Antibiotic Resistance and Biofilm-Forming Ability in Enterococcal Isolates from Red Meat and Poultry Preparations
Abstract
:1. Introduction
2. Results and Discussion
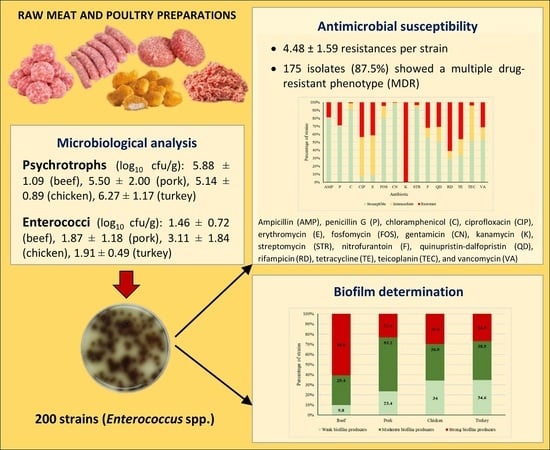
2.1. Microbial Counts
2.2. Antimicrobial Susceptibility
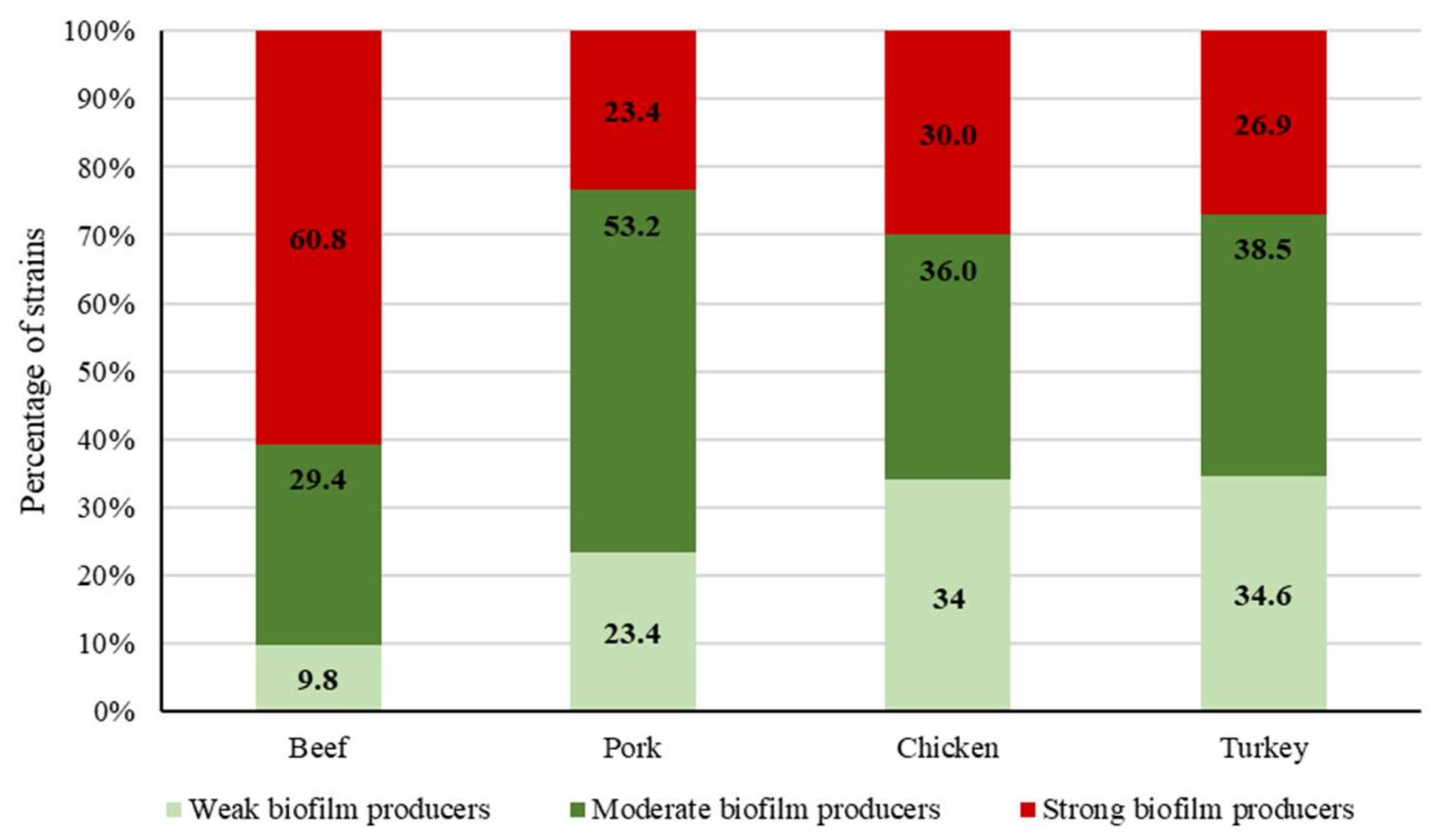
2.3. Biofilm-Forming Ability
3. Materials and Methods
3.1. Samples
3.2. Microbiological Analysis
3.3. Antimicrobial Susceptibility Testing
3.4. Biofilm Determination
3.5. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- FAOSTAT. 2020. Available online: http://www.fao.org/faostat/es/#data (accessed on 9 November 2020).
- Regulation (EC) No 853/2004 of the European Parliament and of the Council of 29 April 2004 laying down specific hygiene rules for food of animal origin. Off. J. Eur. Union 2004, L139, 55.
- Capita, R.; Alonso-Calleja, C. Antibiotic-resistant bacteria: A challenge for the food industry. Crit. Rev. Food Sci. Nutr. 2013, 53, 11–48. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections globally: Final Report and Recommendations. The Review on Antimicrobial Resistance. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 9 November 2020).
- OECD. Antimicrobial Resistance. Tackling the Burden in the European Union. Organization for Economic Coordination and Development (OECD), 2019. Available online: https://www.oecd.org/health/health-systems/AMR-Tackling-the-Burden-in-the-EU-OECD-ECDC-Briefing-Note-2019.pdf (accessed on 9 November 2020).
- Guerrero-Ramos, E.; Molina-González, D.; Blanco-Morán, S.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C.; Capita, R. Prevalence, antimicrobial resistance, and genotypic characterization of vancomycin-resistant enterococci in meat preparations. J. Food Prot. 2016, 79, 748–756. [Google Scholar] [CrossRef] [PubMed]
- SCENIHR. Assessment of the Antibiotic Resistance Effects of Biocides. Scientific Committee on Emerging and Newly Identified Health Risks, 19 January 2009. Available online: http://ec.europa.eu/health/ph_risk/committees/04_scenihr/docs/scenihr_o_021.pdf (accessed on 2 July 2020).
