CRISPR-cas3 of Salmonella Upregulates Bacterial Biofilm Formation and Virulence to Host Cells by Targeting Quorum-Sensing Systems
Abstract
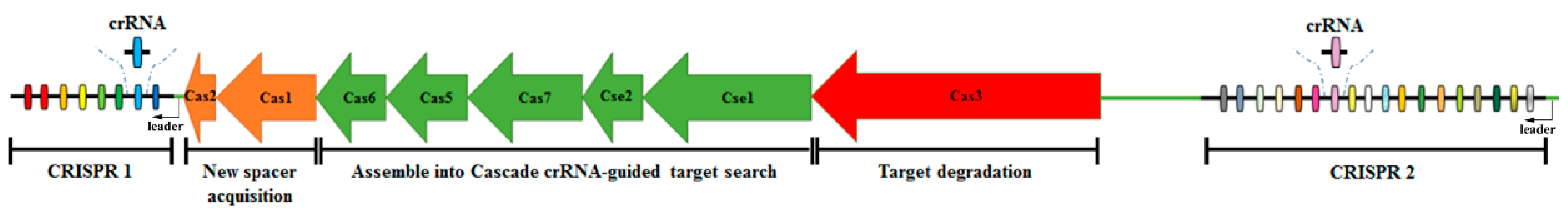
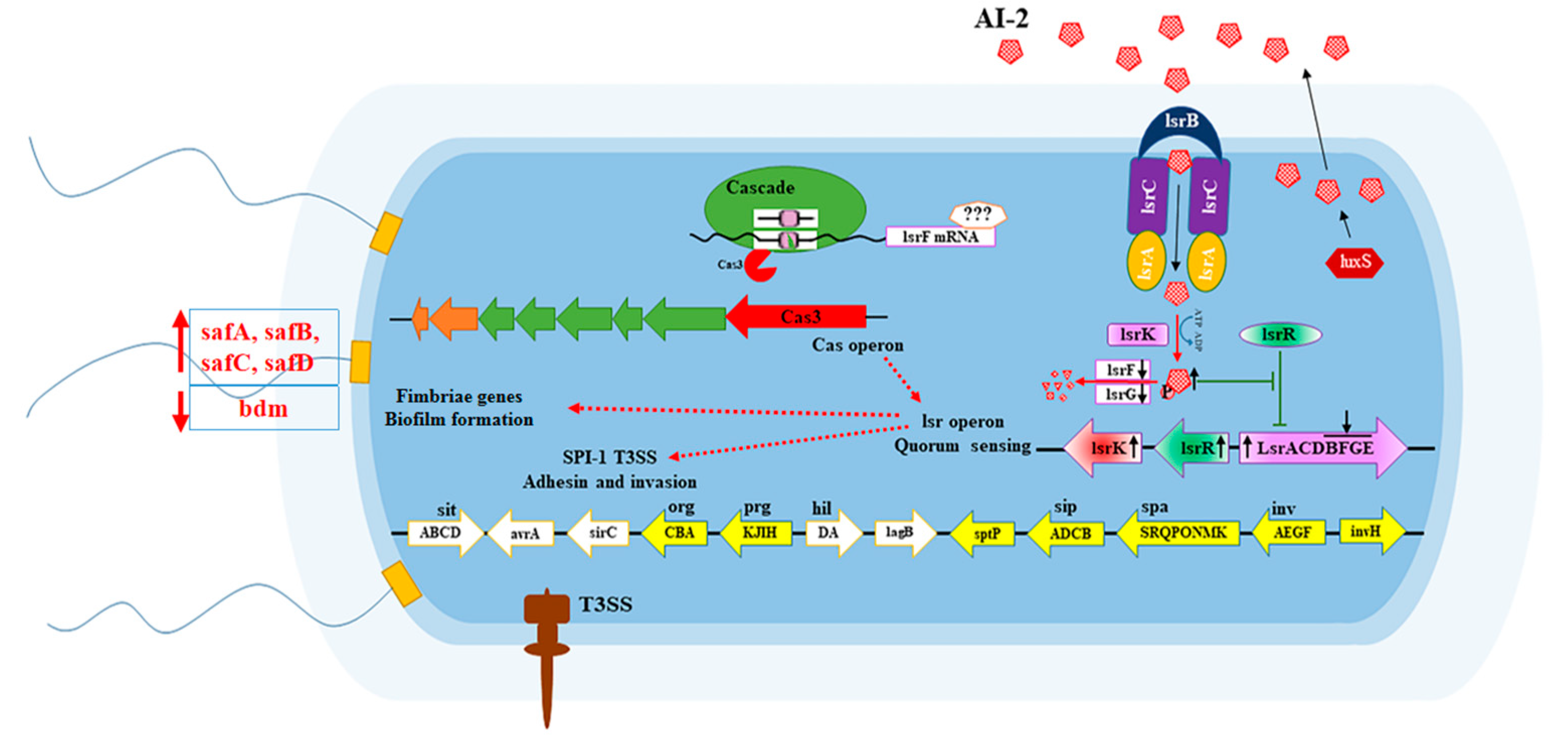
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Primers, and Growth Conditions
2.2. Construction of Cas3 Gene Deletion Strain Δcas3
2.3. Construction of Cas3 Gene Complementary Strain Δcas3/pBAD3-CM-cas3
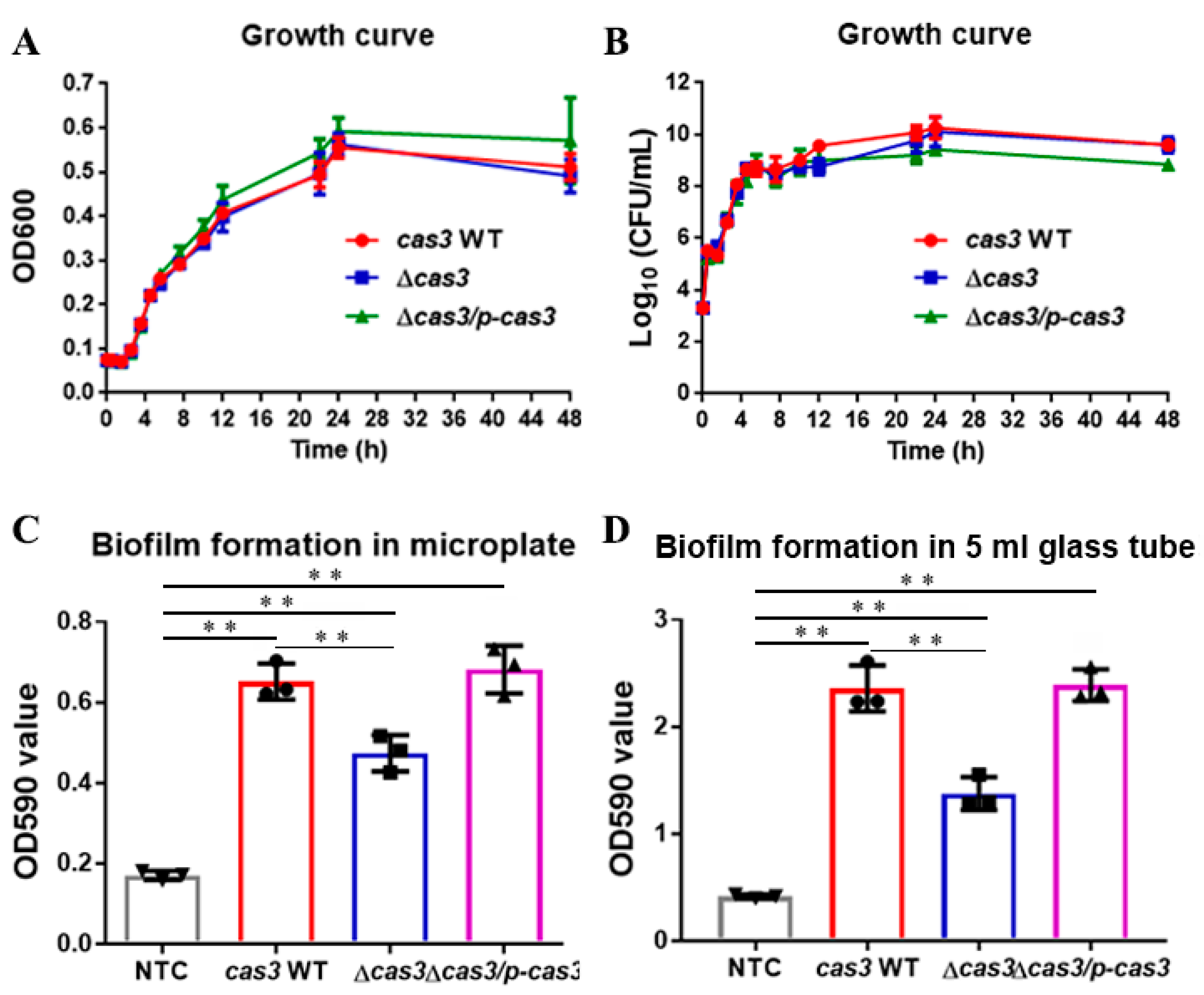
2.4. Determination of Standard Growth Curve
2.5. Biofilm Assays
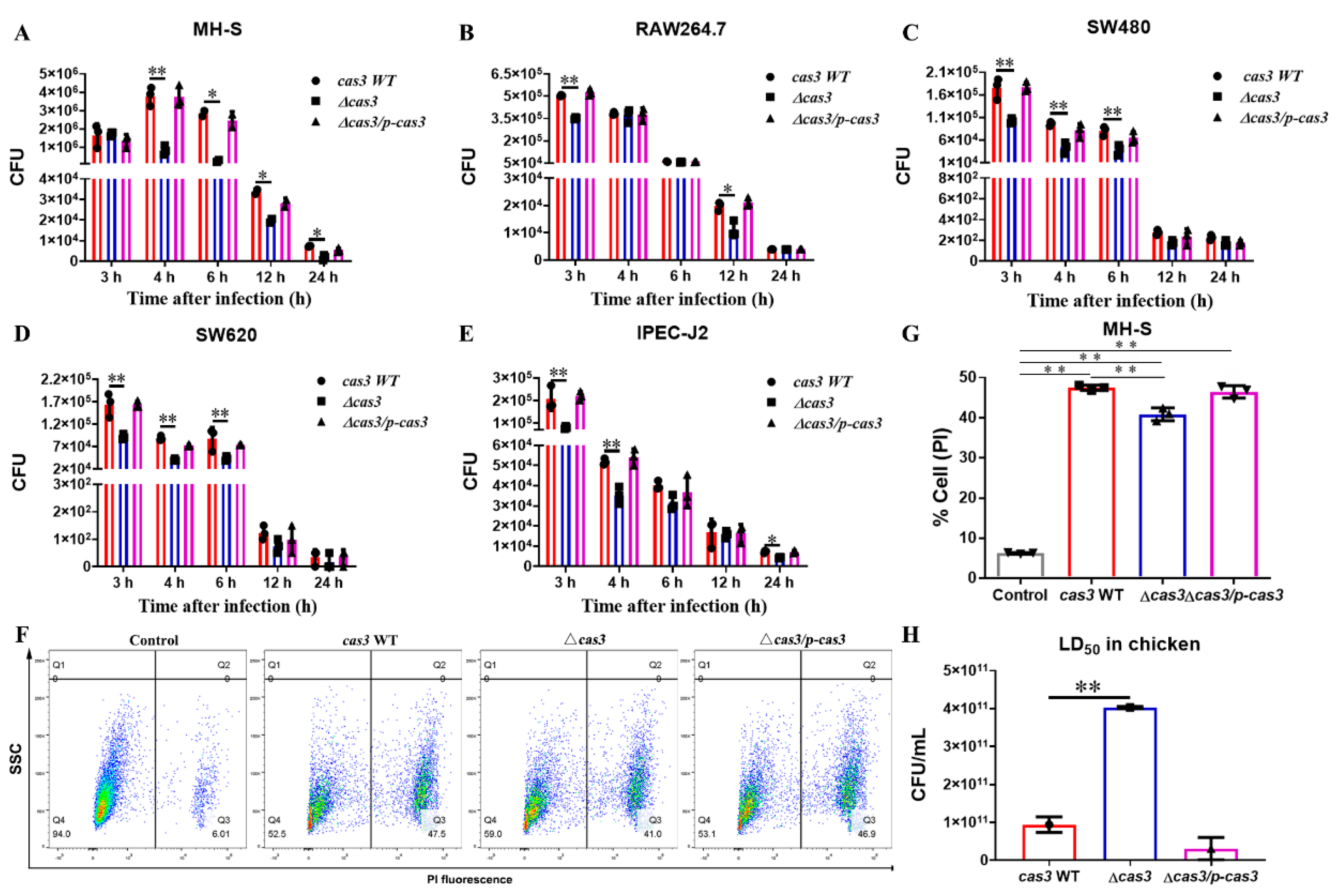
2.6. Cell Culture Infection Assays
2.7. Determination of the 50% Lethal Dose (LD50) for SPF Chicken during Salmonella Oral Infection
2.8. Transcriptome Analysis by RNA-Seq
2.8.1. RNA Isolation, Library Construction, Sequencing, and Sequence Data Filtering
2.8.2. Differentially Gene Expression (DEGs), Clustering Analysis, and Functional Analysis of DEGs
2.8.3. Data Validation by RT-qPCR
2.9. Statistical Analysis
3. Results
3.1. Deletion of Cas3 has No Effect on Bacterial Growth
3.2. Cas3 Deletion Impacts Bacterial Biofilm Formation and Cell Infection
3.3. Deletion of Cas3 Increased the LD50 Dose for SPF Chickens during Oral Infection
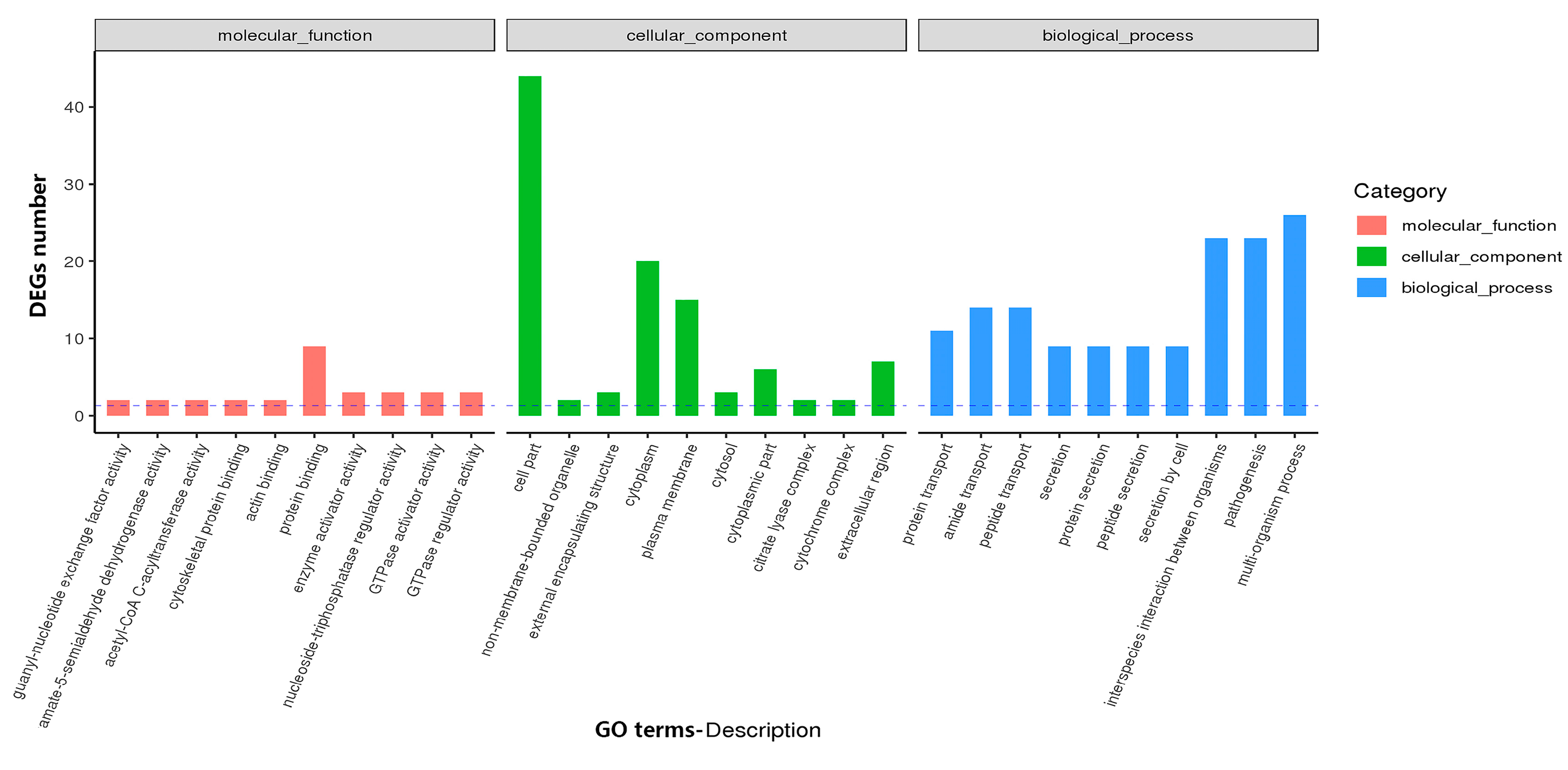
3.4. Transcriptomics Analysis Reveals Differentially Expressed Genes between Cas3 WT and Δcas3 Strains
3.4.1. GO and KEGG Analysis Identifies Functional Relevance to DEGs
3.4.2. The Patterns of DEGs are Similarly Revealed by RT-qPCR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fierer, J.; Guiney, D.G. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J. Clin. Investig. 2001, 107, 775–780. [Google Scholar] [CrossRef]
- Foley, S.L.; Lynne, A.M. Food animal-associated Salmonella challenges: Pathogenicity and antimicrobial resistance. J. Anim. Sci. 2008, 86, E173–E187. [Google Scholar] [CrossRef]
- Hohmann, E.L. Nontyphoidal salmonellosis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2001, 32, 263–269. [Google Scholar]
- Grissa, I.; Vergnaud, G.; Pourcel, C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 2007, 8, 172. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, K.; Yin, H.; Li, Q.; Wang, L.; Liu, Z. Detection of Salmonella and several common Salmonella serotypes in food by loop-mediated isothermal amplification method. Food Sci. Hum. Wellness 2015, 4, 75–79. [Google Scholar] [CrossRef]
- Thompson, C.P.; Doak, A.N.; Amirani, N.; Schroeder, E.A.; Wright, J.; Kariyawasam, S.; Lamendella, R.; Shariat, N.W. High-Resolution Identification of Multiple Salmonella Serovars in a Single Sample by Using CRISPR-SeroSeq. Appl. Environ. Microbiol. 2018, 84, e01859. [Google Scholar] [CrossRef] [PubMed]
- Rauch, H.E.; Vosik, D.; Kariyawasam, S.; M’Ikanatha, N.; Shariat, N.W. Prevalence of Group I Salmonella Kentucky in domestic food animals from Pennsylvania and overlap with human clinical CRISPR sequence types. Zoonoses Public Health 2018, 65, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, X.; Yin, K.; Hu, Y.; Xu, H.; Xie, X.; Xu, L.; Fei, X.; Chen, X.; Jiao, X. Genetic analysis and CRISPR typing of Salmonella enterica serovar Enteritidis from different sources revealed potential transmission from poultry and pig to human. Int. J. Food Microbiol. 2018, 266, 119–125. [Google Scholar] [CrossRef]
- Mohammed, M. Phage typing or CRISPR typing for epidemiological surveillance of Salmonella Typhimurium? BMC Res. Notes 2017, 10, 578. [Google Scholar] [CrossRef]
