New Species of the Genus Curvularia: C. tamilnaduensis and C. coimbatorensis from Fungal Keratitis Cases in South India
Abstract
1. Introduction
2. Results
2.1. Strain Selection and Case Details
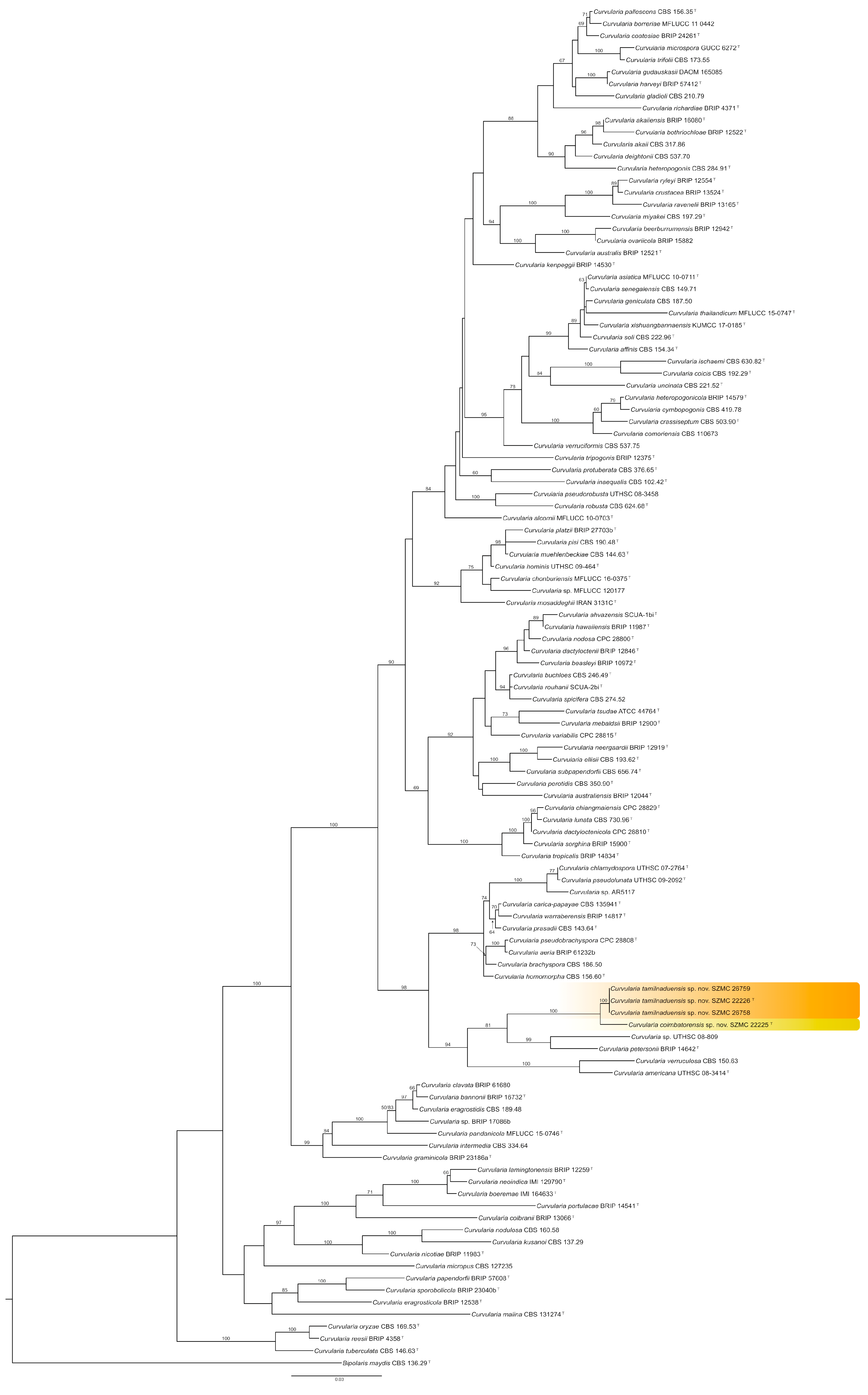
2.2. Updated Phylogeny of the Genus Curvularia
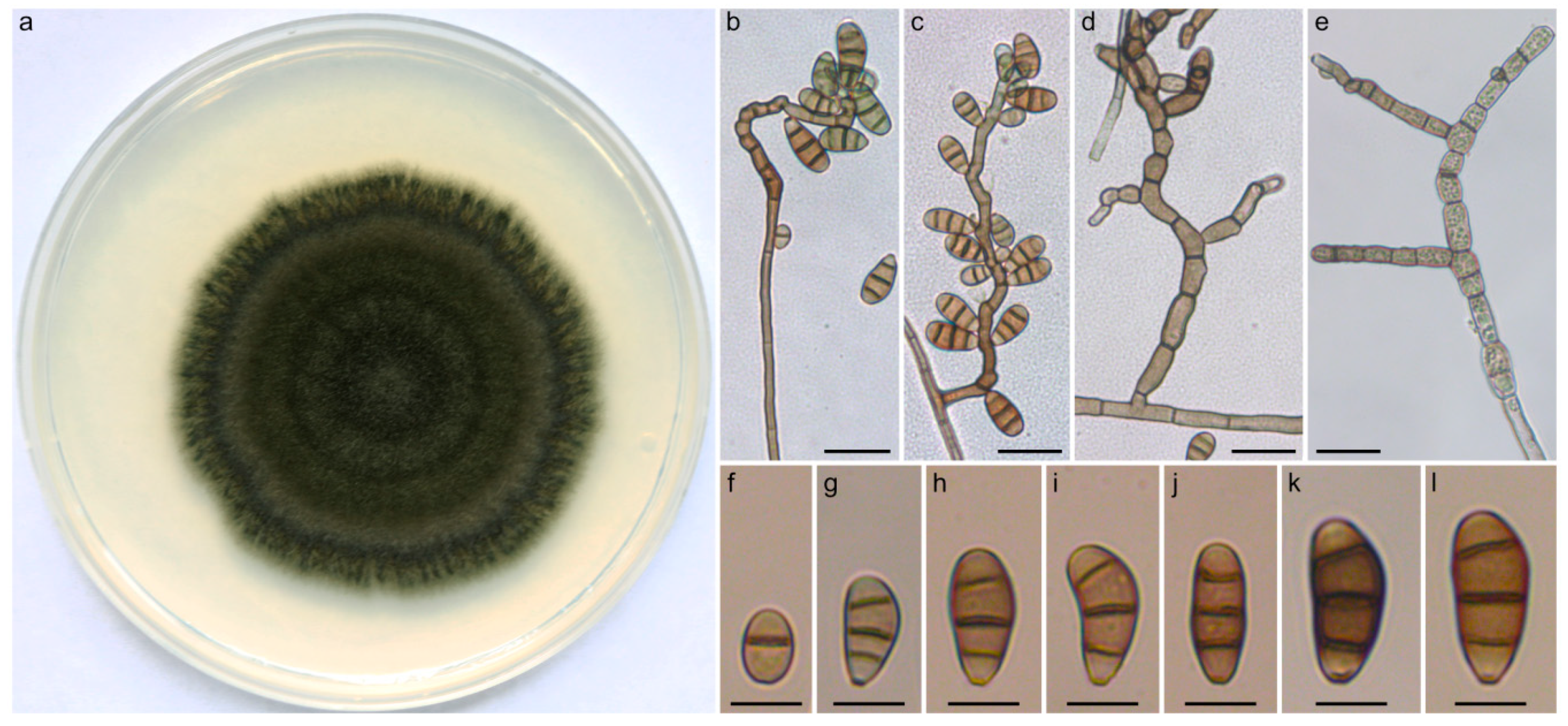
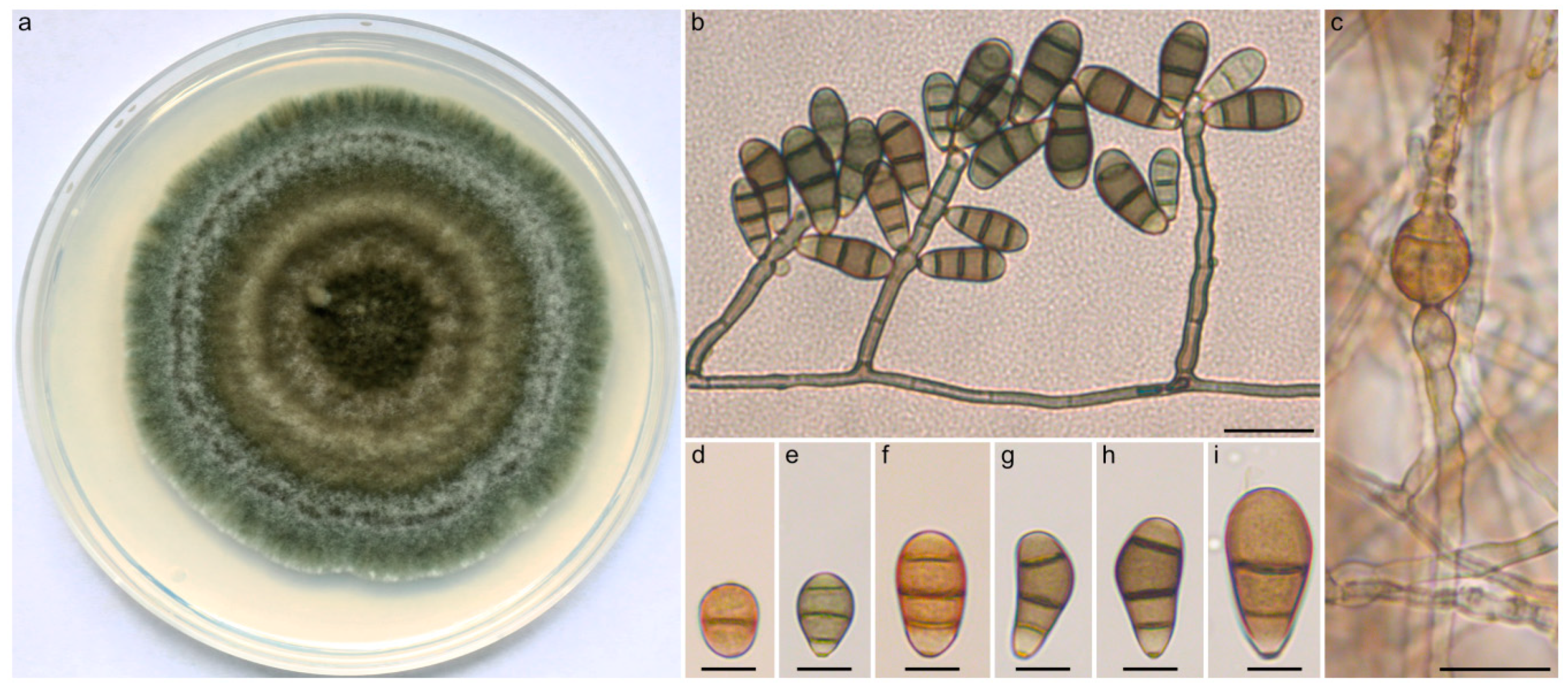
2.3. Taxonomy and Related Information
2.4. Antifungal Susceptibilities of Curvularia Strains Isolated from Fungal Keratitis
3. Discussion
4. Materials and Methods
4.1. Curvularia Strains, Culture Conditions, and Morphological Examination
4.2. DNA Extraction, Amplification, Sequencing, and Phylogenetic Analysis
4.3. Antifungal Susceptibility Testing
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bengyella, L.; Iftikhar, S.; Nawaz, K.; Fonmboh, D.J.; Yekwa, E.L.; Jones, R.C.; Njanu, Y.M.T.; Roy, P. Biotechnological application of endophytic filamentous Bipolaris and Curvularia: A review on bioeconomy impact. World J. Microbiol. Biotechnol. 2019, 35, 69. [Google Scholar] [CrossRef] [PubMed]
- Kusai, N.A.; Azmi, M.M.Z.; Zulkifly, S.; Yusof, M.T.; Zainudin, N.A.I.M. Morphological and molecular characterization of Curvularia and related species associated with leaf spot disease of rice in Peninsular Malaysia. Rend. Lincei Sci. Fis. Nat. 2016, 27, 205–214. [Google Scholar] [CrossRef]
- Krizsán, K.; Papp, T.; Manikandan, P.; Shobana, C.S.; Chandrasekaran, M.; Vágvölgyi, C.; Kredics, L. Clinical Importance of the Genus Curvularia. In Medical Mycology: Current Trends and Future Prospects; Razzaghi-Abyaneh, M., Shams-Ghahfarokhi, M., Rai, M., Eds.; CRC Press: Boca Raton, FL, USA, 2016; pp. 147–204. [Google Scholar] [CrossRef]
- Yanagihara, M.; Kawasaki, M.; Ishizaki, H.; Anzaw, K.; Udagawa, S.; Mochizuki, T.; Sato, Y.; Tachikawa, N.; Hanakawa, H. Tiny keratotic brown lesions on the interdigital web between the toes of a healthy man caused by Curvularia species infection and a review of cutaneous Curvularia infections. Mycoscience 2010, 51, 224–233. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Cai, L.; McKenzie, E.H.C.; Crous, P.W.; Madrid, H.; Chukeatirote, E.; Shivas, R.G.; Tan, Y.P.; Hyde, K.D. A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Divers. 2012, 56, 131–144. [Google Scholar] [CrossRef]
