Extracellular DNA (eDNA): Neglected and Potential Sources of Antibiotic Resistant Genes (ARGs) in the Aquatic Environments
Abstract
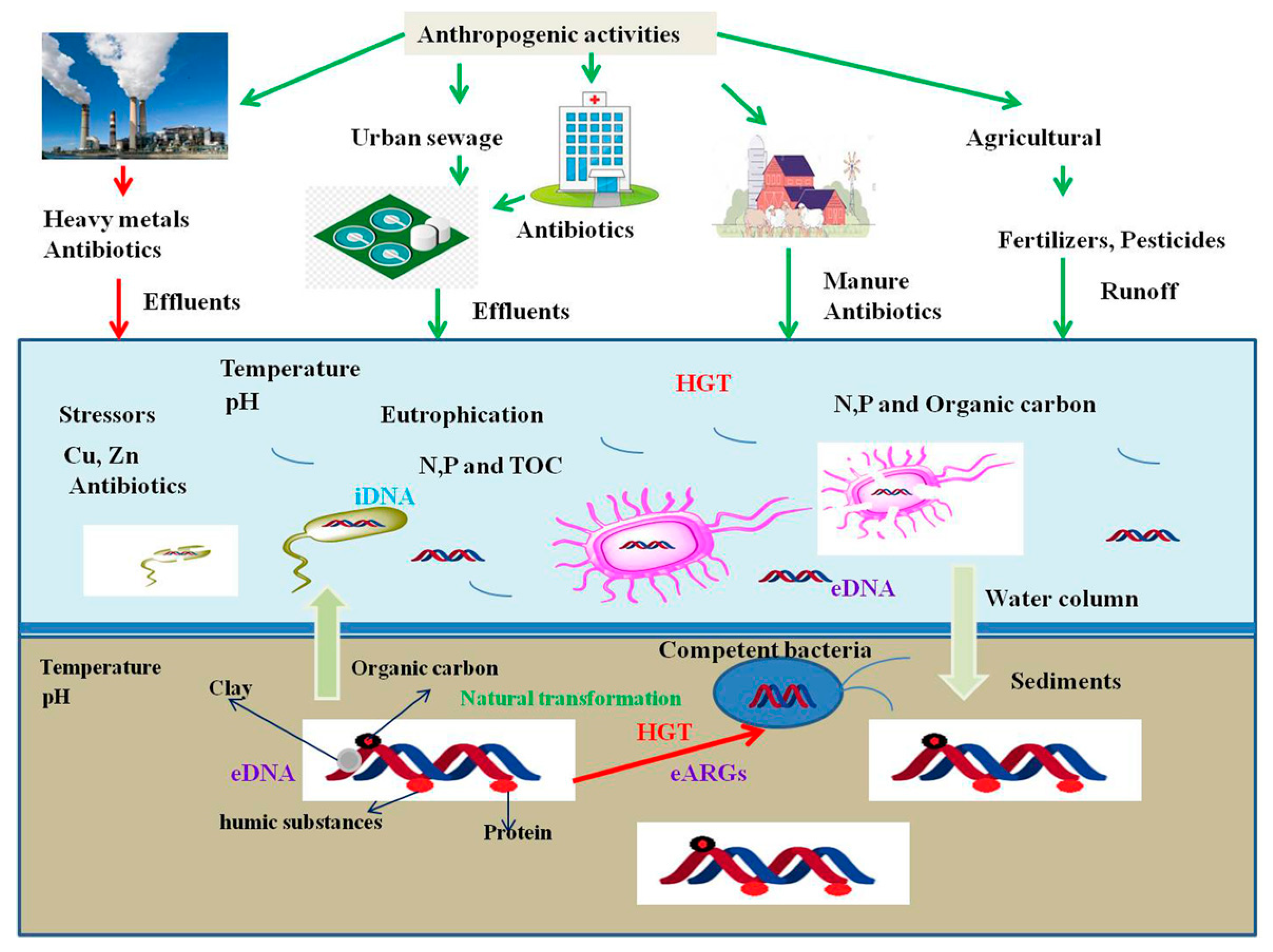
1. Introduction
2. eDNA Persistence and Natural Transformation
3. Recent Advancements in eDNA Isolation from Water, Sludge and Sediment Samples
3.1. Cetyl Trimethyl Ammonium Bromide (CTAB) based Method
3.2. Nucleic Acid Adsorption Particles (NAAPs)
3.3. Magnetic Beads Method for eDNA Extraction
4. eDNA as Sources of Antibiotic Resistance in the Environment
5. Abiotic Factors Influence spread of ARGs in the Environment
6. Treatment Technologies to Remove eDNA and eARGs
7. eDNA and Environmental Microbiomes
8. Conclusion and Future Remarks
Author Contributions
Funding
Conflicts of Interest
References
- WHO. Global Action Plan on Antimicrobial Resistance; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States; USA Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013.
