Abstract
Morels (Morchella spp.) are popular edible fungi with significant economic and scientific value. However, white mold disease, caused by Paecilomyces penicillatus, can reduce morel yield by up to 80% in the main cultivation area in China. Paecilomyces is a polyphyletic genus and the exact phylogenetic placement of P. penicillatus is currently still unclear. Here, we obtained the first high-quality genome sequence of P. penicillatus generated through the single-molecule real-time (SMRT) sequencing platform. The assembled draft genome of P. penicillatus was 40.2 Mb, had an N50 value of 2.6 Mb and encoded 9454 genes. Phylogenetic analysis of single-copy orthologous genes revealed that P. penicillatus is in Hypocreales and closely related to Hypocreaceae, which includes several genera exhibiting a mycoparasitic lifestyle. CAZymes analysis demonstrated that P. penicillatus encodes a large number of fungal cell wall degradation enzymes. We identified many gene clusters involved in the production of secondary metabolites known to exhibit antifungal, antibacterial, or insecticidal activities. We further demonstrated through dual culture assays that P. penicillatus secretes certain soluble compounds that are inhibitory to the mycelial growth of Morchella sextelata. This study provides insights into the correct phylogenetic placement of P. penicillatus and the molecular mechanisms that underlie P. penicillatus pathogenesis.
1. Introduction
Morels (Morchella spp.) are valuable edible fungi that belong within the order Pezizales. There is a long history of morel consumption in Asia, Europe, and North America [1]. The increasing demand for dried and fresh morels has driven the growth of the industry and the expansion of suitable morel cultivation areas [2,3]. In China, the morel cultivation area reached approximately 9000 ha in the production season of 2018–2019 and continues to expand [4]. With the expanding cultivation range and density of production, disease has become a major limiting factor for morel production. Typical diseases include stipe rot disease caused by the Fusarium incarnatum—F. equiseti species complex and pileus rot disease caused by Diploöspora longispora [5,6]. White mold disease, caused by Paecilomyces penicillatus, is another major disease affecting morels. This disease was first reported in Sichuan, China in 2013 [7]. P. penicillatus infection in one location can quickly spread to neighboring morel cultivation areas and can account for up to 80% of morel yield losses. Currently, no commercial fungicide has been registered to control for morel white mold disease, and the management of this disease relies primarily on the removal of the symptomatic fruiting bodies. The molecular basis of P. penicillatus pathogenicity is, to the best of our knowledge, unknown. This makes it difficult to develop tailored solutions to control this disease.
Paecilomyces is a polyphyletic genus, and Paecilomyces spp. are classified within Sordariomycetes or Eurotiomycetes based on phylogenetic analyses of 18S rRNA sequences, internal transcribed spacer (ITS) sequences and 5.8S rDNA sequences [8,9]. P. penicillatus was first isolated from rotten mushrooms by Samson in 1974 and was originally placed in Trichocomaceae, order Eurotiales, class Eurotiomycetes based on the morphological characteristics of this pathogen [10]. However, molecular phylogenetic analyses based on β-tubulin and 18S rRNA sequences later placed P. penicillatus within the order Hypocreales of Sordariomycetes, and revealed an uncertain affinity with Clavicipitaceae [8,11]. In addition to Clavicipitaceae, Hypocreales also contains seven other families, namely Hypocreaceae, Bionectriaceae, Cordycipitaceae, Nectriaceae, Niessliaceae, Ophiocordycipitaceae, and Stachybotryaceae. With the rapid development of sequencing technologies, an increasing number of fungal genomes have become available [12,13,14]. Phylogenetic analysis of the single-copy orthologous genes has been widely used for evolutionary studies and provides improved resolution for elucidating the phylogenetic relationships of organisms compared to the use of sequences from one or a few genes [13,15,16,17]. Due to the lack of P. penicillatus genomic resources, a comprehensive phylogenetic analysis based on single-copy orthologous genes is not available.
The molecular mechanisms of mycoparasitism in P. penicillatus remain unclear. Though not known from P. penicillatus, lysis of the host cell wall is a common mechanism of mycoparasitism [18]. Many mycoparasitic fungi have been identified in Hypocreaceae, including Cladobotryum dendroides and Cladobotryum protrusum, which can cause cobweb disease, as well as Hypomyces perniciosus, the causative agent of wet bubble disease [13,15,16]. In particular, Trichoderma spp. have been used as model systems to study the mechanisms of mycoparasitism [19]. For instance, Trichoderma harzianum, the causative agent of green mold disease on white button mushroom, produces various fungal cell wall-degradation enzymes that are being used in biotechnology and in biological control [20,21,22]. Genome analysis of Trichoderma atroviride and Trichoderma virens indicated that they contain a large numbers of genes encoding fungal cell wall-degradation enzymes compared to non-mycoparasitic fungi [18].
In this study, we obtained the first genome sequence of P. penicillatus using the single-molecule real-time (SMRT) sequencing platform [23]. Through phylogenetic analysis of single-copy orthologous genes, we determined the phylogenetic placement of P. penicillatus. Based on carbohydrate-active enzymes (CAZymes) analysis, secondary metabolite analysis, and virulence factor analysis, we demonstrated the putative molecular mechanisms of mycoparasitism of P. penicillatus. This study and the genomic resources made available herein provide a basis for the future functional characterization of the pathogenesis-related genes in P. penicillatus as well as the development of morel white mold disease management strategies.