- Díez-García, M.; Capita, R.; Alonso-Calleja, C. Influence of serotype on the growth kinetics and the ability to form biofilms of Salmonella isolates from poultry. Food Microbiol. 2012, 31, 173–180. [Google Scholar] [CrossRef]
- Rodríguez-Melcón, C.; Alonso-Calleja, C.; Capita, R. Architecture and viability of the biofilms formed by nine Listeria strains on various hydrophobic and hydrophilic materials. Appl. Sci. 2019, 9, 5256. [Google Scholar] [CrossRef] [Green Version]
- Capita, R.; Riesco-Peláez, F.; Alonso-Hernando, A.; Alonso-Calleja, C. Exposure of Escherichia coli ATCC 12806 to sublethal concentrations of food-grade biocides influences its ability to form biofilm, resistance to antimicrobials, and ultrastructure. Appl. Environ. Microbiol. 2014, 80, 1268–1280. [Google Scholar] [CrossRef] [Green Version]
- Piercey, M.J.; Hingston, P.A.; Hansen, L.T. Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15 °C. Int. J. Food Microbiol. 2016, 223, 63–74. [Google Scholar] [CrossRef]
- Buzón-Durán, L.; Capita, R.; Alonso-Calleja, C. Microbial loads and antibiotic resistance patterns of Staphylococcus aureus in different types of raw poultry-based meat preparations. Poult. Sci. 2017, 96, 4046–4052. [Google Scholar] [CrossRef]
- González-Gutiérrez, M.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Microbial load and antibiotic resistance in raw beef preparations from northwest Spain. Food Sci. Nutr. 2020, 8, 777–785. [Google Scholar] [CrossRef]
- Pascual-Anderson, M.R. Microbiología Alimentaria: Metodología Analítica para Alimentos y Bebidas; Díaz de Santos: Madrid, Spain, 1992. [Google Scholar]
- Dainty, R.H.; Mackey, B.M. The relationship between the phenotypic properties of bacteria from chill-stored meat and spoilage processes. J. Appl. Bacteriol. Symp. Suppl. 1992, 73, 103S–114S. [Google Scholar] [CrossRef]
- Álvarez-Astorga, M.; Capita, R.; Alonso-Calleja, C.; Moreno, B.; García-Fernández, M.C. Microbiological quality of retail chicken by-products in Spain. Meat Sci. 2002, 62, 45–50. [Google Scholar] [CrossRef]
- Alonso-Calleja, C.; Martínez-Fernández, B.; Prieto, M.; Capita, R. Microbiological quality of vacuum-packed retail ostrich meat in Spain. Food Microbiol. 2004, 21, 241–246. [Google Scholar] [CrossRef]
- Franz, C.M.; Holzapfel, W.H.; Stiles, M.E. Enterococci at the crossroads of food safety. Int. J. Food Microbiol. 1999, 47, 1–24. [Google Scholar] [CrossRef]
- Thian, T.S.; Hartman, P.A. Gentamicin- thallous-carbonate medium for isolation of fecal streptococci from foods. Appl. Environ. Microbiol. 1981, 41, 724–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero-Ramos, E.; Cordero, J.; Molina-González, D.; Poeta, P.; Igrejas, G.; Alonso-Calleja, C.; Capita, R. Antimicrobial resistance and virulence genes in enterococci from wild game meat in Spain. Food Microbiol. 2016, 53, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Cordero, J.; Alonso-Calleja, C.; García-Fernández, C.; Capita, R. Microbial load and antibiotic resistance patterns of Escherichia coli and Enterococcus faecalis isolates from the meat of wild and domestic pigeons. Foods 2019, 8, 536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Álvarez-Fernández, E.; Alonso-Calleja, C.; García-Fernández, C.; Capita, R. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from poultry in Spain: Comparison between 1993 and 2006. Int. J. Food Microbiol. 2012, 153, 281–287. [Google Scholar] [CrossRef]