- Sanderson, H.; Fricker, C.; Brown, R.S.; Majury, A.; Liss, S.N. Antibiotic resistance genes as an emerging environmental contaminant. Environ. Rev. 2016, 24, 205–218. [Google Scholar] [CrossRef]
- Guo, X.; Yang, Y.; Lu, D.-P.; Niu, Z.-S.; Feng, J.-N.; Chen, Y.-R.; Tou, F.-Y.; Garner, E.; Xu, J.; Liu, M.; et al. Biofilms as a sink for antibiotic resistance genes (ARGs) in the Yangtze Estuary. Water Res. 2018, 129, 277–286. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infectiong globally: Final Report and Recommendations. The Review on Antimicrobial Resistance. 2016. Available online: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed on 30 August 2019).
- WHO. Antimicrobial resistance: Global report on surveillance. World Health Organization. 2015. Available online: http://apps.who.Int/iris/bitstream/10665/112642/1/9789241564748eng.pdf (accessed on 15 August 2020).
- WHO. World Health Organization. One health policy. WHO: Geneva, 2015. Available online: http://www.euro.who.int/en/health-topics/disease-prevention/antimicrobial-resistance/about-amr/one-health (accessed on 30 August 2020).
- Paul, R.J.; Varghese, D. AMR in Animal Health: Issues and One Health Solutions for LMICs. In Antimicrobial Resistance; Thomas, S., Ed.; Springer: Singapore, 2020; Available online: http://doi-org-443.webvpn.fjmu.edu.cn/10.1007/978-981-15-3658-8_6 (accessed on 11 August 2020).
- Di Cesare, A.; Eckert, E.M.; Teruggi, A.; Fontaneto, D.; Bertoni, R.; Callieri, C.; Corno, G. Constitutive presence of antibiotic resistance genes within the bacterial community of a large subalpine lake. Mol. Ecol. 2015, 24, 3888–3900. [Google Scholar] [CrossRef] [PubMed]
- Czekalski, N.; Sigdel, R.; Birtel, J.; Matthews, B.; Bürgmann, H. Does human activity impact the natural antibiotic resistance background? Abundance of antibiotic resistance genes in 21 Swiss lakes. Environ. Int. 2015, 81, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Corinaldesi, C.; Beolchini, F.; Dell’Anno, A. Damage and degradation rates of extracellular DNA in marine sediments: Implications for the preservation of gene sequences. Mol. Ecol. 2008, 17, 3939–3951. [Google Scholar] [CrossRef] [PubMed]
- Torti, A.; Lever, M.A.; Jørgensen, B.B. Origin, dynamics, and implications of extracellular DNA pools in marine sediments. Mar. Genom. 2015, 24, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Nagler, M.; Marie, P.S.; Griffith, G.W.; Insam, H.; Ascher, J. The use of extracellular DNA as a proxy for specific microbial activity. Appl. Microbiol. Biotechnol. 2018, 102, 2885–2898. [Google Scholar] [CrossRef]
- Whitchurch, C.B. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef]
- Nielsen, K.M.; Johnsen, P.J.; Bensasson, D.; Daffonchio, D. Release and persistence of extracellular DNA in the environment. Environ. Biosaf. Res. 2007, 6, 37–53. [Google Scholar] [CrossRef]
- Tani, K.; Nasu, M. Roles of Extracellular DNA in Bacterial Ecosystem. In Extracellular Nucleic Acids. Nucleic Acids and Molecular Biology; Kikuchi, Y., Rykova, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 25. [Google Scholar] [CrossRef]
- Nagler, M.; Insam, H.; Pietramellara, G.; Ascher-Jenull, J. Extracellular DNA in natural environments: Features, relevance and applications. Appl. Microbiol. Biotechnol. 2018, 102, 6343–6356. [Google Scholar] [CrossRef]
- Dong, P.; Wang, H.; Fang, T.; Wang, Y.; Ye, Q. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG. Environ. Int. 2019, 125, 90–96. [Google Scholar] [CrossRef]