2. Materials and Methods
2.1. Fungal Material
The P. penicillatus strain CCMJ2836 was provided by the Engineering Research Center of the Chinese Ministry of Education for Edible and Medicinal Fungi, Jilin Agricultural University (Changchun, China), which was isolated from white mold-diseased morel fruiting bodies in Sichuan Province, China. The mycelial plugs of strain CCMJ2836 were grown on potato glucose agar (PDA) medium overlaid with cellophane sheets for 10 d at 25 °C under a light/dark photoperiod (12/12 h).
2.2. Genome Sequencing and Assembly
P. penicillatus CCMJ2836 genomic DNA was extracted from the mycelia using NuClean Plant Genomic DNA Kits (CWBIO, Beijing, China) following the manufacturer’s instructions. The 20 kb-gDNA library was prepared and sequenced on two SMRT cells with a PacBio Sequel system. Raw data were de novo assembled via SMARTdenovo (https://github.com/ruanjue/smartdenovo). Genome assembly completeness was assessed with both the Core Eukaryotic Genes Mapping Approach (CEGMA) [24] and Benchmarking Universal Single-Copy Orthologs (BUSCO; [25]). Protein-encoding genes were predicted with GLEAN [26] (http://sourceforge.net/projects/glean-gene) through both extrinsic and ab initio algorirhms.
2.3. Gene Prediction and Genome Annotation
Annotations of the predicted genes were obtained through BLASTp (e-value ≤ 1 × 10−5) referencing the following databases: InterPro, TrEMBL, National Center for Biotechnology Information non-redundant (NCBI-nr), Swiss-Prot [27], Gene Ontology (GO) [28], Clusters of Orthologous Groups (COG) [29], and Kyoto Encyclopedia of Genes and Genomes (KEGG) [30]. Transposon elements and tandem repeat sequences (TRF) were predicted with RepeatMasker (http://www.repeatmasker.org) and Tandem Repeat Finder [31], respectively. Transfer RNAs (tRNAs) were predicted with tRNAscan-SE [32]. Ribosomal RNA (rRNA) sequences were identified with rRNAmmer [33] following both evidence-based and ab initio approaches. Non-coding small RNAs were annotated with the Rfam database [34].
2.4. Phylogenetic Analysis of Single-Copy Orthologs
Reciprocal BLASTp searches were conducted (e-value ≤ 1 × 10−7) with protein sequences from the representative species of Eurotiomycetes (four Eurotiales species: Paecilomyces variotii [35], Penicillium chrysogenum [36], Aspergillus nidulans [37], and A. niger [38]) and Sordariomycetes (Hypocreales species: Metarhizium robertsii [39], T. atroviride [40], T. harzianum [41], Trichoderma reesei [42], T. virens [43], C. dendroides [16], C. protrusum [13], H. perniciosus [15], Cordyceps militaris [44], Fusarium graminearum [45], and Niesslia exilis (https://mycocosm.jgi.doe.gov/Nieex1), Ophiocordyceps unilateralis [46], Stachybotrys chartarum [47], and one Sordariales species: Neurospora crassa [48]) were included to obtain single-copy ortholog sequences. Schizosaccharomyces pombe [49] in Schizosaccharomycetes was used as an outgroup for this analysis. Alignments with high-score pairs (alignment ratio ≥ 30%) were conjoined by SOLAR (V0.0.19) and used for the subsequent phylogenetic analysis. Sequences of single-copy orthologs were aligned with MUSCLE [50], and the phylogenetic relationships of the species were estimated using the maximum likelihood (ML) model in RA × ML [51] with 1000 bootstrap replicates. Schizosaccharomyces pombe was included in this analysis as an outgroup taxon.
2.5. Carbohydrate-Active Enzyme (CAZyme) Family Analysis
The deduced amino acid sequences of the P. penicillatus genes were searched against the CAZy database (http://www.cazy.org/) [52] with BLASTp (e-value ≤ 1 × 10−5, identity ≥ 40% and coverage ≥ 40%), and were further grouped into auxiliary activities (AAs), carbohydrate-binding modules (CBMs), carbohydrate esterases (CEs), glycoside hydrolases (GHs), glycosyl transferases (GTs), and polysaccharide lyases (PLs). Modular analysis of GH family 18 was carried out with InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/). Signal peptides of GH family 18 were predicted using the SignalP 5.0 server (http://www.cbs.dtu.dk/services/SignalP-5.0/).
2.6. Secondary Metabolite Gene Cluster Analysis and Pathogenicity-Related Genes
Gene clusters encoding secondary metabolite enzymes were analyzed with AntiSMASH fungal version 3.0 (https://fungismash.secondarymetabolites.org/) with the following built-in features activated: ActiveSiteFinder, ClusterBlast, Cluster Pfam analysis, KnownClusterBlast, Pfam-based GO term annotation, and SubClusterBlast. The cutoff similarity value for a known cluster was 40%. Genes within the clusters were further searched against the Pathogen-Host Interactions database (PHI-base) and the database of fungal virulence factor (DFVF) by BLASTp (e-value ≤ 1 × 10−5, identity ≥ 40% and coverage ≥ 40%) to predict the putative involvement of the gene clusters in the pathogenesis of P. penicillatus. Pathogenic genes were identified using the PHI (http://www.phi-base.org/) [53] and DFVF (http://sysbio.unl.edu/DFVF/Download.php) [54] databases.
2.7. Dual Culture Assay
Cultures of P. penicillatus strain CCMJ2836 and M. sextelata strain GB-Ch-102 were inoculated on HIMEDIA corn meal peptone yeast agar (CMA) and incubated at room temperature for 5 d. P. penicillatus agar discs were inoculated on one side of regular or two-compartment CMA dishes. After 6 d, M. sextelata agar discs were inoculated on the other side of the petri dish at about 4 cm from the P. penicillatus agar disc and then incubated for two more days. Each confrontation culture was replicated 10 times and incubated at room temperature. Cultures were photographed with an Epson Perfection V700 Photo scanner.