- Buzón-Durán, L.; Capita, R.; Alonso-Calleja, C. Antibiotic susceptibility of methicillin-resistant staphylococci (MRS) of food origin: A comparison of agar disc diffusion method and a commercially available miniaturized test. Food Microbiol. 2018, 72, 220–224. [Google Scholar] [CrossRef]
- Capita, R.; Álvarez-Fernández, E.; Fernández-Buelta, E.; Manteca, J.; Alonso-Calleja, C. Decontamination treatments can increase the prevalence of resistance to antibiotics of Escherichia coli naturally present on poultry. Food Microbiol. 2013, 34, 112–117. [Google Scholar] [CrossRef]
- Davis, G.S.; Waits, K.; Nordstrom, L.; Grande, H.; Weaver, B.; Papp, K.; Horwinski, J.; Koch, B.; Hungate, B.A.; Liu, C.M.; et al. Antibiotic-resistant Escherichia coli from retail poultry meat with different antibiotic use claims. BMC Microbiol. 2018, 18, 174. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Shaik, S.; Ranjan, A.; Nandanwar, N.; Tiwari, S.K.; Majid, M.; Baddam, R.; Qureshi, I.A.; Semmler, T.; Wieler, L.H.; et al. Risk of transmission of antimicrobial resistant Escherichia coli from commercial broiler and free-range retail chicken in India. Front. Microbiol. 2017, 8, 2120. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Woo, G. Distribution and transferability of tetracycline resistance determinants in Escherichia coli isolated from meat and meat products. Int. J. Food Microbiol. 2011, 145, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Ojer-Usoz, E.; González, D.; Vitas, A.I.; Leiva, J.; García-Jalón, I.; Febles-Casquero, A.; Escolano, M. Prevalence of extended-spectrum-beta-lactamase producing Enterobacteriaceae in meat products sold in Navarra, Spain. Meat Sci. 2013, 93, 316–321. [Google Scholar] [CrossRef]
- Carramiñana, J.J.; Rota, C.; Agustín, I.; Herrera, A. High prevalence of multiple resistance to antibiotics in Salmonella serovars isolated from a poultry slaughterhouse in Spain. Vet. Microbiol. 2004, 104, 133–139. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020, 18, e06007. [Google Scholar]
- Poeta, P.; Costa, D.; Igrejas, G.; Rodrigues, J.; Torres, C. Phenotypic and genotypic characterization of antimicrobial resistance in faecal enterococci from wild boars (Sus scrofa). Vet. Microbiol. 2007, 125, 368–374. [Google Scholar] [CrossRef]
- Radhouani, H.; Igrejas, G.; Gonçalves, A.; Pacheco, R.; Monteiro, R.; Sargo, R.; Brito, F.; Torres, C.; Poeta, P. Antimicrobial resistance and virulence genes in Escherichia coli and enterococci from red foxes (Vulpes vulpes). Anaerobe 2013, 23, 82–86. [Google Scholar] [CrossRef]
- Radimersky, T.; Frolkova, P.; Janoszowska, D.; Dolejska, M.; Svec, P.; Roubalova, E.; Cikova, P.; Cizek, A.; Literak, I. Antibiotic resistance in faecal bacteria (Escherichia coli, Enterococcus spp.) in feral pigeons. J. Appl. Microbiol. 2010, 109, 1687–1695. [Google Scholar] [CrossRef]
- Santos, T.; Silva, N.; Igrejas, G.; Rodrigues, P.; Micael, J.; Rodrigues, T.; Resendes, R.; Goncalves, A.; Marinho, C.; Goncalves, D.; et al. Dissemination of antibiotic resistant Enterococcus spp. and Escherichia coli from wild birds of Azores Archipelago. Anaerobe 2013, 24, 25–31. [Google Scholar] [CrossRef]