- Paredes, K.; Capilla, J.; Sutton, D.A.; Mayayo, E.; Fothergill, A.W.; Guarro, J. Virulence of Curvularia in a murine model. Mycoses 2013, 56, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Madrid, H.; da Cunha, K.C.; Gené, J.; Dijksterhuis, J.; Cano, J.; Sutton, D.A.; Guarro, J.; Crous, P.W. Novel Curvularia species from clinical specimens. Persoonia 2014, 33, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.P.; Crous, P.W.; Shivas, R.G. Cryptic species of Curvularia in the culture collection of the Queensland Plant Pathology Herbarium. MycoKeys 2018, 35, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Marcus, L.; Vismer, H.F.; van der Hoven, H.J.; Gove, E.; Meewes, P. Mycotic keratitis caused by Curvularia brachyspora (Boedjin). A report of the first case. Mycopathologia 1992, 119, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Kwon-Chung, K.J.; Kleiner, D.E.; Geber, A.; Lawson, W.; Pass, H.I.; Henderson, D. Unusual aspects of allergic bronchopulmonary fungal disease: Report of two cases due to Curvularia organisms associated with allergic fungal sinusitis. Hum. Pathol. 1991, 22, 1240–1248. [Google Scholar] [CrossRef]
- Guarro, J.; Akiti, T.; Horta, R.A.; Morizot Leite-Filho, L.A.; Gené, J.; Ferreira-Gomes, S.; Aguilar, C.; Ortoneda, M. Mycotic keratitis due to Curvularia senegalensis and in vitro antifungal susceptibilities of Curvularia spp. J. Clin. Microbiol. 1999, 37, 4170–4173. [Google Scholar] [PubMed]
- Fan, Y.M.; Huang, W.M.; Li, S.F.; Wu, G.F.; Li, W.; Chen, R.Y. Cutaneous phaeohyphomycosis of foot caused by Curvularia clavata. Mycoses 2009, 52, 544–546. [Google Scholar] [CrossRef] [PubMed]
- Vasikasin, V.; Nasomsong, W.; Srisuttiyakorn, C.; Mitthamsiri, W.; Oer-Areemitr, N.; Changpradub, D. Disseminated phaeohyphomycosis caused by Curvularia tuberculata in a previously healthy man. Mycopathologia 2019, 184, 321–325. [Google Scholar] [CrossRef]
- Pimentel, J.D.; Mahadevan, K.; Woodgyer, A.; Sigler, L.; Gibas, C.; Harris, O.C.; Lupino, M.; Athan, E. Peritonitis due to Curvularia inaequalis in an elderly patient undergoing peritoneal dialysis and a review of six cases of peritonitis associated with other Curvularia spp. J. Clin. Microbiol. 2005, 43, 4288–4292. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Scarano, E.; La Sorda, M.; Torelli, R.; De Corso, E.; Mulé, A.; Paludetti, G.; Fadda, G.; Sanguinetti, M. Eosinophilic fungal rhinosinusitis due to the unusual pathogen Curvularia inaequalis. Mycoses 2010, 53, 84–88. [Google Scholar] [CrossRef]
- Cruz, R.; Barthel, E.; Espinoza, J. Allergic rhinosinusitis by Curvularia inaequalis (Shear) Boedijn. Rev. Chil. Infectol. 2013, 30, 319–322. (In Spanish) [Google Scholar] [CrossRef] [PubMed]