- Hao, H.; Shi, D.-Y.; Yang, D.; Yang, Z.-W.; Qiu, Z.-G.; Liu, W.-L.; Shen, Z.-Q.; Yin, J.; Wang, H.-R.; Li, J.-W.; et al. Profiling of intracellular and extracellular antibiotic resistance genes in tap water. J. Hazard. Mater. 2019, 365, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Luo, Y.; Mathieu, J.; Wang, Q.; Feng, L.; Mu, Q.; Feng, C.; Alvarez, P.J.J. Persistence ofextracellular DNA in river sediment facilitates antibiotic resistance gene propagation. Environ. Sci. Technol. 2014, 48, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Wang, X.; Chen, X.; Zhao, Z.; Fang, L.; Chen, B.; Jiang, J.; Luan, T.; Chen, B. Occurrence of antibiotic resistance genes in extracellular and intracellular DNA from sediments collected from two types of aquaculture farms. Chemosphere 2019, 234, 520–527. [Google Scholar] [CrossRef] [PubMed]
- McKinney, C.W.; Dungan, R.S. Influence of environmental conditions on extracellular and intracellular antibiotic resistance genes in manure-amended soil: A microcosm study. Soil Sci. Soc. Am. J. 2020, 84, 747–759. [Google Scholar] [CrossRef]
- Oliveira, M.; Nunes, M.; Crespo, M.T.B.; Silva, A.F. The environmental contribution to the dissemination of carbapenem and (fluoro)quinolone resistance genes by discharged and reused wastewater effluents: The role of cellular and extracellular DNA. Water Res. 2020, 182, 116011. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Snow, D.D.; Parker, D.; Zhou, Z.; Li, X. Intracellular and Extracellular antimicrobial resistance genes in the sludge of livestock waste management structures. Environ. Sci. Technol. 2013, 47, 10206–10213. [Google Scholar] [CrossRef]
- Eichmiller, J.J.; Best, S.E.; Sorensen, P.W. Effects of temperature and Trophic State on degradation of environmental DNA in Lake Water. Environ. Sci. Technol. 2016, 50, 1859–1867. [Google Scholar] [CrossRef]
- Krüger, N.-J.; Stingl, K. Two steps away from novelty—Principles of bacterial DNA uptake. Mol. Microbiol. 2011, 80, 860–867. [Google Scholar] [CrossRef]
- Pietramellara, G.; Ascher, J.; Borgogni, F.; Ceccherini, M.T.; Guerri, G.; Nannipieri, P. Extracellular DNA in soil and sediment: Fate and ecological relevance. Biol. Fertil. Soils 2008, 45, 219–235. [Google Scholar] [CrossRef]
- Corinaldesi, C.; Barucca, M.; Luna, G.M.; Dellanno, A. Preservation, origin and genetic imprint of extracellular DNA in permanently anoxic deep-sea sediments. Mol. Ecol. 2010, 20, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Ju, F.; Beck, K.; Yin, X.; Maccagnan, A.; McArdell, C.S.; Singer, H.P.; Johnson, D.R.; Zhang, T.; Bürgmann, H. Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME J. 2019, 13, 346–360. [Google Scholar] [CrossRef]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Genet. 2005, 3, 711–721. [Google Scholar] [CrossRef]
- Griffith, F. The significance of Pneumococcal Types. J. Hyg. 1928, 27, 113–159. [Google Scholar] [CrossRef] [PubMed]
- Avery, O.T.; MacLeod, C.M.; Mccarty, M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type iii. J. Exp. Med. 1944, 79, 137–158. [Google Scholar] [CrossRef] [PubMed]
- Overballe-Petersen, S.; Harms, K.; Orlando, L.; Moreno-Mayar, J.V.; Rasmussen, S.; Dahl, T.W.; Rosing, M.T.; Poole, A.M.; Sicheritz-Pontén, T.; Brunak, S.; et al. Bacterial natural transformation by highly fragmented and damaged DNA. Proc. Natl. Acad. Sci. USA 2013, 110, 19860–19865. [Google Scholar] [CrossRef]
- Johnston, C.; Martin, B.; Fichant, G.; Polard, P.; Claverys, J.-P. Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Genet. 2014, 12, 181–196. [Google Scholar] [CrossRef]
- Ray, J.L.; Nielsen, K.M. Experimental Methods for assaying natural transformation and inferring horizontal Gene Transfer. Methods Enzymol. 2005, 395, 491–520. [Google Scholar] [CrossRef]