- Silva, N.; Igrejas, G.; Figueiredo, N.; Gonçalves, A.; Radhouani, H.; Rodrigues, J. Molecular characterization of antimicrobial resistance in enterococci and Escherichia coli isolates from European wild rabbit (Oryctolagus cuniculus). Sci. Total Environ. 2010, 408, 4871–4876. [Google Scholar] [CrossRef] [PubMed]
- Zigo, F.; Takac, L.; Zigova, M.; Takacova, J.; Vasi, M. Occurrence of antibiotic-resistant bacterial strains isolated in carrier pigeons during the race season. J. Chem. Pharm. Sci. 2017, 10, 10–13. [Google Scholar]
- Cameron, A.; McAllister, A.A. Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 2016, 7, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Briyne, N.; Atkinson, J.; Pokludová, L.; Borriello, S.P. Antibiotics used most commonly to treat animals in Europe. Vet. Rec. 2014, 175, 325. [Google Scholar] [CrossRef] [Green Version]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Doming, K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef]
- Álvarez-Fernández, E.; Cancelo, A.; Díaz-Vega, C.; Capita, R.; Alonso-Calleja, C. Antimicrobial resistance in E. coli isolates from conventionally and organically reared poultry: A comparison of agar disc diffusion and Sensi Test Gram-negative methods. Food Control 2013, 30, 227–234. [Google Scholar] [CrossRef]
- World Health Organization. Critically Important Antimicrobials for Human Medicine; 6th revision; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- World Organization for Animal Health. OIE List of Antimicrobial Agents of Veterinary Importance; World Organization for Animal Health: Paris, France, 2018. [Google Scholar]
- Barbosa, J.; Gibbs, P.A.; Teixeira, P. Virulence factors among enterococci isolated from traditional fermented meat products produced in the North of Portugal. Food Control 2010, 21, 651–656. [Google Scholar] [CrossRef]
- Ducková, V.; Čanigová, M.; Kročko, M. Enterococci and their resistance to antibiotics and thyme essential oil. J. Microbiol. Biotechnol. Food Sci. 2014, 3, 1–4. [Google Scholar]
- Koreňová, J.; Lopašovská, J.; Kuchta, T. Biofilm forming bacterial contaminants in small and medium-sized ewes’ milk and meat processing enterprises in Slovakia. J. Food Nutr. Res. 2009, 48, 115–120. [Google Scholar]
- Necidová, L.; Janštová, B.; Karpíšková, S.; Cupáková, Š.; Dušková, M.; Karpíšková, R. Importance of Enterococcus spp. for forming a biofilm. Czech J. Food Sci. 2009, 27, 354–356. [Google Scholar] [CrossRef] [Green Version]
- Cepas, V.; López, Y.; Muñoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship between biofilm formation and antimicrobial resistance in Gram-Negative bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, L.; Li, H.; Zhang, C.; Liang, B.; Li, J.; Wang, L.; Du, X.; Liu, X.; Qiu, S.; Song, H. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 2016, 7, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CLSI. M100 Performance Standars for Antimicrobial Susceptibility Testing, 29th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]

| Antibiotic Resistance Pattern | Number of Strains |
|---|---|
| K | 2 |
| CIP/K | 2 |
| E/K | 6 |
| K/RD | 4 |
| K/TE | 9 |
| K/VA | 1 |
| AMP/P/K | 1 |
| AMP/CIP/K | 1 |
| P/CIP/K | 1 |
| P/K/RD | 2 |
| P/K/TE | 2 |
| CIP/K/RD | 3 |