- Flanagan, K.L.; Bryceson, A.D. Disseminated infection due to Bipolaris australiensis in a young immunocompetent man: Case report and review. Clin. Infect. Dis. 1997, 25, 311–313. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Filizzola, M.J.; Martinez, F.; Rauf, S.J. Phaeohyphomycosis of the central nervous system in immunocompetent hosts: Report of a case and review of the literature. Int. J. Infect. Dis. 2003, 7, 282–286. [Google Scholar] [CrossRef][Green Version]
- Gadgil, N.; Kupferman, M.; Smitherman, S.; Fuller, G.N.; Rao, G. Curvularia brain abscess. J. Clin. Neurosci. 2013, 20, 173–175. [Google Scholar] [CrossRef]
- Vachharajani, T.J.; Zaman, F.; Latif, S.; Penn, R.; Abreo, K.D. Curvularia geniculata fungal peritonitis: A case report with review of literature. Int. Urol. Nephrol. 2005, 37, 781–784. [Google Scholar] [CrossRef]
- Diskin, C.J.; Stokes, T.J.; Dansby, L.M.; Radcliff, L.; Carter, T.B. Case report and review: Is the tendency for Curvularia tubular obstruction significant in pathogenesis? Perit. Dial. Int. 2008, 28, 678–679. [Google Scholar]
- Saenz, R.E.; Brown, W.D.; Sanders, C.V. Allergic bronchopulmonary disease caused by Bipolaris hawaiiensis presenting as a necrotizing pneumonia: Case report and review of literature. Am. J. Med. Sci. 2001, 321, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Robson, A.M.; Craver, R.D. Curvularia urinary tract infection: A case report. Pediatr. Nephrol. 1994, 8, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Safdar, A. Curvularia—Favorable response to oral itraconazole therapy in two patients with locally invasive phaeohyphomycosis. Clin. Microbiol. Infect. 2003, 9, 1219–1223. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fernandez, M.; Noyola, D.E.; Rossmann, S.N.; Edwards, M.S. Cutaneous phaeohyphomycosis caused by Curvularia lunata and a review of Curvularia infections in pediatrics. Pediatr. Infect. Dis. J. 1999, 18, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Dave, V.P.; Joseph, J.; Pathengay, A.; Pappuru, R.R.; Das, T. Clinical presentations, diagnosis, and management outcomes of Curvularia endophthalmitis and a review of literature. Retina 2018. [Google Scholar] [CrossRef] [PubMed]
- Tibpromma, S.; Hyde, K.D.; Bhat, J.D.; Mortimer, P.E.; Xu, J.; Promputtha, I.; Doilom, M.; Yang, J.B.; Tang, A.M.C.; Karunarathna, S.C. Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 2018, 33, 25–67. [Google Scholar] [CrossRef] [PubMed]
