Status and Epidemiology of Maize Lethal Necrotic Disease in Northern Tanzania
Abstract
1. Introduction
2. Results
2.1. Screening for MCMV Using RT-PCR
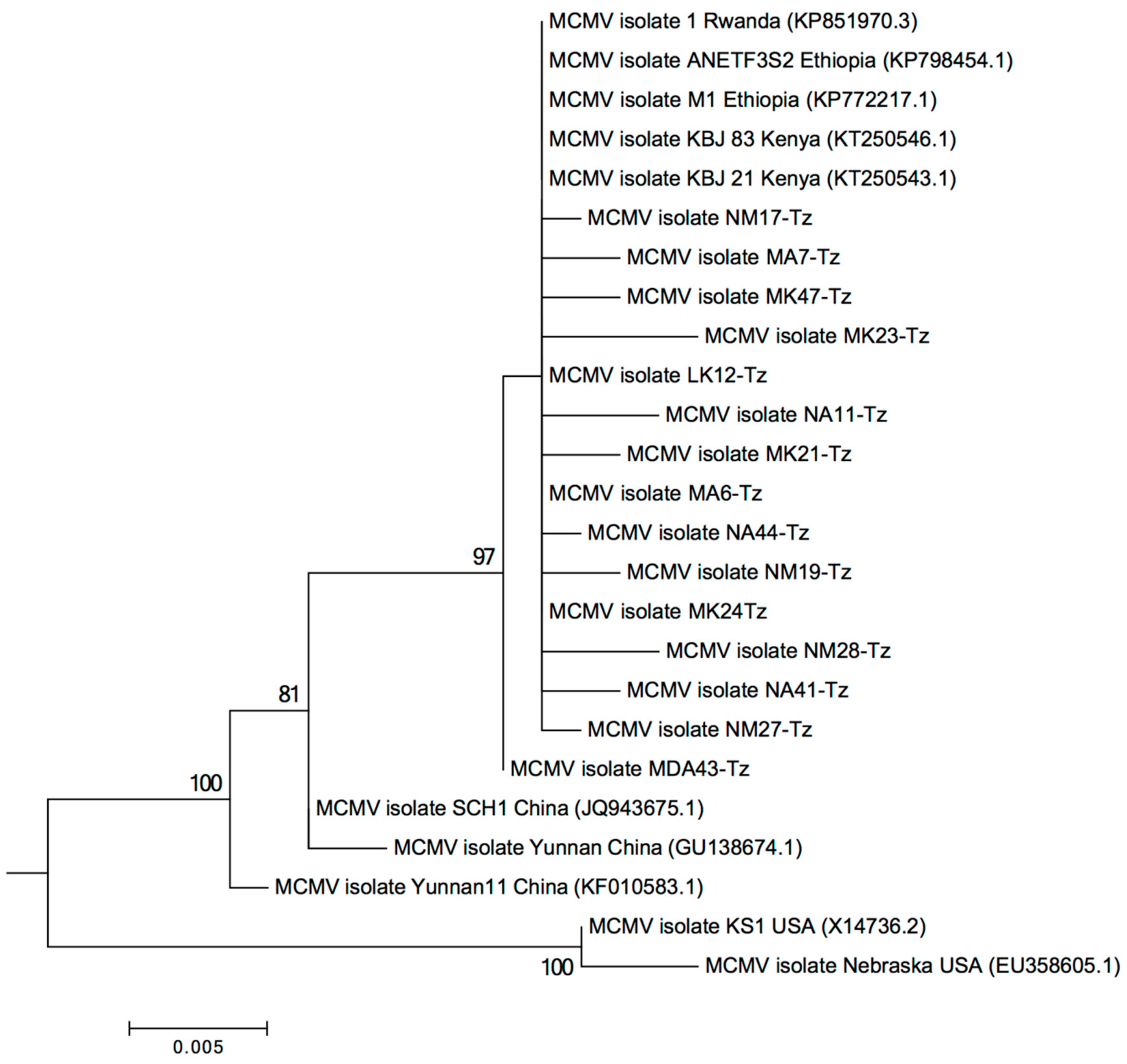
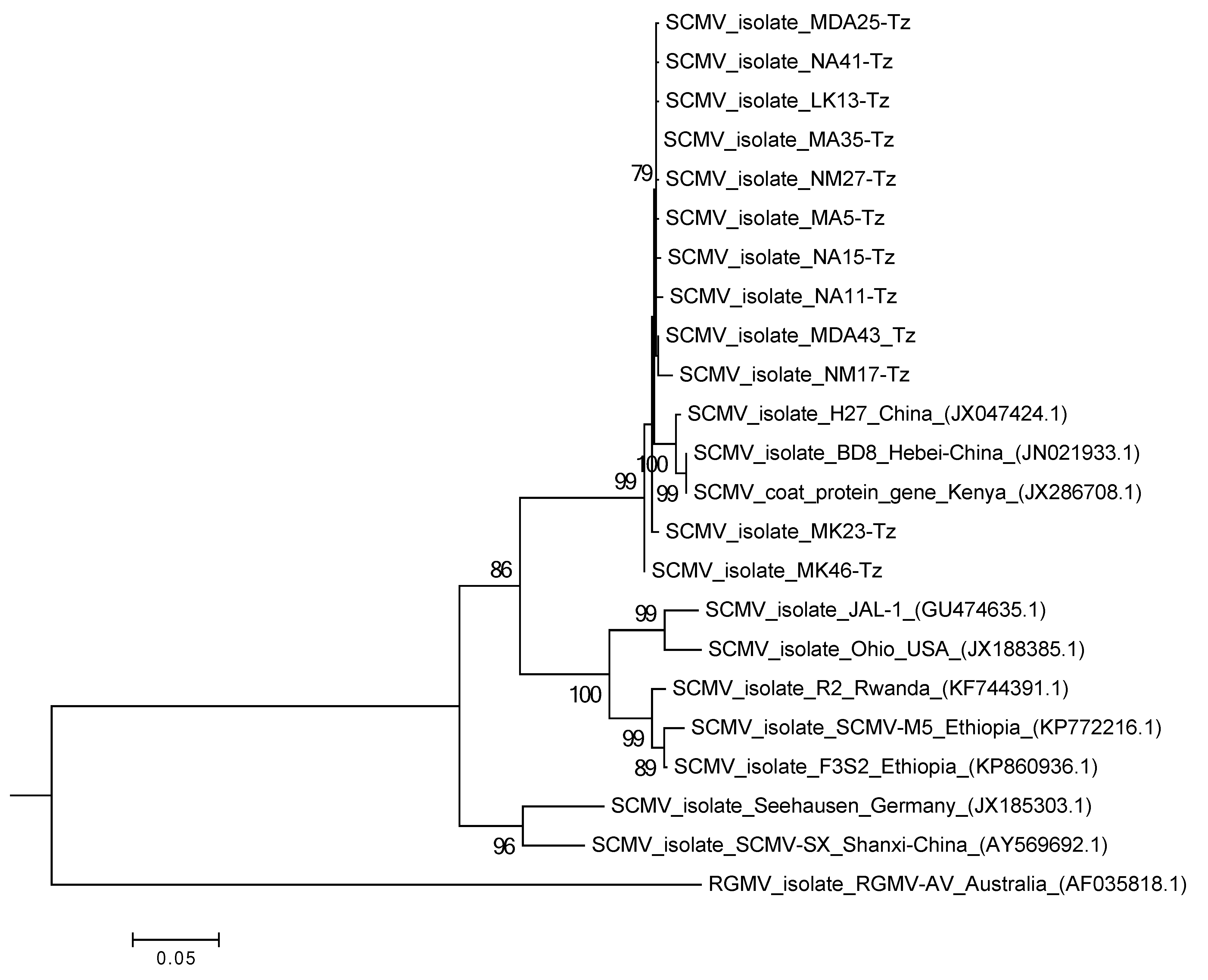
2.2. Characterization of MLN Viruses Using Next-Generation Sequencing
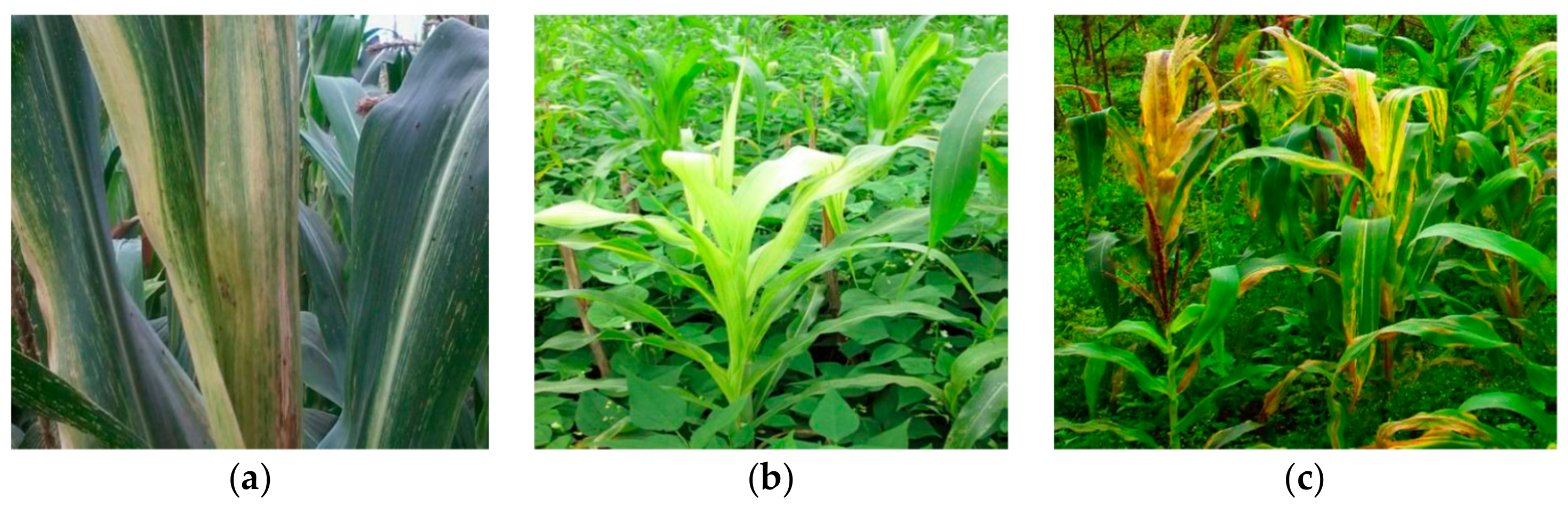
2.3. Prevalence of MLN in Farmers’ Fields in Northern Tanzania
2.4. Farmers’ Experiences on MLN Occurrence and the Associated Yield Loss
3. Discussion
4. Materials and Methods
4.1. Study Site and Design
4.2. Assessment of Prevalence and Collection of Samples
4.3. Detection of MCMV Using Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
4.4. Characterization of MLN Viruses by Next Generation Sequencing