- Vosik, D.; Tewari, D.; Dettinger, L.; M’Ikanatha, N.M.; Shariat, N.W. CRISPR Typing and Antibiotic Resistance Correlates with Polyphyletic Distribution in Human Isolates of Salmonella Kentucky. Foodborne Pathog. Dis. 2018, 15, 101–108. [Google Scholar] [CrossRef]
- Almeida, F.; Medeiros, M.I.C.; Rodrigues, D.D.P.; Allard, M.W.; Falcao, J.P. Molecular characterization of Salmonella Typhimurium isolated in Brazil by CRISPR-MVLST. J. Microbiol. Methods 2017, 133, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Shariat, N.; Timme, R.E.; Pettengill, J.B.; Barrangou, R.; Dudley, E.G. Characterization and evolution of Salmonella CRISPR-Cas systems. Microbiology 2015, 161, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Hiley, L.; Octavia, S.; Tanaka, M.M.; Sintchenko, V.; Lan, R. Comparative genomics of Australian and international isolates of Salmonella Typhimurium: Correlation of core genome evolution with CRISPR and prophage profiles. Sci. Rep. 2017, 7, 9733. [Google Scholar] [CrossRef]
- Xie, X.; Hu, Y.; Xu, Y.; Yin, K.; Li, Y.; Chen, Y.; Xia, J.; Xu, L.; Liu, Z.; Geng, S.; et al. Genetic analysis of Salmonella enterica serovar Gallinarum biovar Pullorum based on characterization and evolution of CRISPR sequence. Vet. Microbiol. 2017, 203, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Medina-Aparicio, L.; Rebollar-Flores, J.E.; Beltran-Luviano, A.A.; Vazquez, A.; Gutierrez-Rios, R.M.; Olvera, L.; Calva, E.; Hernandez-Lucas, I. CRISPR-Cas system presents multiple transcriptional units including antisense RNAs that are expressed in minimal medium and upregulated by pH in Salmonella enterica serovar Typhi. Microbiology 2017, 163, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 2008, 322, 1843–1845. [Google Scholar] [CrossRef]
- Redman, M.; King, A.; Watson, C.; King, D. What is CRISPR/Cas9? Arch. Dis. Child. Educ. Pract. Ed. 2016, 101, 213–215. [Google Scholar] [CrossRef]
- Deveau, H.; Garneau, J.E.; Moineau, S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010, 64, 475–493. [Google Scholar] [CrossRef]
- Mohanraju, P.; Makarova, K.S.; Zetsche, B.; Zhang, F.; Koonin, E.V.; van der Oost, J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 2016, 353. [Google Scholar] [CrossRef]
- Staals, R.H.; Zhu, Y.; Taylor, D.W.; Kornfeld, J.E.; Sharma, K.; Barendregt, A.; Koehorst, J.J.; Vlot, M.; Neupane, N.; Varossieau, K.; et al. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol. Cell 2014, 56, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Pu, Q.; Shen, G.; Li, R.; Guo, K.; Zhou, C.; Liang, H.; Jiang, J.; Wu, M. CdpR Inhibits CRISPR-Cas Adaptive Immunity to Lower Anti-viral Defense while Avoiding Self-Reactivity. iScience 2019, 13, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovic, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef] [PubMed]
- Westra, E.R.; Buckling, A.; Fineran, P.C. CRISPR-Cas systems: Beyond adaptive immunity. Nat. Rev. Microbiol. 2014, 12, 317–326. [Google Scholar] [CrossRef]
- Faucher, S.P.; Curtiss, R.; Daigle, F. Selective capture of Salmonella enterica serovar typhi genes expressed in macrophages that are absent from the Salmonella enterica serovar Typhimurium genome. Infect. Immun. 2005, 73, 5217–5221. [Google Scholar] [CrossRef]
- Gunderson, F.F.; Cianciotto, N.P. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. MBio 2013, 4, e00074-13. [Google Scholar] [CrossRef]