3. Results and Discussion
3.1. Genome Sequencing and Assembly of P. penicillatus
The genome of P. penicillatus CCMJ2836 strain was sequenced on the PacBio SMRT Sequel platform using two SMRT cells. A total of 4,266 Mb clean data (~105 × coverage) was generated (Table S1). The de novo assembly of the P. penicillatus genome was ~40.20 Mb, consisting of 52 scaffolds with 2.60 Mb in N50 value and 44.70% in guanine-cytosine (GC) content (Table 1). The completeness of the P. penicillatus genome was assessed through the CEGMA and BUSCOs analyses, with completeness scores of 95.56% and 97.20%, respectively. The whole genomic sequence of P. penicillatus CCMJ2836 has been deposited at GenBank and is available under accession number JACGSR000000000. The version described in this paper is version JACGSR010000000.

Table 1.
Summary of the P. penicillatus CCMJ2836 genome.
3.2. Gene Prediction and Annotation
Combining both homology-based and ab initio gene prediction approaches, 9454 protein-coding genes were predicted in P. penicillatus, with an average sequence length of 1694.45 bp. On average, each gene contains 2.61 exons in 557.93 bp. The putative protein-coding genes were functionally annotated using the following databases: NCBI nr (95.75%), Swiss-Prot (63.48%), KEGG (62.07%), COG (44.16%), TrEMBL (95.65%), InterPro (70.85%), and GO (49.55%). Overall, 96.34% genes had homologs in at least one database (Table S2 and Text S1). We identified ~3.1 Mbp repeat sequences and 0.12% non-coding RNA species (ncRNA) in the P. penicillatus genome. Among these repeat sequences, long terminal repeats (LTRs) were the most abundant representing 5.64% of the genome (Table S3 and Text S1). The ncRNA species contained 197 transfer RNA (tRNA) genes, 87 microRNA (miRNA) genes, 38 ribosomal RNA (rRNA) genes, and 25 small nuclear RNA (snRNA) genes (Table S4 and Text S1).
3.3. Phylogenomic Analysis
In view of the polyphyletic status of the Paecilomyces genus and the uncertain phylogenetic placement of P. penicillatus, a phylogenetic analysis was conducted using the single-copy orthologous genes (Figure 1). As expected, species in Eurotiomycetes and Sordariomycetes were clearly separated into two phylogenetic clades. In agreement with the conclusion made from both morphological characteristics and ITS sequences [9], P. variotii was placed under Eurotiomycetes. Within Sordariomycetes, Hypocreales species grouped together and were separated from N. crassa in Sordariales. P. penicillatus was placed under Hypocreales and was closest to Hypocreaceae, which includes many mycoparasitic fungi that can infect edible mushrooms. The next closest families were Nectriaceae and Niessliaceae. P. penicillatus is distantly related to Stachybotryaceae, Clavicipitaceae and Ophiocordycipitaceae, and has the most distant relationship with Cordycipitaceae. Our results indicate that Paecilomyces is polyphyletic and that P. penicillatus and P. variotii are clearly distantly related. We conclude that P. penicillatus is in the order of Hypocreales but not Eurotiales, which was previously concluded based on morphological characters. Our findings support a previous study based on 18S rRNA and β-tubulin gene sequences [8,11]. Our phylogenetic analysis further demonstrated that P. penicillatus is closely related to Hypocreaceae. A previous phylogenetic analysis of the 5.8S rDNA and ITS sequences of entomopathogenic Paecilomyces spp. demonstrated that most Paecilomyces species are Hypocreales and a few Paecilomyces species are Eurotiales. Within Hypocreales, Paecilomyces spp. was further classified into three subgroups. One subgroup includes Paecilomyces viridis, P. penicillatus, and Paecilomyces carneus. Most strains of Paecilomyces lilacinus and Paecilomyces marquandii are closely related and are classified as a distinct subgroup. The other subgroup contains Paecilomyces farinosus, Paecilomyces fumosoroseus, and other Paecilomyces species [9]. Due to the lack of genomic resources of most Paecilomyces spp., a single-copy orthologous genes-based phylogenetic analysis is not technically possible yet. Our study, therefore, lays the foundation for further phylogenetic classification of Paecilomyces spp. by deploying the single-copy orthologous gene sequences.
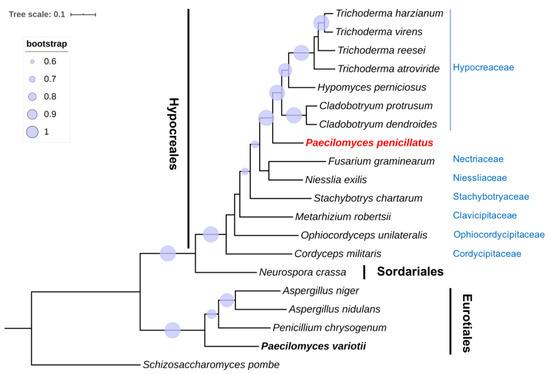
Figure 1.
Phylogenetic analysis of single-copy orthologous genes of P. penicillatus and 13 representative species of Hypocreales, one Sordariales species, and four Eurotiales species. S. pombe was used as an outgroup for this analysis. P. variotii is indicated in bold font, and P. pencililatus is highlighted in red font. Families of Hypocreales are labeled in blue font. The phylogenetic tree was generated using the maximum-likelihood method with 1000 bootstrap replicates. The tree scale indicates residue substitutions per site. Only bootstrap values greater than 0.6 are displayed.