- Manamgoda, D.S.; Rossman, A.Y.; Castlebury, L.A.; Crous, P.W.; Madrid, H.; Chukeatirote, E.; Hyde, K.D. The genus Bipolaris. Stud. Mycol. 2014, 79, 221–288. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Senwanna, C.; Cheewangkoon, R.; Crous, P.W. New species and records of Bipolaris and Curvularia from Thailand. Mycosphere 2017, 8, 1556–1574. [Google Scholar] [CrossRef]
- Sivanesan, A. Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. Mycol. Pap. 1987, 158, 1–261. [Google Scholar]
- Mythili, A.; Babu Singh, Y.R.; Priya, R.; Shafeeq Hassan, A.; Manikandan, P.; Panneerselvam, K.; Narendran, V.; Shobana, C.S. In vitro and comparative study on the extracellular enzyme activity of molds isolated from keratomycosis and soil. Int. J. Ophthalmol. 2014, 7, 778–784. [Google Scholar] [CrossRef]
- Schoch, C.L.; Crous, P.W.; Groenewald, J.Z.; Boehm, E.W.A.; Burgess, T.I.; de Gruyter, J.; de Hoog, G.S.; Dixon, L.J.; Grube, M.; Gueidan, C.; et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009, 64, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Berbee, M.; Pirseyedi, M.; Hubbard, S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, J.W., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Löytynoja, A. Phylogeny-aware alignment with PRANK. Meth. Mol. Biol. 2014, 1079, 155–170. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multigene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Simmons, M.P.; Ochoterena, H. Gaps as characters in sequence-based phylogenetic analysis. Syst. Biol. 2000, 49, 369–381. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Lewis, P.O. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 2001, 50, 913–925. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 3rd ed.; Approved Standard, CLSI Document M27-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
| Strain | Age | Sex | Clinical Diagnosis | Corneal Scraping | Therapy | Outcome |
|---|---|---|---|---|---|---|
| SZMC 22225 | 80 | Male | Fungal corneal ulcer | 11 July 2012 | NAT, ECZ, HTR | Lost to follow up after two visits |
| SZMC 22226 | 66 | Male | Fungal corneal ulcer | 2 March 2013 | NAT, ECZ, HTR | Lost to follow up after one visit |
| SZMC 26758 | 40 | Male | Fungal corneal ulcer | 21 March 2011 | NAT, ECZ, HTR | Lost to follow up after one visit |
| SZMC 26759 | NA | NA | Fungal corneal ulcer | NA | NA | Lost to follow up |
| Curvularia Species | Strain | GenBank Accession Number | ||
|---|---|---|---|---|
| ITS | tef1a | gpdh | ||
| Bipolaris maydis | CBS 136.29 T | KJ909780 | KM093794 | KM034846 |
| Curvularia aeria | BRIP 61232b | KX139029 | KU552155 | KU552162 |