- Medina-Aparicio, L.; Rebollar-Flores, J.E.; Gallego-Hernandez, A.L.; Vazquez, A.; Olvera, L.; Gutierrez-Rios, R.M.; Calva, E.; Hernandez-Lucas, I. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J. Bacteriol. 2011, 193, 2396–2407. [Google Scholar] [CrossRef]
- Deiwick, J.; Nikolaus, T.; Erdogan, S.; Hensel, M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 1999, 31, 1759–1773. [Google Scholar] [CrossRef]
- Hernandez-Lucas, I.; Gallego-Hernandez, A.L.; Encarnacion, S.; Fernandez-Mora, M.; Martinez-Batallar, A.G.; Salgado, H.; Oropeza, R.; Calva, E. The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi. J. Bacteriol. 2008, 190, 1658–1670. [Google Scholar] [CrossRef]
- Baek, C.H.; Wang, S.; Roland, K.L.; Curtiss, R. Leucine-responsive regulatory protein (Lrp) acts as a virulence repressor in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2009, 191, 1278–1292. [Google Scholar] [CrossRef]
- Espinosa, E.; Casadesus, J. Regulation of Salmonella enterica pathogenicity island 1 (SPI-1) by the LysR-type regulator LeuO. Mol. Microbiol. 2014, 91, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- Guadarrama, C.; Villasenor, T.; Calva, E. The Subtleties and Contrasts of the LeuO Regulator in Salmonella Typhi: Implications in the Immune Response. Front. Immunol. 2014, 5, 581. [Google Scholar] [CrossRef] [PubMed]
- Lucchini, S.; Rowley, G.; Goldberg, M.D.; Hurd, D.; Harrison, M.; Hinton, J.C. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006, 2, e81. [Google Scholar] [CrossRef] [PubMed]
- Marshall, D.G.; Sheehan, B.J.; Dorman, C.J. A role for the leucine-responsive regulatory protein and integration host factor in the regulation of the Salmonella plasmid virulence (spv) locus in Salmonella typhimurium. Mol. Microbiol. 1999, 34, 134–145. [Google Scholar] [CrossRef]
- Navarre, W.W.; Porwollik, S.; Wang, Y.; McClelland, M.; Rosen, H.; Libby, S.J.; Fang, F.C. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 2006, 313, 236–238. [Google Scholar] [CrossRef]
- Kaiser, D.; Robinson, M.; Kroos, L. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef]
- Julien, B.; Kaiser, A.D.; Garza, A. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 2000, 97, 9098–9103. [Google Scholar] [CrossRef]
- Sampson, T.R.; Saroj, S.D.; Llewellyn, A.C.; Tzeng, Y.L.; Weiss, D.S. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 2013, 497, 254. [Google Scholar] [CrossRef]
- Sampson, T.R.; Weiss, D.S. Cas9-dependent endogenous gene regulation is required for bacterial virulence. Biochem. Soc. Trans. 2013, 41, 1407–1411. [Google Scholar] [CrossRef]
- Louwen, R.; Horst-Kreft, D.; de Boer, A.G.; van der Graaf, L.; de Knegt, G.; Hamersma, M.; Heikema, A.P.; Timms, A.R.; Jacobs, B.C.; Wagenaar, J.A.; et al. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barre syndrome. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2013, 32, 207–226. [Google Scholar] [CrossRef]
- Aklujkar, M.; Lovley, D.R. Interference with histidyl-tRNA synthetase by a CRISPR spacer sequence as a factor in the evolution of Pelobacter carbinolicus. BMC Evol. Biol. 2010, 10, 230. [Google Scholar] [CrossRef] [PubMed]
- Zegans, M.E.; Wagner, J.C.; Cady, K.C.; Murphy, D.M.; Hammond, J.H.; O’Toole, G.A. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 2009, 191, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Cady, K.C.; O’Toole, G.A. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J. Bacteriol. 2011, 193, 3433–3445. [Google Scholar] [CrossRef] [PubMed]