3.4. The CAZyme
To successfully infect fungal hosts, mycoparasitic fungi commonly produce chitinases and glucanases to aid in the digestion of the fungal host cell wall, which is composed of chitin and glucan [19,55,56]. In light of the mycoparasitic nature of P. penicillatus, we hypothesized that P. penicillatus may also produce enzymes that can degrade the cell wall of its host. To assess this possibility, the genome of P. penicillatus was mapped to the CAZymes database (http://www.cazy.org), which includes all the enzyme families known to function in carbohydrate metabolism [57]. A total of 299 CAZymes were identified including 170 GHs, 67 GTs, 28 AAs, 24 CBMs, seven CEs, and three PLs (Figure 2). The most abundant CAZyme family in P. penicillatus is the GH family 18. The second and the third most abundant CAZyme families are the GH family 16 and the AA family 3, respectively (Figure 2).
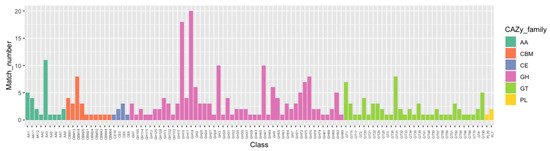
Figure 2.
Annotation of carbohydrate-related genes in the P. penicillatus genome.
The GH family 18 contains all the enzymes that have been previously shown to contribute to the degradation and replenishment of both intrinsic and extrinsic fungal chitin [58] (Figure 2). GH family 18 can be further divided into three subgroups, namely A, B, and C [59,60]. We were particularly interested in subgroup C (sgC) chitinases due to their potential involvement in mycoparasitism and their other interesting features [41,61]. Currently, known sgC chitinases contain CBM 18 (chitin binding) domains and/or CBM 50 (LysM) domains [18,62]. An analysis of sgC chitinases was performed. Based on the domain prediction, out of the 20 chitinases in GH family 18, five were predicted to be in sgC. Three of them have the CBM 18 domain and the other two have both the CBM 18 and the CBM 50 domains. In view of the close phylogenetic relationship between P. penicillatus and Trichoderma spp. and the well-studied production of fungal cell wall-degradation enzymes by Trichoderma spp., a phylogenetic analysis of sgC chitinases of P. penicillatus, Trichoderma atroviride and T. virens was conducted (Figure 3). Interestingly, our results showed that out of five genes, only two orthologue pairs could be found in the sgC chitinases of P. penicillatus and T. virens (PP_10005833/TVC9, PP_10007271/TVC10). The drastic differences in the number and sequence divergence of sgC in these phylogenetically closely-related mycoparasitic organisms suggests a strong evolutionary pressure on this protein family.
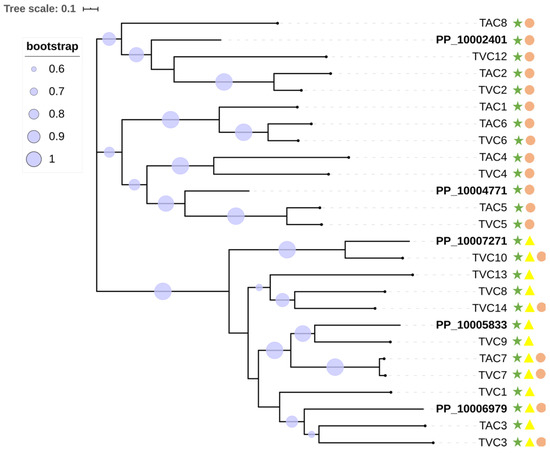
Figure 3.
Phylogenetic relationships of P. penicillatus, T. atroviride, and T. virens sgC chitinases. The presence of GH18 modules (IPR001223) is indicated with green stars, LysMs (IPR018392) are indicated in yellow triangles, and CBM18s (IPR001002) are represented by pink circles.
Beside chitinases, β-(1,3)-glucanases are another enzyme family that decomposes the fungal cell wall and belongs to GH family17, 55, 64, and 81 [41]. Compared with Trichoderma spp. and other representative fungal pathogens of plants or animals, P. penicillatus and Trichoderma spp. encode the highest number of GH family 18 proteins among the fungal organisms included in this study [41]. For GH families 64 and 81, we found a higher number in P. penicillatus and Trichoderma spp. in comparison to the other filamentous fungi. Similarly, P. penicillatus contains the highest number of GH family 75, which could be involved in the decomposition of chitinous carbohydrates [41]. Our results indicated that P. penicillatus produces a plethora of chitinases and β-(1,3)-glucanases in resemblance to the well-characterized mycoparasitic Trichoderma spp.