| Curvularia affinis | CBS 154.34 T | KJ909782 | KM196566 | KM230401 |
| Curvularia ahvazensis | SCUA-1bi T | KJ415539 | MG428686 | MG428693 |
| Curvularia akaii | CBS 317.86 | JX256420 | KM196569 | KM230402 |
| Curvularia akaiiensis | BRIP 16080 T | HE861833 | KJ415453 | KJ415407 |
| Curvularia alcornii | MFLUCC 10-0703 T | JX256424 | JX266589 | JX276433 |
| Curvularia americana | UTHSC 08-3414 T | KJ415540 | - | HF565488 |
| Curvularia asiatica | MFLUCC 10-0711 T | KJ415541 | JX266593 | JX276436 |
| Curvularia australiensis | BRIP 12044 T | KJ415542 | KJ415452 | KJ415406 |
| Curvularia australis | BRIP 12521 T | MH414892 | KJ415451 | KJ415405 |
| Curvularia bannonii | BRIP 16732 T | MH414894 | KJ415450 | KJ415404 |
| Curvularia beasleyi | BRIP 10972 T | MH414911 | MH433654 | MH433638 |
| Curvularia beerburrumensis | BRIP 12942 T | KP400638 | MH433657 | MH433634 |
| Curvularia boeremae | IMI 164633 T | KJ415543 | - | MH433641 |
| Curvularia borreriae | MFLUCC 11-0422 | KJ922372 | KM196571 | KP419987 |
| Curvularia bothriochloae | BRIP 12522 T | KJ909765 | KJ415449 | KJ415403 |
| Curvularia brachyspora | CBS 186.50 | HG778984 | KM230405 | KM061784 |
| Curvularia buchloës | CBS 246.49 T | MF490814 | KM196588 | KM061789 |
| Curvularia carica-papayae | CBS 135941 T | HG779021 | - | HG779146 |
| Curvularia chiangmaiensis | CPC 28829 T | MH275055 | MF490857 | MF490836 |
| Curvularia chlamydospora | UTHSC 07-2764 T | KU552205 | - | HG779151 |
| Curvularia chonburiensis | MFLUCC 16-0375 T | MH414897 | - | MH412747 |
| Curvularia clavata | BRIP 61680b | AF081447 | KU552159 | KU552167 |
| Curvularia coatesiae | BRIP 24261 T | MH414898 | MH433659 | MH433636 |
| Curvularia coicis | CBS 192.29 T | LT631357 | JN601006 | AF081410 |
| Curvularia colbranii | BRIP 13066 T | LT631310 | MH433660 | MH433642 |
| Curvularia comoriensis | CBS 110673 | KJ415544 | - | LT715841 |
| Curvularia crassiseptum | CBS 503.90 T | HG778985 | - | LT715882 |
| Curvularia crustacea | BRIP 13524 T | MF490815 | KJ415448 | KJ415402 |
| Curvularia cymbopogonis | CBS 419.78 | KJ415545 | - | HG779129 |
| Curvularia dactyloctenicola | CPC 28810 T | LT631356 | MF490858 | MF490837 |
| Curvularia dactyloctenii | BRIP 12846 T | JN192375 | KJ415447 | KJ415401 |
| Curvularia deightonii | CBS 537.70 | MH414899 | - | LT715839 |
| Curvularia ellisii | CBS 193.62 T | HG778986 | JN601007 | JN600963 |
| Curvularia eragrosticola | BRIP 12538 T | KJ909781 | MH433661 | MH433643 |
| Curvularia eragrostidis | CBS 189.48 | HG778987 | - | HG779154 |