- Li, R.P.; Fang, L.Z.; Tan, S.R.; Yu, M.; Li, X.F.; He, S.S.; Wei, Y.Q.; Li, G.P.; Jiang, J.X.; Wu, M. Type I CRISPR-Cas targets endogenous genes and regulates virulence to evade mammalian host immunity. Cell Res. 2016, 26, 1273–1287. [Google Scholar] [CrossRef]
- Nealson, K.H.; Platt, T.; Hastings, J.W. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 1970, 104, 313–322. [Google Scholar] [CrossRef]
- Pan, J.; Ren, D. Quorum sensing inhibitors: A patent overview. Expert Opin. Ther. Pat. 2009, 19, 1581–1601. [Google Scholar] [CrossRef]
- Lowery, C.A.; Dickerson, T.J.; Janda, K.D. Interspecies and interkingdom communication mediated by bacterial quorum sensing. Chem. Soc. Rev. 2008, 37, 1337–1346. [Google Scholar] [CrossRef]
- Vendeville, A.; Winzer, K.; Heurlier, K.; Tang, C.M.; Hardie, K.R. Making ‘sense’ of metabolism: Autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 2005, 3, 383–396. [Google Scholar] [CrossRef]
- Taga, M.E.; Semmelhack, J.L.; Bassler, B.L. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 2001, 42, 777–793. [Google Scholar] [CrossRef]
- Taga, M.E.; Miller, S.T.; Bassler, B.L. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 2003, 50, 1411–1427. [Google Scholar] [CrossRef]
- Surette, M.G.; Miller, M.B.; Bassler, B.L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: A new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 1999, 96, 1639–1644. [Google Scholar] [CrossRef] [PubMed]
- Xavier, K.B.; Miller, S.T.; Lu, W.; Kim, J.H.; Rabinowitz, J.; Pelczer, I.; Semmelhack, M.F.; Bassler, B.L. Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. ACS Chem. Biol. 2007, 2, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Park, J.; Ryu, S. Repressed Quorum sensing by overexpressing LsrR Hampers Salmonella evasion from oxidative killing within macrophages. J. Microbiol. Biotechnol. 2010, 20, 1624–1629. [Google Scholar] [PubMed]
- Sperandio, V.; Mellies, J.L.; Nguyen, W.; Shin, S.; Kaper, J.B. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 1999, 96, 15196–15201. [Google Scholar] [CrossRef] [PubMed]
- Galan, J.E. Salmonella interactions with host cells: Type III secretion at work. Annu. Rev. Cell Dev. Biol. 2001, 17, 53–86. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; He, X.; Liu, Q.; Hu, C. Developing Universal Genetic Tools for Rapid and Efficient Deletion Mutation in Vibrio Species Based on Suicide T-Vectors Carrying a Novel Counterselectable Marker, vmi480. PLoS ONE 2015, 10, e0144465. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ji, L.; Wang, X.; Li, J.; Zhu, J.; Sun, A. Role of RpoS in stress resistance, quorum sensing and spoilage potential of Pseudomonas fluorescens. Int. J. Food Microbiol. 2018, 270, 31–38. [Google Scholar] [CrossRef]
- Song, D.; Li, X.; Cheng, Y.; Xiao, X.; Lu, Z.; Wang, Y.; Wang, F. Aerobic biogenesis of selenium nanoparticles by Enterobacter cloacae Z0206 as a consequence of fumarate reductase mediated selenite reduction. Sci. Rep. 2017, 7, 3239. [Google Scholar] [CrossRef]
- Hua, Y.; Sun, Q.; Wang, X.; Du, Y.; Shao, N.; Zhang, Q.; Zhao, W.; Wan, C. [Construction of enterohemorrhagic Escherichia coli O157:H7 strains with espF gene deletion and complementation]. J. South. Med. Univ. 2015, 35, 1546–1551. [Google Scholar]
- Hussain, H.I.; Iqbal, Z.; Seleem, M.N.; Huang, D.; Sattar, A.; Hao, H.; Yuan, Z. Virulence and transcriptome profile of multidrug-resistant Escherichia coli from chicken. Sci. Rep. 2017, 7, 8335. [Google Scholar] [CrossRef]