3.5. Secondary Metabolites and Pathogenicity-Related Genes
We identified 74 putative secondary metabolite biosynthesis gene clusters in P. penicillatus. Among them, seven gene clusters had significant hits to known secondary metabolite biosynthesis gene clusters (Table 2), including two antifungal (squalestatin 1 and AbT1), one antibacterial (cephalosporin C), one insecticidal (leucinostatin), two mammal-toxic (monascorubrin and 6-methylsalicyclic acid), and one plant-toxic (dimethylcoproge) secondary metabolic compounds. Multiple P. penicillatus genes in these gene clusters showed significant sequence similarity to genes in the Pathogen-Host Interactions database (PHI-base) and/or the database of fungal virulence factor (DFVF), indicative of possible involvement of these gene clusters in the pathogenesis of P. penicillatus. Specifically, Squalestatin 1 (40% similarity) is a potent inhibitor of squalene synthase, an essential enzyme for sterol biosynthesis [63]. Squalestatin 1 exhibits broad-spectrum antifungal activity [64] and was identified in Phoma sp. and several ascomycetes [65,66]. AbT1 (100% similarity) is a precursor of the cyclic peptide antibiotic Aureobasidin A (AbA), known to be produced by Aureobasidium pullulans [67]. AbA has been shown to exhibit inhibitory activity to yeast inositol phosphorylceramide synthase and is toxic to several other fungal organisms, such as Aspergillus nidulans and A. niger [67,68]. The genome of P. penicillatus also contains a gene cluster that has a 40% similarity to that of a gene involved in the biosynthesis of leucinostatin, a compound with antitrypanosomal properties. Interestingly, most Paecilomyces spp. identified thus far are entomophagous [69], and leucinostatin was previously reported to be produced by a Paecilomyces sp. isolated from soil [70]. In addition, several causative gene clusters for mammal or plant mycotoxins or their precursors were also predicted in P. penicillatus with 100% similarity, including dimethylcoproge [71], monascorubrin [72], and 6-methylsalicyclic acid [73]. It remains to be investigated whether these mycotoxic secondary compounds facilitate the infection of P. penicillatus on morel mycelia and fruiting bodies, or whether they coordinate or contribute to interspecies competition against other microbes living in the same environmental niche.

Table 2.
Summary of secondary metabolite biosynthesis gene clusters in P. penicillatus with known functions and their putative involvement in pathogenesis.
3.6. Dual Culture Assay
Based on the above analyses, we hypothesized that P. penicillatus may produce certain inhibitory compounds that can affect the hyphal growth of morels. To test this possibility, we conducted dual culture assays where agar plugs of both P. penicillatus and M. sextelata were inoculated onto the same petri dish. Indeed, M. sextelata had minimum growth in the direction towards the colony of P. penicillatus, and extensive growth of M. sextelata was observed in the direction away from P. penicillatus (Figure 4A), suggesting that P. penicillatus does produce host-inhibitory substances in a contact-independent manner. In an attempt to further investigate whether the substances are volatile or soluble, we also conducted confrontation assays on enclosed two-compartment petri dishes, where P. penicillatus and M. sextelata were inoculated on the center of each compartments. We expected to observe a similar inhibitory effect of P. penicillatus on M. sextelata in the case of volatile compounds being produced by P. penicillatus. However, M. sextelata grew rapidly in all directions at nearly equal speed in these assays (Figure 4B). This indicated that the inhibitory compounds were unlikely to be volatile, and rather, that they must be secreted and diffused in the medium. We are currently investigating whether P. penicillatus exhibits the antagonistic activity against M. sextelata through extracellular fungal cell wall-degradation enzymes or mycotoxins, as predicted in this study.
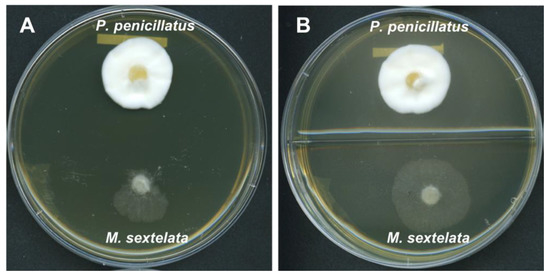
Figure 4.
Confrontation cultures between P. penicillatus and M. sextelata on CMA medium. (A). Normal petri dish (B). Two-compartment petri dish.
4. Conclusions
In this study, we present the first whole-genome sequence of P. penicillatus, the causative agent of the morel white mold disease. The genome of P. penicillatus is ~40.20 Mb in size with an N50 value of 2.59 Mbp and 9454 coding genes. The phylogenetic analysis based on the single-copy orthologous genes demonstrated that P. penicillatus is within Hypocreales and is closely related to Hypocreaceae, a family that is known to include many mycoparasitic species. CAZymes analysis further illustrated that the genome of P. penicillatus has a large number of fungal cell wall degradation enzymes. We identified a plethora of gene clusters that are known to be involved in the biosynthesis of cytotoxic secondary compounds that may function coordinatively during the pathogenesis of P. penicillatus. We also showed that P. penicillatus can produce contact-independent soluble host-inhibitory compounds. Taken together, we provide strong evidence in support of the phylogenetic placement of P. penicillatus within Hypocreales and provide a basis for the future functional characterization of the fungal cell wall-degradation enzymes and secondary metabolites produced by this mycoparasitic fungus.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/10/834/s1, Text S1: Annotation of the genome of P. penicillatus, Table S1: CEGMA analysis on the completion of the P. penicillatus genome, Table S2: Functional annotation of the genes in the P. penicillatus genome against several databases, Table S3: The statistics of the repeat sequences in the P. penicillatus genome, Table S4: Annotation of tRNA, rRNA, miRNA, and snRNA in the P. penicillatus genome.
Author Contributions
Conceptualization and supervision, Y.L. and Y.F.; Formal analysis and writing, X.W.; Methodology, J.P.; Investigation, X.W. and Y.G.; software, J.P., L.S., and X.W.; Writing—review and editing, J.P., Y.F., and G.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (No. 2017YFD0601002); the Special Fund for Agro-Scientific Research in the Public Interest (No. 201503137); the Program of Creation and Utilization of Germplasm of Mushroom Crop of “111” Project (No. D17014); and the Internationally Cooperative Research Center of China for New Germplasm Breeding of Edible Mushroom (2017B01011).