- Nielsen, K.M.; Smalla, K.; Van Elsas, J.D. Natural transformation of acinetobactersp. Strain BD413 with Cell Lysates of Acinetobacter sp.,Pseudomonas fluorescens, and Burkholderia cepaciain soil microcosms. Appl. Environ. Microbiol. 2000, 66, 206–212. [Google Scholar] [CrossRef]
- Williams, H.G.; Day, M.J.; Fry, J.C.; Stewart, G.J. Natural transformation in river epilithon. Appl. Environ. Microbiol. 1996, 62, 2994–2998. [Google Scholar] [CrossRef]
- Paget, E.; Simonet, P. On the track of natural transformation in soil. FEMS Microbiol. Ecol. 1994, 15, 109–117. [Google Scholar] [CrossRef]
- Johnsborg, O.; Eldholm, V.; Håvarstein, L.S. Natural genetic transformation: Prevalence, mechanisms and function. Res. Microbiol. 2007, 158, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, A.; Dai, T.; Li, F.; Xie, H.; Chen, L.; Wen, D. Cell-free DNA: A neglected source for antibiotic resistance genes spreading from WWTPs. Environ. Sci. Technol. 2018, 52, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.J.; Sinigalliano, C.D. Detection of horizontal gene transfer by natural transformation in native and introduced species of bacteria in marine and synthetic sediments. Appl. Environ. Microbiol. 1990, 56, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.J.; Sinigalliano, C.D.; Garko, K.A. Binding of exogenous DNA to marine sediments and the effect of DNA/sediment binding on natural transformation ofPseudomonas stutzeristrain ZoBell in sediment columns. FEMS Microbiol. Lett. 1991, 85, 1–8. [Google Scholar] [CrossRef]
- Chamier, B.; Lorenz, M.G.; Wackernagel, W. Natural transformation of acinetobacter calcoaceticus by plasmid DNA adsorbed on sand and groundwater aquifer material. Appl. Environ. Microbiol. 1993, 59, 1662–1667. [Google Scholar] [CrossRef]
- Lorenz, M.G.; Wackernagel, W. Plasmid transformation of naturally competent Acinetobacter calcoaceticus in non-sterile soil extract and groundwater. Arch. Microbiol. 1992, 157, 355–360. [Google Scholar] [CrossRef]
- Gallori, E.; Franchi, M.; Rinaldi, L.; Vettori, C. Interspecific transformation of Bacillus subtilis by clay-bound DNA in non-sterile soil. Symbiosis 1998, 25, 311–322. [Google Scholar]
- Sikorski, J.; Graupner, S.; Lorenz, M.G.; Wackernagel, W. Natural genetic transformation of Pseudomonas stutzeri in a non-sterile soil. Microbiol. 1998, 144, 569–576. [Google Scholar] [CrossRef][Green Version]
- Yu, W.; Zhan, S.; Shen, Z.; Zhou, Q.; Yang, D. Efficient removal mechanism for antibiotic resistance genes from aquatic environments by graphene oxide nanosheet. Chem. Eng. J. 2017, 313, 836–846. [Google Scholar] [CrossRef]
- McKinney, C.W.; Pruden, A. Ultraviolet disinfection of Antibiotic Resistant Bacteria and their antibiotic resistance genes in water and wastewater. Environ. Sci. Technol. 2012, 46, 13393–13400. [Google Scholar] [CrossRef] [PubMed]
- Breazeal, M.V.R.; Novak, J.T.; Vikesland, P.J.; Pruden, A. Effect of wastewater colloids on membrane removal of antibiotic resistance genes. Water Res. 2013, 47, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhu, Y.; Yan, Y.; Wang, W.; Wang, Y. Deciphering extracellular antibiotic resistance genes (eARGs) in activated sludge by metagenome. Water Res. 2019, 161, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Sui, Q.; Chen, Y.; Yu, D.; Wang, T.; Hai, Y.; Zhang, J.; Chen, M.; Wei, Y. Fates of intracellular and extracellular antibiotic resistance genes and microbial community structures in typical swine wastewater treatment processes. Environ. Int. 2019, 133, 105183. [Google Scholar] [CrossRef] [PubMed]