- Ghaffar, N.M.; Connerton, P.L.; Connerton, I.F. Filamentation of Campylobacter in broth cultures. Front. Microbiol. 2015, 6, 657. [Google Scholar] [CrossRef] [PubMed]
- Solano, C.; Garcia, B.; Valle, J.; Berasain, C.; Ghigo, J.M.; Gamazo, C.; Lasa, I. Genetic analysis of Salmonella enteritidis biofilm formation: Critical role of cellulose. Mol. Microbiol. 2002, 43, 793–808. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Li, Y.; Yue, B.; Wu, M. Genes as early responders regulate quorum-sensing and control bacterial cooperation in Pseudomonas aeruginosa. PLoS ONE 2014, 9, e101887. [Google Scholar] [CrossRef] [PubMed]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, H.; Nishi, J.; Imuta, N.; Tokuda, K.; Miyanohara, H.; Hashiguchi, T.; Zenmyo, M.; Yamamoto, T.; Ijiri, K.; Kawano, Y.; et al. Quantitative analysis of biofilm formation of methicillin-resistant Staphylococcus aureus (MRSA) strains from patients with orthopaedic device-related infections. FEMS Immunol. Med. Microbiol. 2011, 63, 10–15. [Google Scholar] [CrossRef]
- Shabbir, M.A.B.; Tang, Y.; Xu, Z.; Lin, M.; Cheng, G.; Dai, M.; Wang, X.; Liu, Z.; Yuan, Z.; Hao, H. The Involvement of the Cas9 Gene in Virulence of Campylobacter jejuni. Front. Cell. Infect. Microbiol. 2018, 8, 285. [Google Scholar] [CrossRef]
- Crowley, L.C.; Scott, A.P.; Marfell, B.J.; Boughaba, J.A.; Chojnowski, G.; Waterhouse, N.J. Measuring Cell Death by Propidium Iodide Uptake and Flow Cytometry. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef]
- Iqbal, Z.; Seleem, M.N.; Hussain, H.I.; Huang, L.; Hao, H.; Yuan, Z. Comparative virulence studies and transcriptome analysis of Staphylococcus aureus strains isolated from animals. Sci. Rep. 2016, 6, 35442. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Guo, R.; Li, Z.; Zhou, X.; Huang, C.; Hu, Y.; Geng, S.; Chen, X.; Li, Q.; Pan, Z.; Jiao, X. Induction of arthritis in chickens by infection with novel virulent Salmonella Pullorum strains. Vet. Microbiol. 2019, 228, 165–172. [Google Scholar] [CrossRef]
- Djebbi-Simmons, D.; Xu, W.; Janes, M.; King, J. Survival and inactivation of Salmonella enterica serovar Typhimurium on food contact surfaces during log, stationary and long-term stationary phases. Food Microbiol. 2019, 84, 103272. [Google Scholar] [CrossRef] [PubMed]
- Galvao, L.C.; Rosalen, P.L.; Rivera-Ramos, I.; Franco, G.C.; Kajfasz, J.K.; Abranches, J.; Bueno-Silva, B.; Koo, H.; Lemos, J.A. Inactivation of the spxA1 or spxA2 gene of Streptococcus mutans decreases virulence in the rat caries model. Mol. Oral Microbiol. 2017, 32, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.T.; Scott-Anne, K.; Liao, S.; De, A.; Luo, M.; Kovacs, C.; Narvaez, B.S.; Faustoferri, R.C.; Yu, Q.; Taylor, C.M.; et al. Deficiency of BrpA in Streptococcus mutans reduces virulence in rat caries model. Mol. Oral Microbiol. 2018, 33, 353–363. [Google Scholar] [CrossRef]
- Bronnec, V.; Turonova, H.; Bouju, A.; Cruveiller, S.; Rodrigues, R.; Demnerova, K.; Tresse, O.; Haddad, N.; Zagorec, M. Adhesion, Biofilm Formation, and Genomic Features of Campylobacter jejuni Bf, an Atypical Strain Able to Grow under Aerobic Conditions. Front. Microbiol. 2016, 7, 1002. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Gong, T.; Zhou, X.; Lu, M.; Zeng, J.; Peng, X.; Wang, S.; Li, Y. Deletion of cas3 gene in Streptococcus mutans affects biofilm formation and increases fluoride sensitivity. Arch. Oral Biol. 2019, 99, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yin, X.H.; Zhou, Z.T.; Hu, S.S.; Li, S.W.; Liu, M.; Wang, X.L.; Xiao, Y.C.; Shi, D.S.; Bi, D.R.; et al. Cas9 regulated gene expression and pathogenicity in Riemerella anatipestifer. Microb. Pathog. 2019, 136, 103706. [Google Scholar] [CrossRef]