Acknowledgments
We appreciate Xiaolan He, the Soil and Fertilizer Institute, Sichuan Academy of Agricultural Sciences, for providing the P. penicillatus strain CCMJ2836 used in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- He, P.; Wang, K.; Cai, Y.; Liu, W. Live cell confocal laser imaging studies on the nuclear behavior during meiosis and ascosporogenesis in Morchella importuna under artificial cultivation. Micron 2017, 101, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Ower, R. Notes on the Development of the Morel Ascocarp: Morchella Esculenta. Mycologia 1982, 74, 142–144. [Google Scholar] [CrossRef]
- Masaphy, S. Biotechnology of morel mushrooms: Successful fruiting body formation and development in a soilless system. Biotechnol. Lett. 2010, 32, 1523–1527. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Yu, M.; Wang, K.; Cai, Y.; Li, B.; Liu, W. Interspecific hybridization between cultivated morels Morchella importuna and Morchella sextelata by PEG-induced double inactivated protoplast fusion. World J. Microbiol. Biotechnol. 2020, 36, 58. [Google Scholar] [CrossRef]
- Guo, M.P.; Chen, K.; Wang, G.Z.; Bian, Y.B. First Report of Stipe Rot Disease on Morchella importuna Caused by Fusarium incarnatum-F. equiseti Species Complex in China. Plant Dis. 2016, 100, 2530. [Google Scholar] [CrossRef]
- He, P.; Li, C.; Cai, Y.; Zhang, Y.; Bian, Y.; Liu, W. First report of pileus rot disease on cultivated Morchella importuna caused by Diploöspora longispora in China. J. Gen. Plant Pathol. 2018, 84, 65–69. [Google Scholar] [CrossRef]
- He, X.-L.; Peng, W.-H.; Miao, R.-Y.; Tang, J.; Chen, Y.; Liu, L.-X.; Wang, D.; Gan, B.-C. White mold on cultivated morels caused by Paecilomyces Penicillatus. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Luangsa-ard, J.J.; Hywel-Jones, N.L.; Samson, R.A. The polyphyletic nature of Paecilomyces sensu lato based on 18S-generated rDNA phylogeny. Mycologia 2004, 96, 773–780. [Google Scholar] [CrossRef]
- Inglis, P.W.; Tigano, M.S. Identification and taxonomy of some entomopathogenic Paecilomyces spp. (Ascomycota) isolates using rDNA-ITS Sequences. Genet. Mol. Biol. 2006, 29, 132–136. [Google Scholar] [CrossRef]
- Samson, R.A. Paecilomyces and some allied Hyphomycetes. Stud. Mycol. 1974, 6, 1–119. [Google Scholar]
- Luangsa-Ard, J.J.; Hywel-Jones, N.L.; Manoch, L.; Samson, R.A. On the relationships of Paecilomyces sect. Isarioidea species. Mycol. Res. 2005, 109, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Peng, J.; Sun, L.; Bonito, G.; Wang, J.; Cui, W.; Fu, Y.; Li, Y. Genome Sequencing Illustrates the Genetic Basis of the Pharmacological Properties of Gloeostereum incarnatum. Genes 2019, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Sossah, L.F.; Liu, Z.; Yang, C.; Okorley, A.B.; Sun, L.; Fu, Y.; Li, Y. Genome Sequencing of Cladobotryum protrusum Provides Insights into the Evolution and Pathogenic Mechanisms of the Cobweb Disease Pathogen on Cultivated Mushroom. Genes 2019, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Wu, Y.; Jian, Q.; Yin, C.; Li, T.; Gupta, V.K.; Duan, X.; Jiang, Y. Complete genome sequencing of the luminescent bacterium, Vibrio qinghaiensis sp. Q67 using PacBio technology. Sci. Data 2018, 5, 170205. [Google Scholar] [CrossRef]
- Li, D.; Sossah, L.F.; Sun, L.; Fu, Y.; Li, Y. Genome Analysis of Hypomyces perniciosus, the Causal Agent of Wet Bubble Disease of Button Mushroom (Agaricus bisporus). Genes 2019, 10, 417. [Google Scholar] [CrossRef]
- Xu, R.; Liu, X.; Peng, B.; Liu, P.; Li, Z.; Dai, Y.; Xiao, S. Genomic Features of Cladobotryum dendroides, which Causes Cobweb Disease in Edible Mushrooms, and Identification of Genes Related to Pathogenicity and Mycoparasitism. Pathogens 2020, 9, 232. [Google Scholar] [CrossRef]
- Wu, F.; Mueller, L.A.; Crouzillat, D.; Pétiard, V.; Tanksley, S.D. Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: A test case in the euasterid plant clade. Genetics 2006, 174, 1407–1420. [Google Scholar] [CrossRef]
- Gruber, S.; Seidl-Seiboth, V. Self versus non-self: Fungal cell wall degradation in Trichoderma. Microbiolology 2012, 158, 26–34. [Google Scholar] [CrossRef]
- Benítez, T.; Rincón, A.M.; Limón, M.C.; Codón, A.C. Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 2005, 7, 249–260. [Google Scholar]
- Ospina-Giraldo, M.D.; Royse, D.J.; Chen, X.; Romaine, C.P. Molecular Phylogenetic Analyses of Biological Control Strains of Trichoderma harzianum and Other Biotypes of Trichoderma spp. Associated with Mushroom Green Mold. Phytopathology 1999, 89, 308–313. [Google Scholar] [CrossRef]
- Perazzolli, M.; Moretto, M.; Fontana, P.; Ferrarini, A.; Velasco, R.; Moser, C.; Delledonne, M.; Pertot, I. Downy mildew resistance induced by Trichoderma harzianum T39 in susceptible grapevines partially mimics transcriptional changes of resistant genotypes. BMC Genom. 2012, 13, 660. [Google Scholar] [CrossRef] [PubMed]