- Sakcham, B.; Kumar, A.; Cao, B. Extracellular DNA in monochloraminated drinking water and its influence on DNA-Based profiling of a microbial community. Environ. Sci. Technol. Lett. 2019, 6, 306–312. [Google Scholar] [CrossRef]
- Saeki, K.; Kunito, T.; Sakai, M. Effects of pH, ionic strength, and solutes on DNA adsorption by andosols. Biol. Fertil. Soils 2010, 46, 531–535. [Google Scholar] [CrossRef]
- Wang, Z.; Han, M.; Li, E.; Liu, X.; Wei, H.; Yang, C.; Lu, S.; Ning, K. Distribution of antibiotic resistance genes in an agriculturally disturbed lake in China: Their links with microbial communities, antibiotics, and water quality. J. Hazard. Mater. 2020, 393, 122426. [Google Scholar] [CrossRef]
- Wang, D.-N.; Liu, L.; Qiu, Z.-G.; Shen, Z.-Q.; Guo, X.; Yang, D.; Li, J.; Liu, W.; Jin, M.; Li, J. A new adsorption-elution technique for the concentration of aquatic extracellular antibiotic resistance genes from large volumes of water. Water Res. 2016, 92, 188–198. [Google Scholar] [CrossRef]
- Ikner, L.A.; Soto-Beltran, M.; Bright, K.R. New method using a positively charged microporous filter and ultrafiltration for concentration of viruses from tap water. Appl. Environ. Microbiol. 2011, 77, 3500–3506. [Google Scholar] [CrossRef]
- Yuan, Q.-B.; Huang, Y.-M.; Wu, W.-B.; Zuo, P.; Hu, N.; Zhou, Y.-Z.; Alvarez, P.J. Redistribution of intracellular and extracellular free & adsorbed antibiotic resistance genes through a wastewater treatment plant by an enhanced extracellular DNA extraction method with magnetic beads. Environ. Int. 2019, 131, 104986. [Google Scholar] [CrossRef]
- Holben, W.E.; Jansson, J.K.; Chelm, B.K.; Tiedje, J.M. DNA Probe method for the detection of specific microorganisms in the soil bacterial community. Appl. Environ. Microbiol. 1988, 54, 703–711. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, Y.; Yang, A.; Yang, G. The effect of pH on charge inversion and condensation of DNA. Soft Matter 2016, 12, 6669–6674. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, L.P. IPC—Isoelectric Point Calculator. Biol. Direct 2016, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, K.; Wu, N.; Li, W.; Xu, W.; Zhang, Y.; Niu, Z. Estuarine sediments are key hotspots of intracellular and extracellular antibiotic resistance genes: A high-throughput analysis in Haihe Estuary in China. Environ. Int. 2020, 135, 105385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Niu, Z.; Zhang, Y.; Zhang, K. Occurrence of intracellular and extracellular antibiotic resistance genes in coastal areas of Bohai Bay (China) and the factors affecting them. Environ. Pollut. 2018, 236, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Hata, A.; Katayama, H.; Kasuga, I. Consecutive ultrafiltration and silica adsorption for recovery of extracellular antibiotic resistance genes from an urban river. Environ. Pollut. 2020, 260, 114062. [Google Scholar] [CrossRef]
- Huijbers, P.M.; Larsson, D.J.; Flach, C.-F. Surveillance of antibiotic resistant Escherichia coli in human populations through urban wastewater in ten European countries. Environ. Pollut. 2020, 261, 114200. [Google Scholar] [CrossRef]
- Lu, Y.; Xiao, Y.; Zheng, G.; Lu, J.; Zhou, L. Conditioning with zero-valent iron or Fe2+ activated peroxydisulfate at an acidic initial sludge pH removed intracellular antibiotic resistance genes but increased extracellular antibiotic resistance genes in sewage sludge. J. Hazard. Mater. 2020, 386, 121982. [Google Scholar] [CrossRef]
- Liu, S.-S.; Qu, H.-M.; Yang, D.; Hu, H.; Liu, W.-L.; Qiu, Z.-G.; Hou, A.-M.; Guo, J.; Li, J.-W.; Shen, Z.-Q.; et al. Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 2018, 136, 131–136. [Google Scholar] [CrossRef]
