants using next generation sequencing: Computational analysis in practice. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- De Clerck, C.; Crew, K.; Van den houwe, I.; McMichael, L.; Berhal, C.; Lassois, L.; Jijakli, M.H.; Roux, N.; Thomas, J.; Massart, S. Lessons learned from the virus indexing of Musa germplasm: Insights from a multiyear collaboration. Ann. Appl. Biol. 2017, 171, 15–27. [Google Scholar] [CrossRef]
- Sharman, M.; Thomas, J.E.; Dietzgen, R.G. Development of a multiplex immunocapture PCR with colourimetric detection for viruses of banana. J. Virol. Methods 2000, 89, 75–88. [Google Scholar] [CrossRef]
- Gazel, M.; Roumi, V.; Ördek, K.; Maclot, F.; Massart, S.; Çağlayan, K. Identification and molecular characterization of a novel foveavirus from Rubus spp. in Turkey. Virus Res. 2020, 286, 26–29. [Google Scholar] [CrossRef]
- Prime, G. User Manual Geneious Prime. Data Base 2012, 3304, 1–148. [Google Scholar]
- Buzkan, N.; Chiumenti, M.; Massart, S.; Sarpkaya, K.; Karadağ, S.; Minafra, A. A new emaravirus discovered in Pistacia from Turkey. Virus Res. 2019, 263, 159–163. [Google Scholar] [CrossRef]
- Komínek, P.; Massart, S.; Pham, K.; van Leeuwen, P.; Komínková, M. Characterisation of a novel virus infecting orchids of the genus Pleione. Virus Res. 2019, 261, 56–59. [Google Scholar] [CrossRef]
- Da Silva, W.; Kutnjak, D.; Xu, Y.; Xu, Y.; Giovannoni, J.; Elena, S.F.; Gray, S. Transmission modes affect the population structure of potato virus Y in potato. PLoS Pathog. 2020, 16, e1008608. [Google Scholar] [CrossRef]
- Hawash, M.B.F.; Al-Jubury, A.; Sengupta, M.E.; Hansen, T.V.A.; Thamsborg, S.M.; Nejsum, P. Evidence for mitochondrial pseudogenes (numts) as a source of contamination in the phylogeny of human whipworms. Infect. Genet. Evol. 2020, 86, 104627. [Google Scholar] [CrossRef]
- Albuquerque, L.C.; Inoue-Nagata, A.K.; Pinheiro, B.; Resende, R.O.; Moriones, E.; Navas-Castillo, J. Genetic diversity and recombination analysis of sweepoviruses from Brazil. Virol. J. 2012, 9. [Google Scholar] [CrossRef]
- Mavridis, K.; Wipf, N.; Medves, S.; Erquiaga, I.; Müller, P.; Vontas, J. Rapid multiplex gene expression assays for monitoring metabolic resistance in the major malaria vector Anopheles gambiae 06 Biological Sciences 0604 Genetics. Parasites Vectors 2019, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Çağlayan, K.; Roumi, V.; Gazel, M.; Elçi, E.; Acioğlu, M.; Plesko, I.M.; Reynard, J.S.; Maclot, F.; Massart, S. Identification and characterization of a novel Robigovirus species from sweet cherry in turkey. Pathogens 2019, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Ii, P.; Positive, T.; Single, S.; Rna, S. Betaflexiviridae. Virus Taxon. 2012, 920–941. [Google Scholar] [CrossRef]
- Meena, R.P.; Prabha, K.; Baranwal, V.K. Development of RT-PCR degenerate primers for the detection of two mandariviruses infecting citrus cultivars in India. J. Virol. Methods 2020, 275, 113753. [Google Scholar] [CrossRef]
- Caruana, M.L.; Galzi, S. Identification of uncharacterised filamentous viral particles on banana plants. Acta Hortic. 1998, 490, 323–335. [Google Scholar] [CrossRef]
- Kesanakurti, P.; Belton, M.; Saeed, H.; Rast, H.; Boyes, I.; Rott, M. Comparative analysis of cherry virus A genome sequences assembled from deep sequencing data. Arch. Virol. 2017, 162, 2821–2828. [Google Scholar] [CrossRef]
- Lauring, A.S.; Andino, R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog 2010, 6, e1001005. [Google Scholar] [CrossRef]
- Jones, D.R. Handbook of Diseases of Banana, Abacá and Enset; CABI: Wallingford, UK, 2019. [Google Scholar]
- Selvarajan, R.; Balasubramanian, V. First report of banana bract mosaic virus in banana in Assam, India. J. Plant Pathol. 2017, 99, 534. [Google Scholar]
- Noll, L.W.; Stoy, C.P.A.; Wang, Y.; Porter, E.G.; Lu, N.; Liu, X.; Burklund, A.; Peddireddi, L.; Hanzlicek, G.; Henningson, J.; et al. Development of a nested PCR assay for detection of Streptococcus equi subspecies equi in clinical equine specimens and comparison with a qPCR assay. J. Microbiol. Methods 2020, 172, 105887. [Google Scholar] [CrossRef]