- Marzano, M.; Gallo, A.; Altomare, C. Improvement of biocontrol efficacy of Trichoderma harzianum vs. Fusarium oxysporum f. sp. lycopersici through UV-induced tolerance to fusaric acid. Biol. Control. 2013, 67, 397–408. [Google Scholar] [CrossRef]
- Li, C.; Lin, F.; An, D.; Wang, W.; Huang, R. Genome Sequencing and Assembly by Long Reads in Plants. Genes 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Parra, G.; Bradnam, K.; Korf, I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 2007, 23, 1061–1067. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Elsik, C.G.; Mackey, A.J.; Reese, J.T.; Milshina, N.V.; Roos, D.S.; Weinstock, G.M. Creating a honey bee consensus gene set. Genome Biol. 2007, 8, R13. [Google Scholar] [CrossRef]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence data bank and its new supplement TREMBL. Nucleic Acids Res. 1996, 24, 21–25. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2017, 46, D335–D342. [Google Scholar] [CrossRef] [PubMed]
- Urquhart, A.S.; Mondo, S.J.; Mäkelä, M.R.; Hane, J.K.; Wiebenga, A.; He, G.; Mihaltcheva, S.; Pangilinan, J.; Lipzen, A.; Barry, K.; et al. Genomic and Genetic Insights into a Cosmopolitan Fungus, Paecilomyces variotii (Eurotiales). Front. Microbiol. 2018, 9, 3058. [Google Scholar] [CrossRef]
- Van den Berg, M.A.; Albang, R.; Albermann, K.; Badger, J.H.; Daran, J.-M.; M Driessen, A.J.; Garcia-Estrada, C.; Fedorova, N.D.; Harris, D.M.; Heijne, W.H.M.; et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 2008, 26, 1161–1168. [Google Scholar] [CrossRef]
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.-J.; Wortman, J.R.; Batzoglou, S.; Lee, S.-I.; Baştürkmen, M.; Spevak, C.C.; Clutterbuck, J.; et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 2005, 438, 1105–1115. [Google Scholar] [CrossRef]
- Pel, H.J.; de Winde, J.H.; Archer, D.B.; Dyer, P.S.; Hofmann, G.; Schaap, P.J.; Turner, G.; de Vries, R.P.; Albang, R.; Albermann, K.; et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007, 25, 221–231. [Google Scholar] [CrossRef]
- Bischoff, J.F.; Rehner, S.A.; Humber, R.A. A multilocus phylogeny of the Metarhizium anisopliae lineage. Mycology 2009, 101, 512–530. [Google Scholar] [CrossRef]
- Shi-Kunne, X.; Seidl, M.F.; Faino, L.; Thomma, B.P.H.J. Draft Genome Sequence of a Strain of Cosmopolitan Fungus Trichoderma atroviride. Genome Announc. 2015, 3, e00287-15. [Google Scholar] [CrossRef]
- Baroncelli, R.; Piaggeschi, G.; Fiorini, L.; Bertolini, E.; Zapparata, A.; Pè, M.E.; Sarrocco, S.; Vannacci, G. Draft Whole-Genome Sequence of the Biocontrol Agent Trichoderma harzianum T6776. Genome Announc. 2015, 3, e00647-15. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.; Berka, R.M.; Henrissat, B.; Saloheimo, M.; Arvas, M.; Baker, S.E.; Chapman, J.; Chertkov, O.; Coutinho, P.M.; Cullen, D.; et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 2008, 26, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Herrera-Estrella, A.; Seidl-Seiboth, V.; Martinez, D.A.; Druzhinina, I.S.; Thon, M.; Zeilinger, S.; Casas-Flores, S.; Horwitz, B.A.; Mukherjee, P.K.; et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011, 12, R40. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.; Hu, X.; Zhang, S.; Zheng, H.; Huang, Y.; Zhou, Y.; Wang, S.; et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional chinese medicine. Genome Biol. 2011, 12, R116. [Google Scholar] [CrossRef] [PubMed]
- Walkowiak, S.; Rowland, O.; Rodrigue, N.; Subramaniam, R. Whole genome sequencing and comparative genomics of closely related Fusarium Head Blight fungi: Fusarium graminearum, F. meridionale and F. asiaticum. BMC Genom. 2016, 17, 1014. [Google Scholar] [CrossRef]
- De Bekker, C.; Ohm, R.A.; Loreto, R.G.; Sebastian, A.; Albert, I.; Merrow, M.; Brachmann, A.; Hughes, D.P. Gene expression during zombie ant biting behavior reflects the complexity underlying fungal parasitic behavioral manipulation. BMC Genom. 2015, 16, 620. [Google Scholar] [CrossRef]
- Betancourt, D.A.; Dean, T.R.; Kim, J.; Levy, J. Genome Sequence of Stachybotrys chartarum Strain 51-11. Genome Announc. 2015, 3, e01114-15. [Google Scholar] [CrossRef]
- Galagan, J.; Calvo, S.; Borkovich, K.; Selker, E.U.; Read, N.D.; Jaffe, D.; Fitzhugh, W.; Ma, L.-J.; Smirnov, S.; Purcell, S.; et al. The genome sequence of the filamentous fungus Neurospora crassa. Nat. Cell Biol. 2003, 422, 859–868. [Google Scholar] [CrossRef]
