Screening of Fish Cell Lines for Piscine Orthoreovirus-1 (PRV-1) Amplification: Identification of the Non-Supportive PRV-1 Invitrome
Abstract
1. Introduction
2. Results
2.1. Screening 30 Cell Lines with PRV-1 Containing Homogenates for Capacity to Support PRV-1 Amplification In Vitro
2.1.1. Lack of PRV-1 Amplification in 11 Cell Lines Exposed to Chilean PRV-1 (Unknown Subtype)
2.1.2. Lack of PRV-1 Amplification in 26 Cell Lines Belonging to 13 Species Exposed to Canadian and Norwegian PRV-1 RBC Homogenates
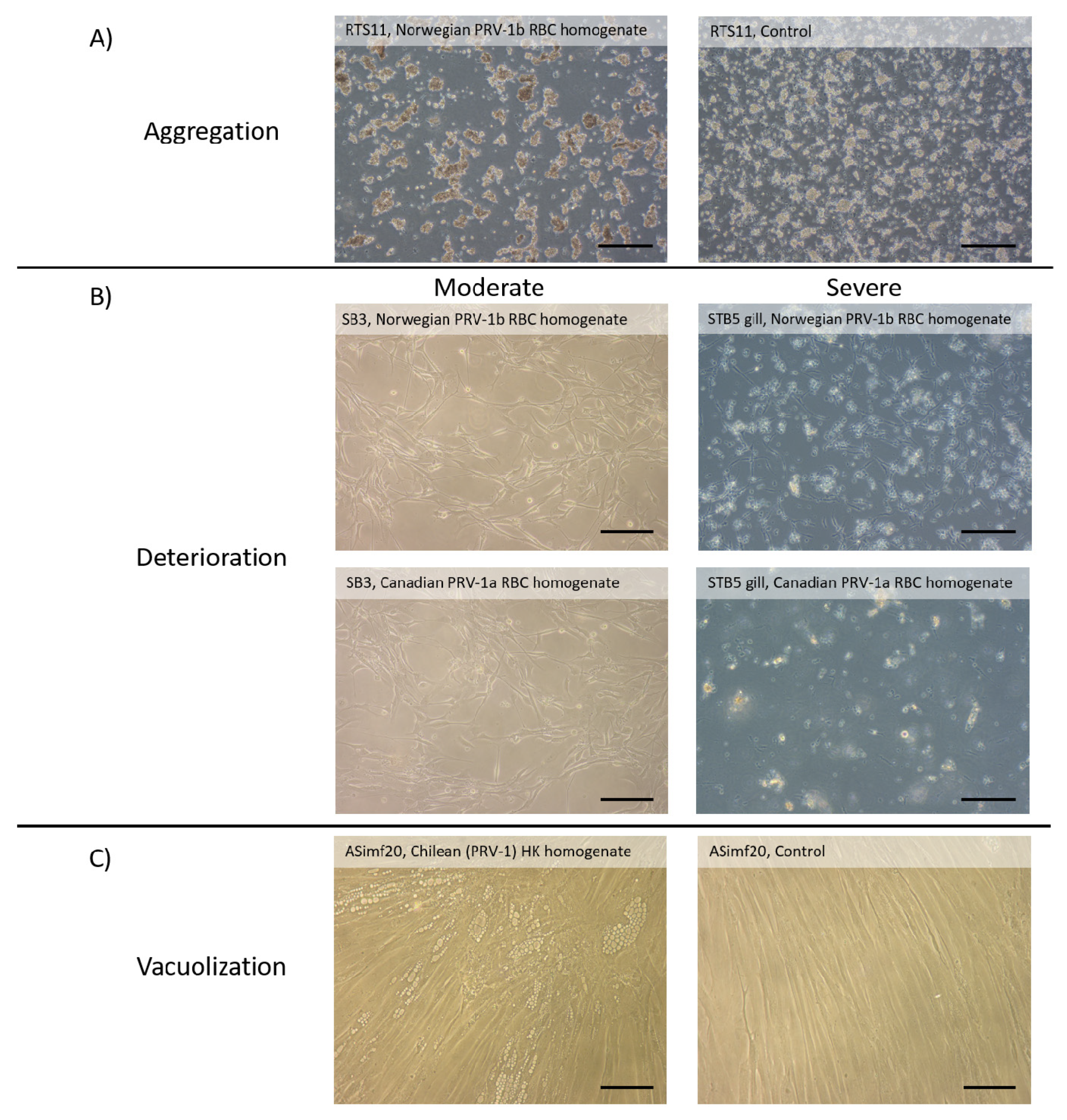
2.1.3. Lack of Consistent Cytopathic Effects (CPE) in Cell Lines Screened for PRV-1 Amplification
2.2. Lack of PRV Amplification in Cell Lines Co-Administered with Either Multiple PRV-1 Isolates, PRV-1 and CSV, or PRV-1 and IPNV
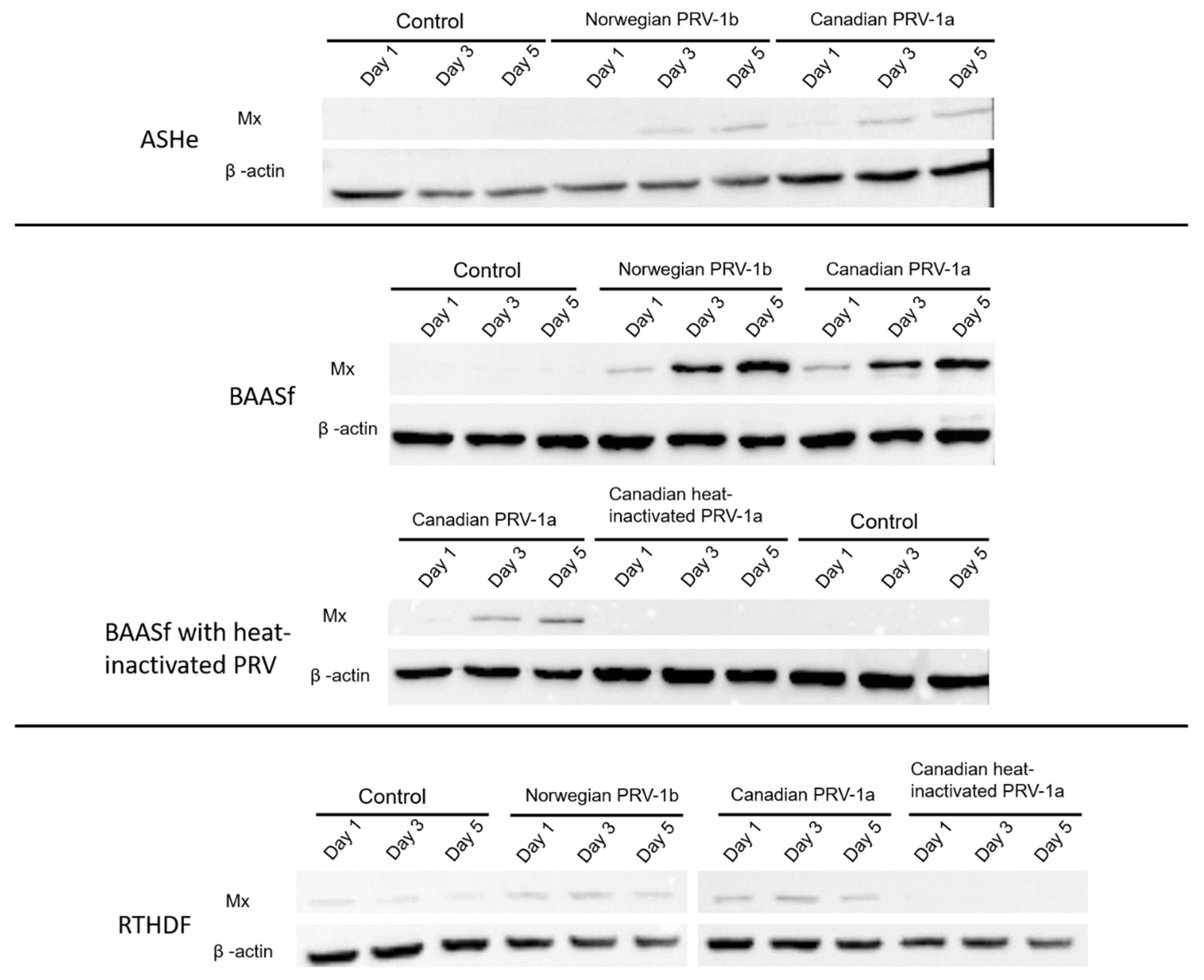
2.3. PRV-1 RBC Homogenates Induce Mx Protein Expression in Atlantic salmon and Rainbow Trout Cells Lines
2.4. Attempts to Establish Cell Lines from Infected Atlantic salmon Resulted in Limited Success but Showed Persistence of PRV-1 RNA in One Culture over Long Periods
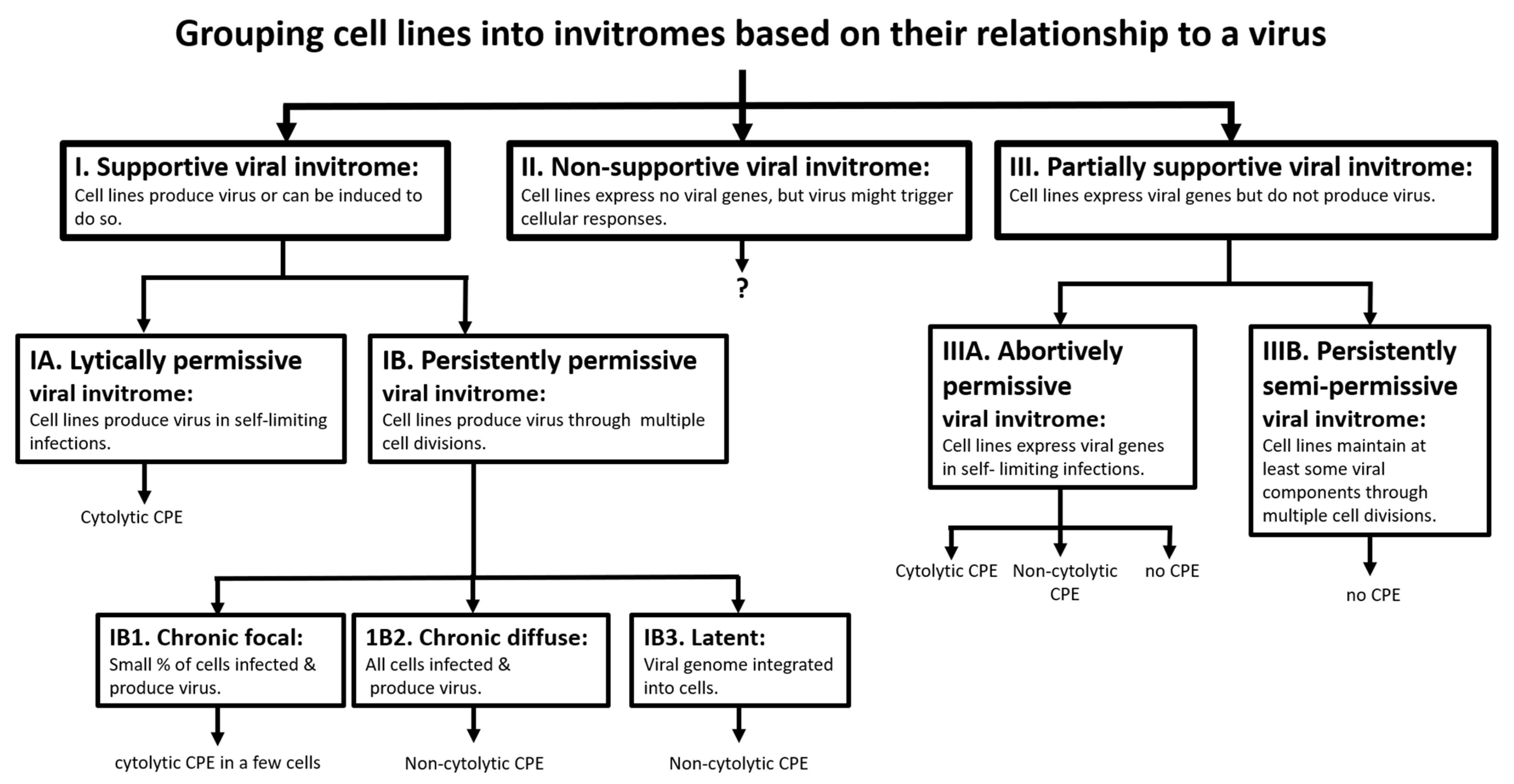
3. Discussion
4. Materials and Methods
4.1. Cell Line Maintenance
4.2. PRV-1 Inoculum Preparation
4.3. Primary Culture Originating from PRV-1 Infected Atlantic salmon
4.4. Monitoring for PRV-1 Amplification in Established Cell Lines Using Five Virus Isolation Assays
4.4.1. Monitoring for PRV-1 Amplification Using Single Cell Culture Passage Isolation Assay
4.4.2. Monitoring for PRV-1 Amplification Using Sub-Cultivation of Exposed Cells with Fresh Cells
4.4.3. Monitoring for PRV-1 Amplification Using Sub-Cultivation of Only Exposed Cells
4.4.4. Monitoring for PRV-1 Amplification Using Cell Lysate Transfer
4.4.5. Monitoring for PRV-1 Amplification Using Supernatant Transfer
4.4.6. Assessing the Appearance of Fish Cell Cultures upon Exposure to PRV-1 Homogenates
4.5. Co-Administration of Cell Cultures with Two Isolates of PRV-1, or with PRV-1 and CSV, or with PRV-1 and IPNV
4.6. RNA Extraction and Real-Time PCR
4.7. Detection of Mx2 Protein Expression in Cell Lines by SDS-Page and Western Blotting
4.8. Ethics Approval
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wessel, Ø.; Braaen, S.; Alarcon, M.; Haatveit, H.; Roos, N.; Markussen, T.; Tengs, T.; Dahle, M.K.; Rimstad, E. Infection with purified Piscine orthoreovirus demonstrates a causal relationship with heart and skeletal muscle inflammation in Atlantic salmon. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Polinski, M.P.; Marty, G.D.; Snyman, H.N.; Garver, K.A. Piscine orthoreovirus demonstrates high infectivity but low virulence in Atlantic salmon of Pacific Canada. Sci. Rep. 2019, 9, 1–22. [Google Scholar] [CrossRef]
- Polinski, M.P.; Vendramin, N.; Cuenca, A.; Garver, K.A. Piscine orthoreovirus: Biology and distribution in farmed and wild fish. J. Fish Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hodinka, R.L.; Kaiser, L. Is the era of viral culture over in the clinical microbiology laboratory? J. Clin. Microbiol. 2013, 51, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.M.; Decker, C.C.; Dao Thi, V.L. Cell Culture Models for Hepatitis E Virus. Viruses 2019, 11, 608. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A. The cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 2018, 29, 25–38. [Google Scholar] [CrossRef]
- Bols, N.C.; Pham, P.H.; Dayeh, V.R.; Lee, L.E.J. Invitromatics, invitrome, and invitroomics: Introduction of three new terms for in vitro biology and illustration of their use with the cell lines from rainbow trout. Vitr. Cell. Dev. Biol.-Anim. 2017, 53, 383–405. [Google Scholar] [CrossRef]