- Wang, L.; Su, H.; Hu, X.; Xu, Y.; Xu, W.; Huang, X.; Li, Z.; Cao, Y.; Wen, G. Abundance and removal of antibiotic resistance genes (ARGs) in the rearing environments of intensive shrimp aquaculture in South China. J. Environ. Sci. Heal. Part B 2019, 54, 211–218. [Google Scholar] [CrossRef]
- Li, W.; Su, H.; Cao, Y.; Wang, L.; Hu, X.; Xu, W.; Xu, Y.; Li, Z.; Wen, G. Antibiotic resistance genes and bacterial community dynamics in the seawater environment of Dapeng Cove, South China. Sci. Total. Environ. 2020, 723, 138027. [Google Scholar] [CrossRef] [PubMed]
- Nõlvak, H.; Truu, M.; Kanger, K.; Tampere, M.; Espenberg, M.; Loit, E.; Raave, H.; Truu, J. Inorganic and organic fertilizers impact the abundance and proportion of antibiotic resistance and integron-integrase genes in agricultural grassland soil. Sci. Total. Environ. 2016, 562, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Sui, Q.; Zhang, J.; Tong, J.; Chen, M.; Wei, Y. Seasonal variation and removal efficiency of antibiotic resistance genes during wastewater treatment of swine farms. Environ. Sci. Pollut. Res. 2015, 24, 9048–9057. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-R.; Guo, X.-P.; Niu, Z.-S.; Lu, D.-P.; Sun, X.-L.; Zhao, S.; Hou, L.; Liu, M.; Yang, Y. Antibiotic resistance genes (ARGs) and their associated environmental factors in the Yangtze Estuary, China: From inlet to outlet. Mar. Pollut. Bull. 2020, 158, 111360. [Google Scholar] [CrossRef]
- Lu, Z.; Na, G.; Gao, H.; Wang, L.; Bao, C.; Yao, Z. Fate of sulfonamide resistance genes in estuary environment and effect of anthropogenic activities. Sci. Total. Environ. 2015, 429–438. [Google Scholar] [CrossRef]
- Mao, D.; Yu, S.; Rysz, M.; Luo, Y.; Yang, F.; Li, F.; Hou, J.; Mu, Q.; Alvarez, P.J.J. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 2015, 85, 458–466. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, L.; Hou, Z.; Wang, L.; Ma, D.; Yang, G.; Guo, S.; Luo, J.; Qi, L.; Luo, Y. Heavy metal copper accelerates the conjugative transfer of antibiotic resistance genes in freshwater microcosms. Sci. Total. Environ. 2020, 717, 137055. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.M.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total. Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Xu, J.; Xu, Y.; Wang, H.; Guo, C.; Qiu, H.; He, Y.; Zhang, Y.; Li, X.; Meng, W. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere 2015, 119, 1379–1385. [Google Scholar] [CrossRef]
- Di Cesare, A.; Eckert, E.M.; Rogora, M.; Corno, G. Rainfall increases the abundance of antibiotic resistance genes within a riverine microbial community. Environ. Pollut. 2017, 226, 473–478. [Google Scholar] [CrossRef]
- Nielsen, K.M.; Calamai, L.; Pietramellara, G. Stabilization of extracellular DNA and proteins by transient binding to various soil components. In Nucleic Acids and Proteins in Soil; Springer: Berlin/Heidelberg, Germany, 2006; pp. 141–157. [Google Scholar]
- Corinaldesi, C.; Danovaro, R.; Dell’Anno, A. Simultaneous Recovery of Extracellular and Intracellular DNA Suitable for Molecular Studies from Marine Sediments. Appl. Environ. Microbiol. 2005, 71, 46–50. [Google Scholar] [CrossRef]