| Curvularia geniculata | CBS 187.50 | JN192376 | KM230410 | KM083609 |
| Curvularia gladioli | CBS 210.79 | KJ415546 | - | HG779123 |
| Curvularia graminicola | BRIP 23186a T | KJ415547 | JN601008 | JN600964 |
| Curvularia harveyi | BRIP 57412 T | KJ415548 | KJ415446 | KJ415400 |
| Curvularia hawaiiensis | BRIP 11987 T | KJ415549 | KJ415445 | KJ415399 |
| Curvularia heteropogonicola | BRIP 14579 T | HG779011 | KJ415444 | KJ415398 |
| Curvularia heteropogonis | CBS 284.91 T | JN192380 | JN601013 | JN600969 |
| Curvularia hominis | CBS 136985 T | KJ922375 | - | HG779106 |
| Curvularia homomorpha | CBS 156.60 T | HG778991 | JN601014 | JN600970 |
| Curvularia inaequalis | CBS 102.42 T | MH861533 | KM196574 | KM061787 |
| Curvularia intermedia | CBS 334.64 | MH414900 | - | HG779155 |
| Curvularia ischaemi | CBS 630.82 T | MH855025 | - | LT715790 |
| Curvularia kenpeggii | BRIP 14530 T | MH414901 | MH433662 | MH433644 |
| Curvularia kusanoi | CBS 137.29 | JX256429 | JN601016 | LT715862 |
| Curvularia lamingtonensis | BRIP 12259 T | JF812154 | MH433663 | MH433645 |
| Curvularia lunata | CBS 730.96 T | MH414902 | JX266596 | JX276441 |
| Curvularia malina | CBS 131274 T | HE792934 | KR493095 | KP153179 |
| Curvularia mebaldsii | BRIP 12900 T | MF139088 | MH433664 | MH433647 |
| Curvularia micropus | CBS 127235 | KJ909770 | - | LT715859 |
| Curvularia microspora | GUCC6272 T | MG846737 | MF139115 | MF139106 |
| Curvularia miyakei | CBS 197.29 T | KP400647 | KM196568 | KM083611 |
| Curvularia mosaddeghii | IRAN 3131C T | KJ415550 | MH392152 | MH392155 |
| Curvularia muehlenbeckiae | CBS 144.63 T | MH414910 | KM196578 | KP419996 |
| Curvularia neergaardii | BRIP 12919 T | KJ415551 | KJ415443 | KJ415397 |
| Curvularia neoindica | IMI 129790 T | MF490816 | MH433667 | MH433649 |
| Curvularia nicotiae | BRIP 11983 T | JN601033 | KJ415442 | KJ415396 |
| Curvularia nodosa | CPC 28800 T | KP400650 | MF490859 | MF490838 |
| Curvularia nodulosa | CBS 160.58 | JN192384 | JN601019 | JN600975 |
| Curvularia oryzae | CBS 169.53 T | KJ922380 | KM196590 | KP645344 |
| Curvularia ovariicola | CBS 470.90 T | MH275056 | JN601020 | JN600976 |
| Curvularia pallescens | CBS 156.35 T | KJ415552 | KM196570 | KM083606 |
| Curvularia pandanicola | MFLUCC 15-0746 T | HG778995 | MH412763 | MH412748 |
| Curvularia papendorfii | CBS 308.67 T | MH414905 | KJ415441 | KJ415395 |
| Curvularia perotidis | CBS 350.90 T | KY905678 | KM230407 | HG779138 |
| Curvularia petersonii | BRIP 14642 T | MH414906 | MH433668 | MH433650 |