- Garver, K.A.; Marty, G.D.; Cockburn, S.N.; Richard, J.; Hawley, L.M.; Müller, A.; Thompson, R.L.; Purcell, M.K.; Saksida, S. Piscine reovirus, but not Jaundice Syndrome, was transmissible to Chinook Salmon, Oncorhynchus tshawytscha (Walbaum), Sockeye Salmon, Oncorhynchus nerka (Walbaum), and Atlantic Salmon, Salmo salar L. J. Fish Dis. 2016, 39, 117–128. [Google Scholar] [CrossRef]
- Kongtorp, R.T.; Taksdal, T. Studies with experimental transmission of heart and skeletal muscle inflammation in Atlantic salmon, Salmo salar L. J. Fish Dis. 2009, 32, 253–262. [Google Scholar] [CrossRef]
- Mikalsen, A.B.; Haugland, O.; Rode, M.; Solbakk, I.T.; Evensen, O. Atlantic Salmon Reovirus Infection Causes a CD8 T Cell Myocarditis in Atlantic Salmon (Salmo salar L.). PLoS ONE 2012, 7, e37269. [Google Scholar] [CrossRef]
- Watanabe, K.; Karlsen, M.; Devold, M.; Isdal, E.; Litlabø, A.; Nylund, A. Virus-like particles associated with heart and skeletal muscle inflammation (HSMI). Dis. Aquat. Organ. 2006, 70, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Bokelmann, M.; Edenborough, K.; Hetzelt, N.; Kreher, P.; Lander, A.; Nitsche, A.; Vogel, U.; Feldmann, H.; Couacy-Hymann, E.; Kurth, A. Utility of primary cells to examine NPC1 receptor expression in Mops condylurus, a potential Ebola virus reservoir. PLoS Negl. Trop. Dis. 2020, 14, e0007952. [Google Scholar] [CrossRef] [PubMed]
- Caì, Y.; Yú, S.; Postnikova, E.N.; Mazur, S.; Bernbaum, J.G.; Burk, R.; Zhāng, T.; Radoshitzky, S.R.; Müller, M.A.; Jordan, I.; et al. CD26/DPP4 Cell-Surface Expression in Bat Cells Correlates with Bat Cell Susceptibility to Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Infection and Evolution of Persistent Infection. PLoS ONE 2014, 9, e112060. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.W.; Yip, C.C.Y.; Tsang, J.O.L.; Tee, K.M.; Cai, J.P.; Chik, K.K.H.; Zhu, Z.; Chan, C.C.S.; Choi, G.K.Y.; Sridhar, S.; et al. Differential cell line susceptibility to the emerging Zika virus: Implications for disease pathogenesis, non-vector-borne human transmission and animal reservoirs. Emerg. Microbes Infect. 2016, 5, e93. [Google Scholar] [CrossRef]
- Lorenzen, E.; Carstensen, B.; Olesen, N.J. Inter-laboratory comparison of cell lines for susceptibility to three viruses: VHSV, IHNV and IPNV. Dis. Aquat. Organ. 1999, 37, 81–88. [Google Scholar] [CrossRef]
- Heaton, N.S. Revisiting the concept of a cytopathic viral infection. PLoS Pathog. 2017, 13, e1006409. [Google Scholar] [CrossRef]
- Boldogh, I.; Albrecht, T.; Porter, D.D. Persistent Viral Infections; University of Texas Medical Branch: Galveston, TX, USA, 1996; ISBN 0963117211. [Google Scholar]
- Frerichs, G.N.; Morgan, D.; Hart, D.; Skerrow, C.; Roberts, R.J.; Onions, D.E. Spontaneously productive C-type retrovirus infection of fish cell lines. J. Gen. Virol. 1991, 72, 2537–2539. [Google Scholar] [CrossRef]
- Kennedy, J.C.; Macdonald, R.D. Persistent infection with infectious pancreatic necrosis virus mediated by defective-interfering (DI) virus particles in a cell line showing strong interference but little DI replication. J. Gen. Virol. 1982, 58, 361–371. [Google Scholar] [CrossRef]
- Batts, W.; Yun, S.; Hedrick, R.; Winton, J. A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii). Virus Res. 2011, 158, 116–123. [Google Scholar] [CrossRef]
- García, I.; Galiana, A.; Falcó, A.; Estepa, A.; Perez, L. Characterization of an infectious pancreatic necrosis (IPN) virus carrier cell culture with resistance to superinfection with heterologous viruses. Vet. Microbiol. 2011, 149, 48–55. [Google Scholar] [CrossRef]