- Fabrega, A.; Vila, J. Salmonella enterica serovar Typhimurium skills to succeed in the host: Virulence and regulation. Clin. Microbiol. Rev. 2013, 26, 308–341. [Google Scholar] [CrossRef]
- Singh, V.; Finke-Isami, J.; Hopper-Chidlaw, A.C.; Schwerk, P.; Thompson, A.; Tedin, K. Salmonella Co-opts Host Cell Chaperone-mediated Autophagy for Intracellular Growth. J. Biol. Chem. 2017, 292, 1847–1864. [Google Scholar] [CrossRef]
- Bourgogne, A.; Garsin, D.A.; Qin, X.; Singh, K.V.; Sillanpaa, J.; Yerrapragada, S.; Ding, Y.; Dugan-Rocha, S.; Buhay, C.; Shen, H.; et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008, 9, R110. [Google Scholar] [CrossRef]
- Kim, K.; Golubeva, Y.A.; Vanderpool, C.K.; Slauch, J.M. Oxygen-dependent regulation of SPI1 type three secretion system by small RNAs in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2019, 111, 570–587. [Google Scholar]
- Fu, Y.; Galan, J.E. A salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 1999, 401, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, K.; Saulnier-Bellemare, J.; Daigle, F. Functional Analysis of the Chaperone-Usher Fimbrial Gene Clusters of Salmonella enterica serovar Typhi. Front. Cell. Infect. Microbiol. 2018, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Prigent-Combaret, C.; Vidal, O.; Dorel, C.; Lejeune, P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 1999, 181, 5993–6002. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Oloketuyi, S.F.; Kim, Y.M. Diversity of bacteria and bacterial products as antibiofilm and antiquorum sensing drugs against pathogenic bacteria. Curr. Drug Targets 2019. [Google Scholar] [CrossRef]
- Dekimpe, V.; Deziel, E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: The transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 2009, 155, 712–723. [Google Scholar] [CrossRef]
- Diggle, S.P.; Cornelis, P.; Williams, P.; Camara, M. 4-quinolone signalling in Pseudomonas aeruginosa: Old molecules, new perspectives. Int. J. Med. Microbiol. IJMM 2006, 296, 83–91. [Google Scholar] [CrossRef]
- Smith, R.S.; Iglewski, B.H.P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 2003, 6, 56–60. [Google Scholar] [CrossRef]
- Tateda, K.; Ishii, Y.; Horikawa, M.; Matsumoto, T.; Miyairi, S.; Pechere, J.C.; Standiford, T.J.; Ishiguro, M.; Yamaguchi, K. The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect. Immun. 2003, 71, 5785–5793. [Google Scholar] [CrossRef]
- DeLisa, M.P.; Wu, C.F.; Wang, L.; Valdes, J.J.; Bentley, W.E. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 2001, 183, 5239–5247. [Google Scholar] [CrossRef]
- Antunes, L.C.; Ferreira, R.B.; Buckner, M.M.; Finlay, B.B. Quorum sensing in bacterial virulence. Microbiology 2010, 156, 2271–2282. [Google Scholar] [CrossRef]
- Marques, J.C.; Oh, I.K.; Ly, D.C.; Lamosa, P.; Ventura, M.R.; Miller, S.T.; Xavier, K.B. LsrF, a coenzyme A-dependent thiolase, catalyzes the terminal step in processing the quorum sensing signal autoinducer-2. Proc. Natl. Acad. Sci. USA 2014, 111, 14235–14240. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wang, B.; Zhou, C.; Lin, P.; Qin, S.; Gao, P.; Wang, Z.; Xia, Z.; Wu, M. Bacterial Type I CRISPR-Cas systems influence inflammasome activation in mammalian host by promoting autophagy. Immunology 2019. [Google Scholar] [CrossRef] [PubMed]