| Curvularia pisi | CBS 190.48 T | KJ415553 | KY905697 | KY905690 |
| Curvularia platzii | BRIP 27703b T | KJ922373 | MH433669 | MH433651 |
| Curvularia portulacae | BRIP 14541 T | KJ922376 | KJ415440 | KJ415393 |
| Curvularia prasadii | CBS 143.64 T | MF490819 | KM230408 | KM061785 |
| Curvularia protuberata | CBS 376.65 T | HE861842 | KM196576 | KM083605 |
| Curvularia pseudobrachyspora | CPC 28808 T | HE861838 | MF490862 | MF490841 |
| Curvularia pseudolunata | UTHSC 09-2092 T | JN192386 | - | HF565459 |
| Curvularia pseudorobusta | UTHSC 08-3458 | MH414907 | - | HF565476 |
| Curvularia ravenelii | BRIP 13165 T | KJ415555 | JN601024 | JN600978 |
| Curvularia reesii | BRIP 4358 T | KJ909783 | MH433670 | MH433637 |
| Curvularia richardiae | BRIP 4371 T | KX139030 | KJ415438 | KJ415391 |
| Curvularia robusta | CBS 624.68 T | KJ415556 | KM196577 | KM083613 |
| Curvularia rouhanii | SCUA-2bi-2 T | HG779001 | MG428687 | MG428694 |
| Curvularia ryleyi | BRIP 12554 T | KY905679 | KJ415437 | KJ415390 |
| Curvularia senegalensis | CBS 149.71 | KJ415558 | - | HG779128 |
| Curvularia soli | CBS 222.96 T | MH414904 | KY905698 | KY905691 |
| Curvularia sorghina | BRIP 15900 T | KP400655 | KJ415435 | KJ415388 |
| Curvularia sp. | BRIP 17068b | KP400654 | MH433666 | MH433648 |
| Curvularia sp. | AR5117 | HE861826 | KP735698 | KP645349 |
| Curvularia sp. | MFLUCC 120177 | JN192387 | KP735697 | KP645348 |
| Curvularia sp. | UTHSC 8809 | MH414908 | - | HF565477 |
| Curvularia spicifera | CBS 274.52 | KJ909777 | JN601023 | JN600979 |
| Curvularia sporobolicola | BRIP 23040b T | MH275057 | MH433671 | MH433652 |
| Curvularia subpapendorfii | CBS 656.74 T | HG779023 | KM196585 | KM061791 |
| Curvularia thailandicum | MFLUCC 15-0747 T | JN192388 | MH412764 | MH412749 |
| Curvularia trifolii | CBS 173.55 | KJ415559 | - | HG779124 |
| Curvularia tripogonis | BRIP 12375 T | KC424596 | JN601025 | JN600980 |
| Curvularia tropicalis | BRIP 14834 T | JX256433 | KJ415434 | KJ415387 |
| Curvularia tsudae | ATCC 44764 T | HG779024 | KC503940 | KC747745 |
| Curvularia tuberculate | CBS 146.63 T | MF490822 | JX266599 | JX276445 |
| Curvularia uncinate | CBS 221.52 T | HG779026 | - | HG779134 |
| Curvularia variabilis | CPC 28815 T | KP400652 | MF490865 | MF490844 |
| Curvularia verruciformis | CBS 537.75 | MH414909 | - | HG779133 |
| Curvularia verruculosa | CBS 150.63 | MH275058 | KP735695 | KP645346 |
| Curvularia warraberensis | BRIP 14817 T | AF071338 | MH433672 | MH433653 |
| Curvularia xishuangbannaensis | KUMCC 17-0185 T | KJ909780 | MH412765 | MH412750 |