- Chi, S.C.; Wu, Y.C.; Cheng, T.M. Persistent infection of betanodavirus in a novel cell line derived from the brain tissue of barramundi Lates calcarifer. Dis. Aquat. Organ. 2005, 65, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Rijji John, K.; Richards, R.H. Characteristics of a new birnavirus associated with a warm-water fish cell line. J. Gen. Virol. 1999, 80, 2061–2065. [Google Scholar] [CrossRef] [PubMed]
- Pham, P.H.; Lai, Y.S.; Lee, F.F.Y.; Bols, N.C.; Chiou, P.P. Differential viral propagation and induction of apoptosis by grouper iridovirus (GIV) in cell lines from three non-host species. Virus Res. 2012, 167, 16–25. [Google Scholar] [CrossRef]
- Pham, P.H.; Huang, Y.J.; Mosser, D.D.; Bols, N.C. Use of cell lines and primary cultures to explore the capacity of rainbow trout to be a host for frog virus 3 (FV3). Vitr. Cell. Dev. Biol.-Anim. 2015, 51, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Pao, H.Y.; Wu, C.Y.; Wen, C.M. Persistent development of adomavirus and aquareovirus in a novel cell line from marbled eel with petechial skin haemorrhage. J. Fish Dis. 2019, 42, 345–355. [Google Scholar] [CrossRef]
- Dabrowska, K.; Switała-Jelen, K.; Opolski, A.; Weber-Dabrowska, B.; Gorski, A. A review: Bacteriophage penetration in vertebrates. J. Appl. Microbiol. 2005, 98, 7–13. [Google Scholar] [CrossRef]
- Sweere, J.M.; Van Belleghem, J.D.; Ishak, H.; Bach, M.S.; Popescu, M.; Sunkari, V.; Kaber, G.; Manasherob, R.; Suh, G.A.; Cao, X.; et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 2019, 363. [Google Scholar] [CrossRef]
- Tian, Y.; Wu, M.; Liu, X.; Liu, Z.; Zhou, Q.; Niu, Z.; Huang, Y. Probing the Endocytic Pathways of the Filamentous Bacteriophage in Live Cells Using Ratiometric pH Fluorescent Indicator. Adv. Healthc. Mater. 2015, 4, 413–419. [Google Scholar] [CrossRef]
- Virgin, H.W. The virome in mammalian physiology and disease. Cell 2014, 157, 142–150. [Google Scholar] [CrossRef]
- Jeon, H.; Lee, J.; Lee, S.; Kang, S.K.; Park, S.J.; Yoo, S.M.; Lee, M.S. Extracellular vesicles from KSHV-infected cells stimulate antiviral immune response through mitochondrial DNA. Front. Immunol. 2019, 10, 876. [Google Scholar] [CrossRef]
- Purcell, M.K.; Powers, R.L.; Taksdal, T.; McKenney, D.; Conway, C.M.; Elliott, D.G.; Polinski, M.; Garver, K.; Winton, J. Consequences of Piscine orthoreovirus genotype 1 (PRV-1) infections in Chinook salmon (Oncorhynchus tshawytscha), coho salmon (O. kisutch) and rainbow trout (O. mykiss). J. Fish Dis. 2020, 43, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Sadler, A.J.; Williams, B.R.G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Druckrey, H.; Schmähl, D.; Steinhoff, D.; Rajewsky, M.; Bannasch, P.; Flaschenträger, T. Cytolysierende Wirkung von Extrakten aus normalen Geweben auf verschiedene Tumoren der Ratte. Z. Krebsforsch. 1959, 63, 28–56. [Google Scholar] [CrossRef]
- Gradwohl, P.R. Mouse epidermal and skin extracts tested for cytostatic activity (“chalones”)-Effects of organ and cell cultures. Arch. Dermatol. Res. 1978, 263, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Lozzio, C.B.; Lozzio, B.B. Brief communication: Cytotoxicity of a factor isolated from human spleen. J. Natl. Cancer Inst. 1973, 50, 535–538. [Google Scholar] [CrossRef]