| CIP/K/TE | 2 |
| E/K/QD | 1 |
| E/K/TE | 8 |
| E/K/VA | 1 |
| FOS/K/VA | 1 |
| P/K/F | 2 |
| K/F/RD | 6 |
| K/F/TE | 1 |
| K/QD/TE | 2 |
| K/RD/VA | 1 |
| AMP/P/K/F | 1 |
| AMP/P/K/RD | 1 |
| AMP/CIP/K/RD | 2 |
| AMP/CIP/K/TE | 1 |
| AMP/E/K/RD | 2 |
| P/CIP/K/F | 2 |
| P/CIP/K/RD | 1 |
| P/CIP/K/TE | 1 |
| P/K/F/RD | 3 |
| P/K/RD/TE | 1 |
| C/E/K/TE | 1 |
| C/K/F/RD | 1 |
| CIP/E/K/F | 1 |
| CIP/E/K/TE | 4 |
| CIP/K/F/RD | 3 |
| CIP/K/F/TE | 2 |
| CIP/K/F/VA | 1 |
| E/K/F/RD | 1 |
| E/K/QD/RD | 3 |
| E/K/QD/VA | 1 |
| K/QD/TE/VA | 4 |
| K/QD/RD/VA | 5 |
| K/RD/TE/VA | 1 |
| AMP/CIP/E/K/RD | 1 |
| AMP/P/CIP/K/F | 2 |
| AMP/P/CIP/K/ RD | 1 |
| AMP/CIP/K/STR/TE | 1 |
| AMP/P/CIP/K/F/TE | 1 |
| AMP/P/K/F/RD | 2 |
| P/CIP/K/F/TE | 1 |
| P/CIP/K/F/RD | 3 |
| P/E/K/F/RD | 1 |
| P/FOS/K/F/RD | 1 |
| P/K/F/RD/VA | 1 |
| P/E/K/RD/TE | 1 |
| CIP/E/K/F/TE | 1 |
| CIP/E/K/RD/TE | 1 |
| CIP/K/QD/RD/VA | 3 |
| CIP/K/F/RD/TE | 1 |
| E/K/F/RD/TE | 3 |
| E/K/STR/QD/RD | 1 |
| E/K/QD/RD/VA | 8 |
| E/K/RD/TEC/VA | 1 |
| AMP/P/CIP/E/K/TE | 1 |
| AMP/P/CIP/K/TE | 2 |
| AMP/P/CIP/K/RD/TE | 1 |
| AMP/CIP/K/STR/QD/TE | 1 |
| AMP/P/CIP/K/F/TE | 2 |
| AMP/P/CIP/K/F/RD | 1 |
| AMP/P/E/K/F/RD | 2 |
| AMP/P/E/K/F/TE | 1 |
| P/CIP/E/K/F/TE | 1 |
| P/CIP/E/K/RD/TE | 1 |
| P/CIP/K/F/RD/TE | 1 |
| P/CIP/K/F/RD/VA | 1 |
| P/E/FOS/K/F/RD | 1 |
| P/E/K/QD/RD/VA | 1 |
| CIP/E/K/F/RD/TE | 1 |
| CIP/E/K/QD/RD/TE | 2 |
| CIP/E/K/QD/RD/VA | 4 |
| CIP/E/K/QD/TE/VA | 1 |
| CIP/K/QD/RD/TE/VA | 6 |
| CIP/E/FOS/K/F/RD | 1 |
| E/K/QD/RD/TE/VA | 3 |
| E/K/QD/RD/TEC/VA | 3 |
| K/QD/RD/TE/VA | 5 |
| AMP/CIP/E/FOS/K/F/RD | 2 |
| AMP/P/CIP/E/FOS/K/TE | 1 |
| AMP/P/CIP/K/F/RD/TE | 3 |
| AMP/P/E/FOS/K/F/RD | 1 |
| P/CIP/E/K/F/RD/TE | 1 |
| P/CIP/E/K/F/RD/VA | 1 |
| P/CIP/K/RD/TE/TEC/VA | 1 |
| CIP/E/K/QD/RD/TE/VA | 1 |
| CIP/K/QD/RD/TE/TEC/VA | 2 |
| E/K/STR/F/QD/RD/TE | 1 |
| E/K/STR/QD/RD/TE/VA | 1 |
| AMP/CIP/E/K/STR/QD/RD/VA | 1 |
| AMP/P/CIP/K/F/RD/TE/VA | 1 |
| CIP/E/CN/K/QD/RD/TE/VA | 1 |
| AMP/P/CIP/E/K/FOS/K/F/RD | 1 |
| CIP/E/K/STR/QD/RD/TE/TEC/VA | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castaño-Arriba, A.; González-Machado, C.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C.; Capita, R. Antibiotic Resistance and Biofilm-Forming Ability in Enterococcal Isolates from Red Meat and Poultry Preparations. Pathogens 2020, 9, 1021. https://doi.org/10.3390/pathogens9121021
Castaño-Arriba A, González-Machado C, Igrejas G, Poeta P, Alonso-Calleja C, Capita R. Antibiotic Resistance and Biofilm-Forming Ability in Enterococcal Isolates from Red Meat and Poultry Preparations. Pathogens. 2020; 9(12):1021. https://doi.org/10.3390/pathogens9121021
Chicago/Turabian StyleCastaño-Arriba, Ana, Camino González-Machado, Gilberto Igrejas, Patrícia Poeta, Carlos Alonso-Calleja, and Rosa Capita. 2020. "Antibiotic Resistance and Biofilm-Forming Ability in Enterococcal Isolates from Red Meat and Poultry Preparations" Pathogens 9, no. 12: 1021. https://doi.org/10.3390/pathogens9121021
APA StyleCastaño-Arriba, A., González-Machado, C., Igrejas, G., Poeta, P., Alonso-Calleja, C., & Capita, R. (2020). Antibiotic Resistance and Biofilm-Forming Ability in Enterococcal Isolates from Red Meat and Poultry Preparations. Pathogens, 9(12), 1021. https://doi.org/10.3390/pathogens9121021