- Rames, E.K.; Macdonald, J. Rapid assessment of viral water quality using a novel recombinase polymerase amplification test for human adenovirus. Appl. Microbiol. Biotechnol. 2019, 103, 8115–8125. [Google Scholar] [CrossRef]
- Ben-Dov, E.; Shapiro, O.H.; Siboni, N.; Kushmaro, A. Advantage of using inosine at the 3′ termini of 16S rRNA gene universal primers for the study of microbial diversity. Appl. Environ. Microbiol. 2006, 72, 6902–6906. [Google Scholar] [CrossRef] [PubMed]
- Ben-Dov, E.; Kushmaro, A. Inosine at Different Primer Positions to Study Structure and Diversity of Prokaryotic Populations. Curr. Issues Mol. Biol. 2015, 17, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.J.; Force, C.R.; Znosko, B.M. Stability of RNA duplexes containing inosine·cytosine pairs. Nucleic Acids Res. 2018, 46, 12099–12108. [Google Scholar] [CrossRef] [PubMed]
| (A) Nucleotide Level | (B) Amino Acid Level | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| MT872724 | MT872725 | MT872724 | MT872725 | ||||||
| CP | RdRp | CP | RdRp | CP | RdRp | CP | RdRp | ||
| BanMMV NC_002729 | CP | 77.6% | n.a. | 76.8% | n.a. | 88% | n.a. | 87.8% | n.a. |
| RdRpp | n.a. | 75.4% | n.a. | 75.4% | n.a. | 81% | n.a. | 81.6% | |
| A- First Novel BanMMV Genome MT872724 | ||||||
| BanMMV New Genome 1 | Total SNPs | RdRp | TGB2 | TGB3 | TGB4 | CP |
| MT872724 | 31 | 20 | 2 | 1 | 2 | 3 |
| Length (nt) | - | 5328 | 675 | 339 | 213 | 678 |
| B- Second Novel BanMMV Genome MT872725 | ||||||
| BanMMV New Genome 2 | Total SNPs | RdRp | TGB2 | TGB3 | TGB4 | CP |
| MT872725 | 14 | 9 | 1 | 3 | 0 | 1 |
| Length (nt) | - | 5307 | 675 | 339 | 213 | 717 |
| Sample | BlastN Results | ||
|---|---|---|---|
| % nt Identity | Sequence ID | «Organism» | |
| ITC 1022 | 84% | AY730737.1 | BanMMV isolate CP2.3 CP gene, partial cds |
| ITC 1709 | 89% | AY730742.1 | BanMMV isolate CP4.2 CP gene, partial cds |
| ITC 1746 | 84% | KT780866.1 | BanMMV isolate TN1 CP gene, complete cds |
| ITC 1518 | 85% | AY730732.1 | BanMMV isolate CP18.1 CP gene, partial cds |
| ITC 1628 | 87% | AY730744.1 | BanMMV isolate CP6.1 CP gene, partial cds |
| ITC 1651 | 83% | AY730748.1 | BanMMV isolate CP77.2 CP gene, partial cds |
| ITC1681 | 87% | AY730743.1 | BanMMV isolate CP4.3 CP gene, partial cds |
| ITC 1733 | 81% | AY730754.1 | BanMMV isolate CP82.2 CP gene, partial cds |
| ITC 1740 | 83% | AY730740.1 | BanMMV isolate CP3.3 CP gene, partial cds |
| ITC 1794 | 77% | AY366187.1 | BanMMV isolate F8 CP gene, complete cds |
| ITC 1929 | 69% | EF143978.1 | BanMMV isolate Q6.2T CP gene, complete cds |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanafi, M.; Tahzima, R.; Ben Kaab, S.; Tamisier, L.; Roux, N.; Massart, S. Identification of Divergent Isolates of Banana Mild Mosaic Virus and Development of a New Diagnostic Primer to Improve Detection. Pathogens 2020, 9, 1045. https://doi.org/10.3390/pathogens9121045
Hanafi M, Tahzima R, Ben Kaab S, Tamisier L, Roux N, Massart S. Identification of Divergent Isolates of Banana Mild Mosaic Virus and Development of a New Diagnostic Primer to Improve Detection. Pathogens. 2020; 9(12):1045. https://doi.org/10.3390/pathogens9121045
Chicago/Turabian StyleHanafi, Marwa, Rachid Tahzima, Sofiene Ben Kaab, Lucie Tamisier, Nicolas Roux, and Sébastien Massart. 2020. "Identification of Divergent Isolates of Banana Mild Mosaic Virus and Development of a New Diagnostic Primer to Improve Detection" Pathogens 9, no. 12: 1045. https://doi.org/10.3390/pathogens9121045
APA StyleHanafi, M., Tahzima, R., Ben Kaab, S., Tamisier, L., Roux, N., & Massart, S. (2020). Identification of Divergent Isolates of Banana Mild Mosaic Virus and Development of a New Diagnostic Primer to Improve Detection. Pathogens, 9(12), 1045. https://doi.org/10.3390/pathogens9121045