- Okudaira, H.; Kataoka, T.; Okada, H.; Furuse-irie, R.; Kawachi, S.; Nojima, S.; Nishioka, K. Cytotoxic factor demonstrated in lymph node extract. J. Biochem. 1970, 68, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Pompeia, C.; Lima, T.; Curi, R. Arachidonic acid cytotoxicity: Can arachidonic acid be a physiological mediator of cell death? Cell Biochem. Funct. 2003, 21, 97–104. [Google Scholar] [CrossRef]
- Henics, T.; Wheatley, D.N. Cytoplasmic vacuolation, adaptation and cell death: A view on new perspectives and features. Biol. Cell 1999, 91, 485–498. [Google Scholar] [CrossRef]
- Dayeh, V.R.; Schirmer, K.; Bols, N.C. Ammonia-containing industrial effluents, lethal to rainbow trout, induce vacuolisation and neutral red uptake in the rainbow trout gill cell line, RTgill-W1. ATLA Altern. Lab. Anim. 2009, 37, 77–87. [Google Scholar] [CrossRef]
- Pham, P.H.; Sokeechand, B.S.H.; Garver, K.A.; Jones, G.; Lumsden, J.S.; Bols, N.C. Fish viruses stored in RNAlater can remain infectious and even be temporarily protected from inactivation by heat or by tissue homogenates. J. Virol. Methods 2018, 253, 31–37. [Google Scholar] [CrossRef]
- Lundgren, E.; Roos, G. Vacuolization in cultured cells induced by amphotericin B. Antimicrob. Agents Chemother. 1978, 14, 267–269. [Google Scholar] [CrossRef]
- Dhamotharan, K.; Bjørgen, H.; Malik, M.S.; Nyman, I.B.; Markussen, T.; Dahle, M.K.; Koppang, E.O.; Wessel, Ø.; Rimstad, E. Dissemination of piscine orthoreovirus-1 (PRV-1) in atlantic salmon (salmo salar) during the early and regenerating phases of infection. Pathogens 2020, 9, 143. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.S.; Bjørgen, H.; Dhamotharan, K.; Wessel, Ø.; Koppang, E.O.; Di Cicco, E.; Hansen, E.F.; Dahle, M.K.; Rimstad, E. Erythroid Progenitor Cells in Atlantic Salmon (Salmo salar) May Be Persistently and Productively Infected with Piscine Orthoreovirus (PRV). Viruses 2019, 11, 824. [Google Scholar] [CrossRef] [PubMed]
- Ganassin, R.C.; Bols, N.C. Development of long-term rainbow trout spleen cultures that are haemopoietic and produce dendritic cells. Fish Shellfish Immunol. 1996, 6, 17–34. [Google Scholar] [CrossRef]
- Ganassin, R.C.; Bols, N.C. Development of a monocyte/macrophage-like cell line, RTS11, from rainbow trout spleen. Fish Shellfish Immunol. 1998, 8, 457–476. [Google Scholar] [CrossRef]
- Key, T.; Read, J.; Nibert, M.L.; Duncan, R. Piscine reovirus encodes a cytotoxic, non-fusogenic, integral membrane protein and previously unrecognized virion outer-capsid proteins. J. Gen. Virol. 2013, 94, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Kongtorp, R.T.; Taksdal, T.; Lyngøy, A. Pathology of heart and skeletal muscle inflammation (HSMI) in farmed Atlantic salmon Salmo salar. Dis. Aquat. Organ. 2004, 59, 217–224. [Google Scholar] [CrossRef]
- Wessel, Ø.; Olsen, C.M.; Rimstad, E.; Dahle, M.K. Piscine orthoreovirus (PRV) replicates in Atlantic salmon (Salmo salar L.) erythrocytes ex vivo. Vet. Res. 2015, 46. [Google Scholar] [CrossRef]
- Zhang, Y.; Polinski, M.P.; Morrison, P.R.; Brauner, C.J.; Farrell, A.P.; Garver, K.A. High-load reovirus infections do not imply physiological impairment in salmon. Front. Physiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- DeWitte-Orr, S.J.; Bols, N.C. Cytopathic effects of chum salmon reovirus to salmonid epithelial, fibroblast and macrophage cell lines. Virus Res. 2007, 126, 159–171. [Google Scholar] [CrossRef]
- Sanjuán, R.; Thoulouze, M.-I. Why viruses sometimes disperse in groups†. Virus Evol. 2019, 5. [Google Scholar] [CrossRef]