| Curvularia gudauskasii | DAOM 165085 | KX139029 | KM093794 | AF081393 |
| Curvularia tamilnaduensis sp. nov. | SZMC 22226 T * | MN628311 | MN628303 | MN628307 |
| SZMC 26758 * | MN628308 | MN628300 | MN628304 | |
| SZMC 26759 * | MN628309 | MN628301 | MN628305 | |
| Curvularia coimbatorensis sp. nov. | SZMC 22225 T * | MN628310 | MN628302 | MN628306 |
| Strain | Antifungal Agent | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMB | CLT | ECN | FLC | ITC | KTC | MCZ | NTM | TRB | |
| C. australiensis CBS 172.57 T | 0.25 | 0.25 | 0.125 | 16 | 0.03 | 0.25 | 0.25 | 2 | 0.25 |
| C. hawaiiensis CBS 173.57 T | 0.25 | 0.06 | 0.06 | 4 | 0.03 | 0.06 | 0.125 | 2 | 0.25 |
| C. spicifera CBS 274.52 T | 0.5 | 4 | 2 | >32 | 0.25 | 2 | 2 | 2 | 1 |
| C. coimbatorensis SZMC 22225 T | 0.5 | 0.5 | 1 | 32 | 0.25 | 0.25 | 1 | 2 | 1 |
| C. tamilnaduensis SZMC 22226 T | 1 | 0.125 | 0.125 | 8 | 0.03 | 0.06 | 0.25 | 2 | 0.25 |
| C. tamilnaduensis SZMC 26758 | 0.5 | 0.125 | 0.125 | 16 | 0.03 | 0.25 | 0.25 | 2 | 0.125 |
| C. tamilnaduensis SZMC 26759 | 1 | 0.125 | 0.125 | 16 | 0.25 | 0.25 | 0.25 | 2 | 0.25 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiss, N.; Homa, M.; Manikandan, P.; Mythili, A.; Krizsán, K.; Revathi, R.; Varga, M.; Papp, T.; Vágvölgyi, C.; Kredics, L.; et al. New Species of the Genus Curvularia: C. tamilnaduensis and C. coimbatorensis from Fungal Keratitis Cases in South India. Pathogens 2020, 9, 9. https://doi.org/10.3390/pathogens9010009
Kiss N, Homa M, Manikandan P, Mythili A, Krizsán K, Revathi R, Varga M, Papp T, Vágvölgyi C, Kredics L, et al. New Species of the Genus Curvularia: C. tamilnaduensis and C. coimbatorensis from Fungal Keratitis Cases in South India. Pathogens. 2020; 9(1):9. https://doi.org/10.3390/pathogens9010009
Chicago/Turabian StyleKiss, Noémi, Mónika Homa, Palanisamy Manikandan, Arumugam Mythili, Krisztina Krizsán, Rajaraman Revathi, Mónika Varga, Tamás Papp, Csaba Vágvölgyi, László Kredics, and et al. 2020. "New Species of the Genus Curvularia: C. tamilnaduensis and C. coimbatorensis from Fungal Keratitis Cases in South India" Pathogens 9, no. 1: 9. https://doi.org/10.3390/pathogens9010009
APA StyleKiss, N., Homa, M., Manikandan, P., Mythili, A., Krizsán, K., Revathi, R., Varga, M., Papp, T., Vágvölgyi, C., Kredics, L., & Kocsubé, S. (2020). New Species of the Genus Curvularia: C. tamilnaduensis and C. coimbatorensis from Fungal Keratitis Cases in South India. Pathogens, 9(1), 9. https://doi.org/10.3390/pathogens9010009