- Cui, G.; Bhat, S.A.; Li, W.; Wei, Y.; Kui, H.; Fu, X.; Gui, H.; Wei, C.; Li, F. Gut digestion of earthworms significantly attenuates cell-free and -associated antibiotic resistance genes in excess activated sludge by affecting bacterial profiles. Sci. Total. Environ. 2019, 691, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Slipko, K.; Reif, D.; Wögerbauer, M.; Hufnagl, P.; Krampe, J.; Kreuzinger, N. Removal of extracellular free DNA and antibiotic resistance genes from water and wastewater by membranes ranging from microfiltration to reverse osmosis. Water Res. 2019, 164, 114916. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-H.; Yuan, L.; Gao, S.-X.; Wang, L.; Sheng, G.-P. Mitigated membrane fouling and enhanced removal of extracellular antibiotic resistance genes from wastewater effluent via an integrated pre-coagulation and microfiltration process. Water Res. 2019, 159, 145–152. [Google Scholar] [CrossRef]
- Ahmed, Y.; Lu, J.; Yuan, Z.; Bond, P.L.; Guo, J.-H. Efficient inactivation of antibiotic resistant bacteria and antibiotic resistance genes by photo-Fenton process under visible LED light and neutral pH. Water Res. 2020, 179, 115878. [Google Scholar] [CrossRef]
- Schloss, P.D.; Handelsman, J. Metagenomics for studying unculturable microorganisms: Cutting the Gordian knot. Genome Biol. 2005, 6, 229. [Google Scholar] [CrossRef] [PubMed]
- I Amann, R.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.; A Stahl, D. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990, 56, 1919–1925. [Google Scholar] [CrossRef]
- Cullen, C.M.; Aneja, K.K.; Beyhan, S.; Cho, C.E.; Woloszynek, S.; Convertino, M.; McCoy, S.J.; Zhang, Y.; Anderson, M.Z.; Alvarez-Ponce, D.; et al. Emerging priorities for microbiome research. Front. Microbiol. 2020, 11, 136. [Google Scholar] [CrossRef]
- Corinaldesi, C.; Tangherlini, M.; Manea, E.; Dell’Anno, A. Extracellular DNA as a genetic recorder of microbial diversity in benthic deep-sea ecosystems. Sci. Rep. 2018, 8, 1839. [Google Scholar] [CrossRef]
| eDNA Isolation Method | Environment Sample Type | Filter Size and Type | Buffer Used | References |
|---|---|---|---|---|
| CTAB | river sediment | 0.22 μm pore size, Polyvinylidene Fluoride (PVDF), Osmonics, U.S | NaH2PO4 (0.12 M, pH 8.0) and 0.2 g of polyvinyl polypyrrolidone (PVPP) | [20] |
| CTAB | activated sludge | 0.02µm pore size, PVDF; Millipore, Bedford, MA, USA | NaH2PO4 (0.12 M, pH 8.0) and 0.1 g of PVPP | [50] |
| CTAB | estuarine sediments | 0.02 µm membrane filter | NaH2PO4 (0.12 M, pH 8.0) and 0.2 g of PVPP | [61] |
| CTAB | coastal area: surface sea water and sediments | 0.02 µm membrane filter | n.a | [62] |
| CTAB | sludge of livestock waste management structures | 0.2 µm pore membrane | 0.1 M phosphate buffer (PB, 0.093 M Na2HPO4 and 0.007 M NaH2PO4, pH = 8.0 | [24] |
| CTAB | WWTP influents and effluents | 0.02 µm membrane filter | n.a | [40] |
| CTAB | sediments of aquaculture farms | 0.22 μm pore size, PVDF, Osmonics, U.S | NaH2PO4 buffer (0.12 M, pH = 8.0) containing 0.2 g PPVP | [21] |
| magnetic beads | water samples and activated sludge in WWTP | 0.22 μm (Millipore, USA). | phosphate buffer (0.12M NaH2PO4, 0.12M Na2HPO4, pH = 4), 0.2g PVPP | [57] |
| NAAPs | water samples from river, lake and reservoir and drinking water | elute was filtered with polyethersulfone (PES) filter (0.45 μm, Millipore, USA). | eluent (15 g/L NaCl, 30 g/L tryptone, 15 g/L beef extract, 3.75 g/L Gly, 0.28 g/L Na(OH), pH = 9.3 ± 0.2) | [55] |
| NAAPs | tap water | elute was filtered using PES microporous membrane filter (0.45 μm, Millipore, USA) | n.a | [19] |
| hollow fiber ultrafiltration (HFUF) and silica binding | urban river water | polyestersulfone syringe filters with a pore size of 0.22 μm (Merck, Darmstadt, Germany) | sodium phosphate buffer (0.12 M Na2HPO4, 0.12 M NaH2PO4, 2% NaCl, pH = 8 | [63] |
| Antibiotic Class | Resistance Genes | Environments | Geographical Locations | References |
|---|---|---|---|---|
| Carbapenem Fluoro quinolones | blaKPC, blaOXA-48, blaNDM, blaIMP, blaVIM qnrA, qnrB and qnrS | WWTP | Portugal | [23] |