- Benton, W.H.; Hurst, C.J. Evaluation of mixed cell types and 5-iodo-2′-deoxyuridine treatment upon plaque assay titers of human enteric viruses. Appl. Environ. Microbiol. 1986, 51, 1036–1040. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, S.H.; Bae, S.Y.; Lim, C.S.; Kim, Y.K.; Lee, K.N.; Lee, C.K. Enhanced detection of respiratory viruses using cryopreserved R-Mix ReadyCells. J. Clin. Virol. 2008, 42, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Wiik-Nielsen, C.R.; Løvoll, M.; Sandlund, N.; Faller, R.; Wiik-Nielsen, J.; Bang Jensen, B. First detection of piscine reovirus (PRV) in marine fish species. Dis. Aquat. Organ. 2012, 97, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Finstad, Ø.W.; Dahle, M.K.; Lindholm, T.H.; Nyman, I.B.; Løvoll, M.; Wallace, C.; Olsen, C.M.; Storset, A.K.; Rimstad, E. Piscine orthoreovirus (PRV) infects Atlantic salmon erythrocytes. Vet. Res. 2014, 45. [Google Scholar] [CrossRef]
- Dahle, M.K.; Wessel, Ø.; Timmerhaus, G.; Nyman, I.B.; Jørgensen, S.M.; Rimstad, E.; Krasnov, A. Transcriptome analyses of Atlantic salmon (Salmo salar L.) erythrocytes infected with piscine orthoreovirus (PRV). Fish Shellfish Immunol. 2015, 45, 780–790. [Google Scholar] [CrossRef]
- Nombela, I.; Carrion, A.; Puente-Marin, S.; Chico, V.; Mercado, L.; Perez, L.; Coll, J.; del Ortega-Villaizan, M.M. Piscine birnavirus triggers antiviral immune response in trout red blood cells, despite not being infective. F1000Research 2017, 6, 1968. [Google Scholar] [CrossRef]
- Garver, K.A.; Johnson, S.C.; Polinski, M.P.; Bradshaw, J.C.; Marty, G.D.; Snyman, H.N.; Morrison, D.B.; Richard, J. Piscine Orthoreovirus from Western North America Is Transmissible to Atlantic Salmon and Sockeye Salmon but Fails to Cause Heart and Skeletal Muscle Inflammation. PLoS ONE 2016, 11, e0146229. [Google Scholar] [CrossRef]
- Palacios, G.; Lovoll, M.; Tengs, T.; Hornig, M.; Hutchison, S.; Hui, J.; Kongtorp, R.-T.; Savji, N.; Bussetti, A.V.; Solovyov, A.; et al. Heart and Skeletal Muscle Inflammation of Farmed Salmon Is Associated with Infection with a Novel Reovirus. PLoS ONE 2010, 5, e11487. [Google Scholar] [CrossRef]
- Pham, P.H.; Tong, W.W.L.; Misk, E.; Jones, G.; Lumsden, J.S.; Bols, N.C. Atlantic salmon endothelial cells from the heart were more susceptible than fibroblasts from the bulbus arteriosus to four RNA viruses but protected from two viruses by dsRNA pretreatment. Fish Shellfish Immunol. 2017, 70, 214–227. [Google Scholar] [CrossRef]
| Cell Line Designation | Grouping Cell Lines Based on the Properties of Either I or II | |||||
|---|---|---|---|---|---|---|
| I. Sample from Which the Cell Line Was Developed | II. Cell Line Once Developed | |||||
| I.A. Species Properties | I.B. Anatomy | II.A. General Properties | ||||
| I.A1. Common Name | I.A2. Habitat | I.A3. Natural Geography | I.B1. Organ | II.A1. Cell Shape | II.A2. Availability | |
| ACBA | Arctic Char | Anadromous | Arctic | Bulbous Arteriosus | Fibroblast | Informal 1 |
| ASHe 3 | Atlantic Salmon | Anadromous | North Atlantic | Heart | Endothelial | Informal |
| ASCF 3 | Atlantic Salmon | Anadromous | North Atlantic | Caudal Fin | Fibroblast | Informal |
| ASimf20 | Atlantic Salmon | Anadromous | North Atlantic | Gut | Myofibroblast | Informal |
| ASP309 | Atlantic Salmon | Anadromous | North Atlantic | Pituitary | Epithelial | Informal |
| BAASf 3 | Atlantic Salmon | Anadromous | North Atlantic | Bulbous Arteriosus | Fibroblast | Informal |
| HK13 *,3 | Atlantic Salmon | Anadromous | North Atlantic | Kidney | Epithelial | Informal |
| CHSE-214 ** | Chinook Salmon | Anadromous | Pacific Northwest | Embryo | Epithelial | Curated 2 |
| RTG-2 ** | Rainbow Trout | Anadromous | Pacific Northwest | Gonad | Fibroblast | Curated |