- Hoyland-Kroghsbo, N.M.; Paczkowski, J.; Mukherjee, S.; Broniewski, J.; Westra, E.; Bondy-Denomy, J.; Bassler, B.L. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc. Natl. Acad. Sci. USA 2017, 114, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Patterson, A.G.; Jackson, S.A.; Taylor, C.; Evans, G.B.; Salmond, G.P.C.; Przybilski, R.; Staals, R.H.J.; Fineran, P.C. Quorum Sensing Controls Adaptive Immunity through the Regulation of Multiple CRISPR-Cas Systems. Mol. Cell 2016, 64, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Pu, Q.; Wu, Q.; Zhou, C.; Wang, B.; Schettler, J.; Wang, Z.; Qin, S.; Gao, P.; Li, R.; et al. High-throughput screen reveals sRNAs regulating crRNA biogenesis by targeting CRISPR leader to repress Rho termination. Nat. Commun. 2019, 10, 3728. [Google Scholar] [CrossRef] [PubMed]

| Locus Tag | Gene | Protein | Fold Change (Δcas3/cas3 WT) | p-Value |
|---|---|---|---|---|
| AV79_RS14235 | cas3 | type I CRISPR-associated protein DNA helicase Cas3 | −301.27 | 5.331 × 10−11 |
| AV79_RS14230 | cse1 | type I-E CRISPR-associated protein Cse1 | 20.47 | 2.96 × 10−5 |
| AV79_RS14210 | cas6 | type I-E CRISPR-associated protein Cas6 | 9.11 | 6.002 × 10−5 |
| AV79_RS14200 | cas2 | CRISPR-associated protein Cas2 | 8.66 | 0.0003026 |
| AV79_RS14215 | cas5 | type I-E CRISPR-associated protein Cas5 | 8.34 | 0.0002544 |
| AV79_RS14205 | cas1 | CRISPR-associated protein Cas1 | 8.25 | 0.0001503 |
| AV79_RS14220 | cas7 | type I-E CRISPR-associated protein Cas7 | 7.68 | 0.0004107 |
| AV79_RS14225 | cse2 | type I-E CRISPR-associated protein Cse2 | 6.98 | 0.0006198 |
| Gene | Protein | Fold Change (Δcas3/cas3 WT) | p-Value | Function |
|---|---|---|---|---|
| lsrF | putative aldolase, 3-hydroxy-5-phosphonooxypentane-2, 4-dione thiolase LsrF | 13.48 | 0.01 | Involved in the degradation of phospho-AI-2, thereby terminating induction of the lsr operon and closing the AI-2 signaling cycle. Catalyzes the transfer of an acetyl moiety from 3-hydroxy-5-phosphonooxypentane-2, 4-dione to CoA to form glycerone phosphate and acetyl-CoA. |
| lsrG | (4S)-4-hydroxy-5-phosphonooxypentane-2, 3-dione isomerase, autoinducer-2 (AI-2) modifying protein LsrG | 7.90 | 0.02 | Involved in the degradation of phospho-AI-2, thereby terminating induction of the lsr operon and closing the AI-2 signaling cycle. Catalyzes the conversion of (4S)-4-hydroxy-5-phosphonooxypentane-2, 3-dione (P-DPD) to 3-hydroxy-5-phosphonooxypentane-2, 4-dione (P-HPD). |
| lsrE | Ribulose-phosphate 3-epimerase, Putative epimerase LsrE | 7.47 | 0.03 | Cofactor. |
| lsrB | Autoinducer 2-binding protein LsrB | 6.88 | 0.03 | Part of the ABC transporter complex LsrABCD involved in autoinducer 2 (AI-2) import. Binds AI-2 and delivers it to the LsrC and LsrD permeases. |
| lsrA | putative ABC transporter ATP-binding protein, Autoinducer 2 import ATP-binding protein LsrA | −2.60 | 0.15 | Part of the ABC transporter complex LsrABCD involved in autoinducer 2 (AI-2) import. Responsible for energy coupling to the transport system. |
| lsrR | transcriptional repressor LsrR | −1.39 | 0.36 | In the absence of autoinducer 2 (AI-2), represses transcription of the lsrACDBFGE operon and its own transcription. In the presence of AI-2, LsrR is inactivated by binding phospho-AI-2, leading to the transcription of the lsr genes |