- Wood, V.; Gwilliam, R.; Rajandream, M.-A.; Lyne, M.; Lyne, R.; Stewart, A.; Sgouros, J.; Peat, N.; Hayles, J.; Baker, S.; et al. The genome sequence of Schizosaccharomyces pombe. Nature 2002, 415, 871–880. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.; Cuzick, A.; Rutherford, K.; Irvine, A.; Pedro, H.; Pant, R.; Sadanadan, V.; Khamari, L.; Billal, S.; Mohanty, S.; et al. PHI-base: A new interface and further additions for the multi-species pathogen-host interactions database. Nucleic Acids Res. 2017, 45, D604–D610. [Google Scholar] [CrossRef]
- Lu, T.; Yao, B.; Zhang, C. DFVF: Database of fungal virulence factors. Database 2012. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.J. Fungal cell wall chitinases and glucanases. Microbiology 2004, 150, 2029–2035. [Google Scholar] [CrossRef]
- Lima, S.L.; Colombo, A.L.; de Almeida Junior, J.N. Fungal Cell Wall: Emerging Antifungals and Drug Resistance. Front. Microbiol. 2019, 10, 2573. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2008, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Funkhouser, J.D.; Aronson, N.N. Chitinase family GH18: Evolutionary insights from the genomic history of a diverse protein family. BMC Evol. Biol. 2007, 7, 96. [Google Scholar] [CrossRef]
- Seidl, V. Chitinases of filamentous fungi: A large group of diverse proteins with multiple physiological functions. Fungal Biol. Rev. 2008, 22, 36–42. [Google Scholar] [CrossRef]
- Seidl, V.; Huemer, B.; Seiboth, B.; Kubicek, C.P. A complete survey of Trichoderma chitinases reveals three distinct subgroups of family 18 chitinases. FEBS J. 2005, 272, 5923–5939. [Google Scholar] [CrossRef]
- Gruber, S.; Kubicek, C.P.; Seidl-Seiboth, V. Differential regulation of orthologous chitinase genes in mycoparasitic Trichoderma species. Appl. Environ. Microbiol. 2011, 77, 7217–7226. [Google Scholar] [CrossRef] [PubMed]
- Gruber, S.; Vaaje-Kolstad, G.; Matarese, F.; López-Mondéjar, R.; Kubicek, C.P.; Seidl-Seiboth, V. Analysis of subgroup C of fungal chitinases containing chitin-binding and LysM modules in the mycoparasite Trichoderma Atroviride. Glycobiology 2010, 21, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Fitzgerald, B.J.; Hutson, J.L.; McCarthy, A.D.; Motteram, J.M.; Ross, B.C.; Sapra, M.; Snowden, M.A.; Watson, N.S.; Williams, R.J. Squalestatin 1, a potent inhibitor of squalene synthase, which lowers serum cholesterol in vivo. J. Biol. Chem. 1992, 267, 11705–11708. [Google Scholar] [PubMed]
- Hasumi, K.; Tachikawa, K.; Sakai, K.; Murakawa, S.; Yoshikawa, N.; Kumazawa, S.; Endo, A. Competitive inhibition of squalene synthetase by squalestatin 1. J. Antibiot. 1993, 46, 689–691. [Google Scholar] [CrossRef] [PubMed]
- Bills, G.F.; Peláez, F.; Polishook, J.D.; Diez-Matas, M.T.; Harris, G.H.; Clapp, W.H.; Dufresne, C.; Byrne, K.M.; Nallin-Omstead, M.; Jenkins, R.G.; et al. Distribution of zaragozic acids (squalestatins) among filamentous ascomycetes. Mycol. Res. 1994, 98, 733–739. [Google Scholar] [CrossRef]
- Blows, W.; Foster, G.; Lane, S.; Noble, D.; Piercey, J.; Sidebottom, P.; Webb, G. The squalestatins, novel inhibitors of squalene synthase produced by a species of Phoma. V. Minor metabolites. J. Antibiot. 1994, 47, 740–754. [Google Scholar] [CrossRef][Green Version]
- Slightom, J.L.; Metzger, B.P.; Luu, H.T.; Elhammer, A.P. Cloning and molecular characterization of the gene encoding the Aureobasidin A biosynthesis complex in Aureobasidium pullulans BP-1938. Gene 2009, 431, 67–79. [Google Scholar] [CrossRef]
- Zhong, W.; Jeffries, M.W.; Georgopapadakou, N.H. Inhibition of Inositol Phosphorylceramide Synthase by Aureobasidin A in Species. Antimicrob. Agents Chemother. 2000, 44, 651. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S. Commercialisation of Microbial Biopesticides for the Management of Pests and Diseases. In Recent Advances in the Diagnosis and Management of Plant Diseases; Awasthi, L.P., Ed.; Springer: New Delhi, India, 2015; pp. 1–10. [Google Scholar]
- Ishiyama, A.; Otoguro, K.; Iwatsuki, M.; Namatame, M.; Nishihara, A.; Nonaka, K.; Kinoshita, Y.; Takahashi, Y.; Masuma, R.; Shiomi, K.; et al. In vitro and in vivo antitrypanosomal activities of three peptide antibiotics: Leucinostatin A and B, alamethicin I and tsushimycin. J. Antibiot. 2009, 62, 303–308. [Google Scholar] [CrossRef]
- Chen, L.-H.; Lin, C.-H.; Chung, K.-R. A nonribosomal peptide synthetase mediates siderophore production and virulence in the citrus fungal pathogen Alternaria alternata. Mol. Plant Pathol. 2013, 14, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lam, C.-W.; Tam, E.W.T.; Lee, K.-C.; Yung, K.K.Y.; Leung, C.K.F.; Sze, K.-H.; Lau, S.K.P.; Yuen, K.-Y. The biosynthetic pathway for a thousand-year-old natural food colorant and citrinin in Penicillium marneffei. Sci. Rep. 2014, 4, 6728. [Google Scholar] [CrossRef] [PubMed]
- Puel, O.; Galtier, P.; Oswald, P.I. Biosynthesis and Toxicological Effects of Patulin. Toxins 2010, 2, 613. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).