| RTgill-W1 | Rainbow Trout | Anadromous | Pacific Northwest | Gill | Epithelial | Curated |
| RTH-149 | Rainbow Trout | Anadromous | Pacific Northwest | Hepatoma | Epithelial | Curated |
| RTL-W1 | Rainbow Trout | Anadromous | Pacific Northwest | Liver | Epithelial | Curated |
| RTgutGC | Rainbow Trout | Anadromous | Pacific Northwest | Intestine | Epithelial | Informal |
| RTHDF | Rainbow Trout | Anadromous | Pacific Northwest | Skin | Fibroblast | Informal |
| RTS11 | Rainbow Trout | Anadromous | Pacific Northwest | Spleen | Macrophage | Informal |
| STB5 Gill 3 | Threespined Stickleback | Anadromous | Pacific Northwest | Gill | Epithelial | Informal |
| STB6 Gill 3 | Threespined Stickleback | Anadromous | Pacific Northwest | Gill | Epithelial | Informal |
| STBH 3 | Threespined Stickleback | Anadromous | Pacific Northwest | Heart | Epithelial | Informal |
| PHL | Pacific Herring | Marine | North Pacific | Larvae | Epithelial | Curated |
| EPC ** | Fathead Minnow | Freshwater | Great Lakes Basin | Skin | Epithelial | Curated |
| GarL | Gar | Freshwater | Great Lakes Basin | Liver | Epithelial | Informal |
| SB3 | Lake Sturgeon | Freshwater | Great Lakes Basin | Brain | Epithelial | Informal |
| WE-Cfin11f | Walleye | Freshwater | Great Lakes Basin | Caudal Fin | Fibroblast | Informal |
| WE-Skin11f | Walleye | Freshwater | Great Lakes Basin | Skin | Fibroblast | Informal |
| WEBA | Walleye | Freshwater | Great Lakes Basin | Bulbous Arteriosus | Endothelial | Informal |
| YFP5 | Yellow Perch | Freshwater | Great Lakes Basin | Fin | Fibroblasts | Informal |
| EelB | American Eel | Catadromous | North Atlantic | Brain | Endothelial | Informal |
| PBLE | American Eel | Catadromous | North Atlantic | Blood | Epithelial | Informal |
| HEW | Haddock | Marine | North Atlantic | Embryo | Epithelial | Informal |
| ZEB2J | Zebrafish | Freshwater | South Asia | Embryo | Epithelial | Informal |
| ZSSJ | Zebrafish | Freshwater | South Asia | Spleen | Epithelial | Informal |
| PRV-1 Ct * in Cultures at Four Time Points (Days) after Exposure | |||||
|---|---|---|---|---|---|
| Species | Cells Lines | 7 | 14 | 21 | 28 |
| Atlantic salmon | ASP309 | 32.9 | Negative | Negative | Negative |
| ASCF | 32.5 | Negative | Negative | Negative | |
| ASimf20 | 32.6 | Negative | Negative | 33.4 | |
| Fathead minnow | EPC | 32.1 | Negative | Negative | Negative |
| Haddock | HEW | 33.7 | Negative | Negative | Negative |
| Rainbow trout | RTgutGC | 27.3 | 29.1 | 35.0 | 35.0 |
| RTG-2 | 35.0 | 35.0 | 34.2 | 31.1 ** | |
| RTL-W1 | Negative | Negative | Negative | Negative | |
| RTHDF | Negative | 26.4 ** | Negative | Negative | |
| Walleye | WE-skin11f | 29.5 | Negative | Negative | Negative |
| WEBA | 30.6 | Negative | 34.6 | Negative | |
| Species | Cell Line | Virus Isolation Assays | Passage Number of Collected Samples (Initial–Final) | Cumulative Days Post-Inoculation (Initial–Final) | Ct Value (Initial–Final) | |
|---|---|---|---|---|---|---|
| Canada PRV | Norway PRV | |||||
| American Eel | EelB | Single cell culture passage | 1 | 2–14 | 30.7–30.9 | 32.6–32.7 |
| Sub-cultivating only exposed cells | 1–2 | 22–43 | 27.6–28.9 | 27.2–30.1 | ||
| PBLE | Single cell culture passage | 1 | 2–14 | 30.5–29.4 * | 33.7–29.2 * | |
| Sub-cultivating only exposed cells | 1–2 | 22–43 | 27.4–27.2 | 26.9–29.4 | ||
| Arctic Charr | ACBA | Supernatant transfer | 1–2 | 14–28 | 25.2–34.2 | 27.6 *–N |
| Sub-cultivating only exposed cells | 1–2 | 22–43 | 28.2–29.1 | 26.4–30.4 | ||
| Atlantic Salmon | ASCF | Single cell culture passage | 1 | 2–14 | 29.9–29.4 | 31.9–32.4 |