| lsrD | ABC transporter membrane protein, Autoinducer 2 import system permease protein LsrD | 1.55 | 0.38 | Part of the ABC transporter complex LsrABCD involved in autoinducer 2 (AI-2) import. Probably responsible for the translocation of the substrate across the membrane. |
| lsrC | sugar transport protein, Autoinducer 2 import system permease protein LsrC | −1.61 | 0.40 | Part of the ABC transporter complex LsrABCD involved in autoinducer 2 (AI-2) import. Probably responsible for the translocation of the substrate across the membrane. |
| lsrK | autoinducer-2 (AI-2) kinase | 1.32 | 0.48 | Catalyzes the phosphorylation of autoinducer-2 (AI-2) to phospho-AI-2, which subsequently inactivates the transcriptional regulator LsrR and leads to the transcription of the lsr operon. Phosphorylates the ring-open form of (S)-4, 5-dihydroxypentane-2, 3-dione (DPD), which is the precursor to all AI-2 signaling molecules, at the C5 position. |
| luxS | S-ribosylhomocysteinase | 1.36 | 0.40 | Involved in the synthesis of autoinducer 2 (AI-2) which is secreted by bacteria and is used to communicate both the cell density and the metabolic potential of the environment. The regulation of gene expression in response to changes in cell density is called quorum sensing. Catalyzes the transformation of S-ribosylhomocysteine (RHC) to homocysteine (HC) and 4, 5-dihydroxy-2, 3-pentadione (DPD). |
| safA | Lipoprotein, Saf-pilin pilus formation protein safA | −3.79 | 0.02 | One of major fimbrial subunits. |
| safB | pili assembly chaperone protein SafB | −3.28 | 0.02 | Involved in the organization of pilus and the chaperone-mediated protein folding. |
| safC | atypical fimbria outer membrane usher SafC | −2.30 | 0.02 | Involved in pilus assembly and positive to fimbrial usher porin activity. |
| safD | fimbrial structural subunit SafD | −2.72 | 0.01 | Part of fimbrial structures. |
| bdm | biofilm-dependent modulation protein BDM | 3.49 | 0.04 | Bdm acts as a transcriptional activator for genes that are involved in the flagella formation and was shown to be downregulated in biofilms. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, L.; Wang, X.; Huang, D.; Zhao, Y.; Feng, J.; Lu, Q.; Pu, Q.; Wang, Y.; Cheng, G.; Wu, M.; et al. CRISPR-cas3 of Salmonella Upregulates Bacterial Biofilm Formation and Virulence to Host Cells by Targeting Quorum-Sensing Systems. Pathogens 2020, 9, 53. https://doi.org/10.3390/pathogens9010053
Cui L, Wang X, Huang D, Zhao Y, Feng J, Lu Q, Pu Q, Wang Y, Cheng G, Wu M, et al. CRISPR-cas3 of Salmonella Upregulates Bacterial Biofilm Formation and Virulence to Host Cells by Targeting Quorum-Sensing Systems. Pathogens. 2020; 9(1):53. https://doi.org/10.3390/pathogens9010053
Chicago/Turabian StyleCui, Luqing, Xiangru Wang, Deyu Huang, Yue Zhao, Jiawei Feng, Qirong Lu, Qinqin Pu, Yulian Wang, Guyue Cheng, Min Wu, and et al. 2020. "CRISPR-cas3 of Salmonella Upregulates Bacterial Biofilm Formation and Virulence to Host Cells by Targeting Quorum-Sensing Systems" Pathogens 9, no. 1: 53. https://doi.org/10.3390/pathogens9010053
APA StyleCui, L., Wang, X., Huang, D., Zhao, Y., Feng, J., Lu, Q., Pu, Q., Wang, Y., Cheng, G., Wu, M., & Dai, M. (2020). CRISPR-cas3 of Salmonella Upregulates Bacterial Biofilm Formation and Virulence to Host Cells by Targeting Quorum-Sensing Systems. Pathogens, 9(1), 53. https://doi.org/10.3390/pathogens9010053