| Sulfonamides Tetracycline β-lactams Chloramphenicol Erythromycin | sul1, sul2, tetW, tetX, blaTEM, blaSHV, ampC, cat, cmr, ermA, ermB, | Sludge sample WWTP, hospital, pharmaceutical industry, sediment samples from Aoyun lake and swine manure | China | [18] |
| Tetracycline, Sulfonamides, β-lactams | TetC, tetM, sul1, sul2, blaTEM, qnrA and ampC. | Tap water | China | [19] |
| Sulfonamides Tetracycline β-lactams | sul1, sul2 tetA, tetC, tetO and tetS and blaTEM-1 and blanps-1 | Sediments from bullfrog farm and polyculture farm | China | [21] |
| tetracyclines, sulfonamides Fluoro quinolones | sul1, sul2 tetA, tetW acc(6′)-Ib, qnrS | Biofilm, and sediment from estuary | China | [4] |
| tetracyclines, sulfonamides, β-lactams, macrolide | TetA, tetC, tetM, tetX, sulI, sulII,blaTEM,ereA, ermB | Wastewater treatment plant | China | [57] |
| tetracyclines, sulfonamides, macrolides, β-lactams | tetC, sulII, ermB, BlaPSE-1 | WWTP | China | [40] |
| Sulfonamides Tetracyclines Macrolides β-lactams Quinolones | sul1, tet(A), ere(A), blaTEM, qnrD | Urban river water | Japan | [63] |
| tetracycline macrolide sulfonamide β-lactam | tetM, tetW, tetG, tetX, ermB, ermF, mefA, ereA, sul1 sul2, and blaTEM | Swine wastewater | China | [51] |
| sulfonamide, tetracycline | sul(I), sul(II), tet(O), tet(Q) and tet(X) | Sludge of livestock waste management structures | USA | [24] |
| Aminoglycoside, tetracycline, chloramphenicol, sulphonamide, vancomycin, multidrug, beta-lactamase | aac(6′)-Ib−03, aac(6′)-II, aadA-02, aadA-01, aadA2 - 02, aadA2 - 01, aadA2 - 03, strB, aadA1, and aadA5 - 01), (tetG-01 and tetM-01), (catB3 and floR), (catB3 and floR), (sul2), (VanC-03), (mexF), (blaVEB) | Municipal sewage sludge | China | [65] |
| sulfonamides tetracyclines β-lactams fluoroquinolones | (sul1, sul2), (tetB and tetM), (blaTEM and blaOXA-1), and (qnrS and oqxB). | Surface sea water from Coastal Bay | China | [62] |
| sulfonamides Tetracyclines | sul1- 3’ CS-TnAs3, sul2-intI1-ISVsa3, and tetX-p63039 | Sludge from WWTP | China | [50] |
| Macrolide, tetracycline, sulfonamide, β-lactam, aminoglycosides, rifampicin and vancomycin. | ermB, tetA, tetB and tetC, sul1, sul2 and sul3, ampC, aph(2’)-Id, katG and vanA. | Water sample from WWTP | China | [66] |
| sulfonamides Tetracyclines | sul1 and sul2 tetW, and tetT | Surface water and superficial sediment from river | China | [20] |
| ClassI integrons-integrase Aminoglycoside tetracycline, multidrug. | intI-1(clinic) aadA01, aadA1,aadA-02,tetG-01, tetX, qacEdelta1-01, qacH-01 | Sediment samples from estuary | China | [61] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sivalingam, P.; Poté, J.; Prabakar, K. Extracellular DNA (eDNA): Neglected and Potential Sources of Antibiotic Resistant Genes (ARGs) in the Aquatic Environments. Pathogens 2020, 9, 874. https://doi.org/10.3390/pathogens9110874
Sivalingam P, Poté J, Prabakar K. Extracellular DNA (eDNA): Neglected and Potential Sources of Antibiotic Resistant Genes (ARGs) in the Aquatic Environments. Pathogens. 2020; 9(11):874. https://doi.org/10.3390/pathogens9110874
Chicago/Turabian StyleSivalingam, Periyasamy, John Poté, and Kandasamy Prabakar. 2020. "Extracellular DNA (eDNA): Neglected and Potential Sources of Antibiotic Resistant Genes (ARGs) in the Aquatic Environments" Pathogens 9, no. 11: 874. https://doi.org/10.3390/pathogens9110874
APA StyleSivalingam, P., Poté, J., & Prabakar, K. (2020). Extracellular DNA (eDNA): Neglected and Potential Sources of Antibiotic Resistant Genes (ARGs) in the Aquatic Environments. Pathogens, 9(11), 874. https://doi.org/10.3390/pathogens9110874