| Sub-cultivating only exposed cells | 1–2 | 15–44 | 25.7–24.8 | 27.8–31.5 | ||
| ASHe | Single cell culture passage | 1 | 2–14 | 30.8–30.9 | 33.5–32.1 | |
| Sub-cultivating with fresh cells | 1–2 | 14–28 | 31.4–31.3 | No data | ||
| Supernatant transfer | 1–2 | 26–40 | 31.4–N | 35.5 *–37.4 * | ||
| Cell lysate transfer | 1–2 | 14–28 | 22.2–25.4 ** | 25.7–27.1 | ||
| Sub-cultivating only exposed cells | 1–2 | 15–44 | 28.8–31.0 | 28.8–32.8 | ||
| ASimf20 | Single cell culture passage | 1 | 2–14 | 28.4–28.4 | 32.1–32.2 | |
| Supernatant transfer | 1–2 | 26–40 | 25.0–37.1 * | 27.2–N | ||
| Cell lysate transfer | 1–2 | 14–28 | 20.9–25.3 | 25.5–27.1 | ||
| Sub-cultivating only exposed cells | 1–2 | 15–43 | 25.8–27.5 | 25.2–31.2 | ||
| ASP309 | Supernatant transfer | 1–2 | 26–40 | 24.4–31.2 | 28.2–34.9 | |
| BAASf | Single cell culture passage | 1 | 2–14 | 28.7–28.7 | 31.6–31.6 | |
| Sub-cultivating with fresh cells | 1–2 | 14–28 | 27.0–30.4 | No data | ||
| Cell lysate transfer | 1–2 | 14–28 | 22.1–26.7 | 25.1–27.6 * | ||
| Sub-cultivating only exposed cells | 1–2 | 15–44 | 27.6–29.8 | 27.2–31.4 | ||
| Chinook Salmon | CHSE-214 | Supernatant transfer | 1–2 | 14–28 | 27.3–33.2 | 30.5–N |
| Cell lysate transfer | 1–2 | 14–28 | 24.0–27.0 * | 24.9–N | ||
| Fathead Minnow | EPC | Supernatant transfer | 1–2 | 14–28 | 24.9–33.9 * | 29.2–33.9 |
| Cell lysate transfer | 1–2 | 14–28 | 25.5–28.2 | 27.1–31.9 | ||
| Gar | GarL | Single cell culture passage | 1 | 2–14 | 29.7 *–27.6 | 33.2 *–30.2 |
| Pacific herring | PHL | Sub-cultivating with fresh cells | 1–2 | 14–28 | 26.2–30.2 | No data |
| Rainbow Trout | RTgill-W1 | Supernatant transfer | 1–2 | 14–28 | 26.9–32.4 | 29.8–33.8 |
| Cell lysate transfer | 1–2 | 14–28 | 25.5–26.5 | 27.5–30.0 | ||
| Sub-cultivating only exposed cells | 1–2 | 14–44 | 26.7–28.1 | N–N | ||
| RTgutGC | Single cell culture passage | 1 | 2–14 | 30.8–28.3 | 31.0–32.0 | |
| Sub-cultivating with fresh cells | 1–2 | 14–28 | 26.0–N | No data | ||
| Cell lysate transfer | 1–2 | 14–28 | 24.6–28.1 | 26.9 *–30.2 | ||
| Sub-cultivating only exposed cells | 1–2 | 14–44 | 27.4–29.2 | 28.5–33.2 | ||
| RTHDF | Single cell culture passage | 1 | 2–14 | 31.7–30.2 | 34.0–34.9 * | |
| Sub-cultivating with fresh cells | 1–2 | 14–28 | 31.6–31.3 | No data | ||
| Supernatant transfer | 1–2 | 14–28 | 26.3 *–33.8 | 29.2–33.1 | ||
| Cell lysate transfer | 1–2 | 14–28 | 22.9–26.6 | 26.5–28.7 | ||
| Sub-cultivating only exposed cells | 1–2 | 14–44 | 28.1–29.4 | 27.0–31.6 | ||
| RTH-149 | Single cell culture passage | 1 | 2–14 | 29.5–26.6 | 33.0 *–N | |
| Sub-cultivating only exposed cells | 1–2 | 15–43 | 24.7–24.7 | 28.9–32.4 | ||
| RTL-W1 | Supernatant transfer | 1–2 | 14–28 | 28.2 *–33.6 | 27.0–N | |
| Cell lysate transfer | 1–2 | 14–28 | 24.9–27.0 | 26.9–N | ||
| Sub-cultivating only exposed cells | 1–2 | 14–44 | 28.0–28.5 | 28.7–33.5 | ||
| RTS11 | Cell lysate transfer | 1–2 | 14–28 | 22.6–27.0 | 26.9–28.8 | |
| Sub-cultivating only exposed cells | 1–2 | 15–43 | 22.6–23.8 | 25.8–26.0 | ||
| Stickleback (Threespined) | STB5 Gill | Single cell culture passage | 1 | 2–14 | 31.7–30.5 | 31.6–33.8 |
| Sub-cultivating with fresh cells | 1–2 | 14–28 | 27.1–32.7 | No data | ||
| STB6 Gill | Sub-cultivating with fresh cells | 1–2 | 14–28 | 23.8–28.3 | No data | |
| STBH | Sub-cultivating with fresh cells | 1–2 | 14–38 | 25.0–27.2 | No data | |
| Sturgeon | SB3 | Supernatant transfer | 1–2 | 14–28 | 24.4–32.7 * | 28.7–N |
| Walleye | WE-cfin11f | Sub-cultivating only exposed cells | 1–2 | 16–43 | 23.7–25.6 | 24.9–29.4 |
| Yellow Perch | YPF5 | Sub-cultivating only exposed cells | 1–2 | 22–43 | 24.3–28.6 | 28.7–31.2 |
| Zebrafish | ZEB2J | Single cell culture passage | 1 | 2–14 | 27.7 *–27.6 | 31.0–32.2 |
| Sub-cultivating only exposed cells | 1–2 | 16–43 | 26.1–28.4 | 27.1–30.8 | ||
| ZSSJ | Sub-cultivating with fresh cells | 1–2 | 14–28 | 26.2–30.1 | No data | |
| Cell Culture | Ct 1 after Exposures for | |||
|---|---|---|---|---|
| Cell Line Designation | Species (Common Name) | Morphology | 2 Days | 14 Days |
| EelB | American eel | endothelial | 27.7 | 28.5 |
| PBLE | American eel | fibroblast | 24.8 | 25.7 |
| ASCF | Atlantic salmon | fibroblast | 26.8 | 26.3 |
| ASHe | Atlantic salmon | endothelial | 27.7 | 29.4 |
| ASimf20 | Atlantic salmon | myofibroblast | 27.5 | 27.7 |
| BAASf | Atlantic salmon | fibroblast | 27.6 | 27.3 |
| RTgill-W1 | Rainbow trout | epithelial | 29.5 | 30.4 |
| Cell Cultures | Ct 3 in Cultures at 2 Times after Initiation of Exposure | |||
|---|---|---|---|---|
| Cell Line | RBC Homogenates | Co-Infected Virus | Passage 1 (13 to 18 Days 4) | Passage 2 [26 to 32 Cumulative Days 5 (12 to 14 Days after Passaging)] |
| EPC | Canada | CSV | 22.4 * | 32.0 * |
| Norway | CSV | 24.5 | 33.7 | |
| Canada | IPNV | 26.8 | 32.4 | |
| Norway | IPNV | 29.9 | Negative | |
| ASHe | Canada | IPNV | 25.3 | 27.8 |
| Norway | IPNV | 25.2 | 27.5 | |
| BAASf | Canada | IPNV | 26.7 | 29.0 |
| Norway | IPNV | 26.6 | 30.0 | |
| CHSE-214 | Canada | CSV | 21.5 | 30.4 |
| Norway | CSV | 23.4 | 30.2 | |
| Canada | IPNV | 25.1 | 31.8 | |
| Norway | IPNV | 26.1 | 30.2 | |
| RTHDF | Canada | CSV | 26.8 | 32.2 |
| Norway | CSV | 29.3 | Negative | |
| Canada | IPNV | 27.9 | 32.6 | |
| Norway | IPNV | 28.0 | 32.8 | |
| RTgill-W1 | Canada | CSV | 24.7 | 28.8 |
| Norway | CSV | 28.0 | 33.1 | |
| Canada | IPNV | 25.0 | 30.7 | |
| Norway | IPNV | 26.9 | 28.6 | |
| RTL-W1 | Canada | CSV | 24.1 | 31.6 |
| Norway | CSV | 28.5 | Negative | |
| RTS11 | Canada | CSV | Negative | 32.8 ** |
| Norway | CSV | 25.8 ** | 33.6 ** | |
| Canada | IPNV | 23.9 | 30.4 ** | |
| Norway | IPNV | 24.9 | 30.4 ** | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, P.H.; Misk, E.; Papazotos, F.; Jones, G.; Polinski, M.P.; Contador, E.; Russell, S.; Garver, K.A.; Lumsden, J.S.; Bols, N.C. Screening of Fish Cell Lines for Piscine Orthoreovirus-1 (PRV-1) Amplification: Identification of the Non-Supportive PRV-1 Invitrome. Pathogens 2020, 9, 833. https://doi.org/10.3390/pathogens9100833
Pham PH, Misk E, Papazotos F, Jones G, Polinski MP, Contador E, Russell S, Garver KA, Lumsden JS, Bols NC. Screening of Fish Cell Lines for Piscine Orthoreovirus-1 (PRV-1) Amplification: Identification of the Non-Supportive PRV-1 Invitrome. Pathogens. 2020; 9(10):833. https://doi.org/10.3390/pathogens9100833
Chicago/Turabian StylePham, Phuc H., Ehab Misk, Fotini Papazotos, Ginny Jones, Mark P. Polinski, Elena Contador, Spencer Russell, Kyle A. Garver, John S. Lumsden, and Niels C. Bols. 2020. "Screening of Fish Cell Lines for Piscine Orthoreovirus-1 (PRV-1) Amplification: Identification of the Non-Supportive PRV-1 Invitrome" Pathogens 9, no. 10: 833. https://doi.org/10.3390/pathogens9100833
APA StylePham, P. H., Misk, E., Papazotos, F., Jones, G., Polinski, M. P., Contador, E., Russell, S., Garver, K. A., Lumsden, J. S., & Bols, N. C. (2020). Screening of Fish Cell Lines for Piscine Orthoreovirus-1 (PRV-1) Amplification: Identification of the Non-Supportive PRV-1 Invitrome. Pathogens, 9(10), 833. https://doi.org/10.3390/pathogens9100833