4.5. Assessment of Farmers’ Awareness and Experiences on MLN
4.6. Data Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Suleiman, R.A.; Rosentrater, K.A. Current Maize Production, Postharvest Losses and the Risk of Mycotoxins Contamination in Tanzania. In Proceedings of the 2015 ASABE Annual International Meeting, New Orleans, LA, USA, 26–29 July 2015. [Google Scholar] [CrossRef][Green Version]
- Nkonya, E.; Xavery, P.; Akonaay, H.; Mwangi, W.; Anandajayasekeram, P.; Verkuijl, H.; Martella, D.; Moshi, A. Adoption of Maize Production Technologies in Northern Tanzania Sustainable Maize and Wheat Systems for the Poor; International Maize and Wheat Improvement Center (CIMMYT): Mexico City, Mexico, 1998. [Google Scholar]
- Wangai, A.W.; Redinbaugh, M.G.; Kinyua, Z.M.; Miano, D.W.; Leley, P.K.; Kasina, M.; Mahuku, G.; Scheets, K.; Jeffers, D. First Report of Maize Chlorotic Mottle Virus and Maize Lethal Necrosis in Kenya. Plant Dis. 2012, 96, 1582. [Google Scholar] [CrossRef] [PubMed]
- Makumbi, D.; Wangai, A. Maize Lethal Necrosis (MLN) Disease in Kenya and Tanzania: Facts and Actions; CIMMYT: Mexico City, Mexico, 2012; p. 4. [Google Scholar]
- Adams, I.P.; Harju, V.A.; Hodges, T.; Hany, U.; Skelton, A.; Rai, S.; Deka, M.K.; Smith, J.; Fox, A.; Uzayisenga, B.; et al. First Report of Maize Lethal Necrosis Disease in Rwanda. New Dis. Rep. 2014, 29, 22. [Google Scholar] [CrossRef]
- Lukanda, M.; Owati, A.; Ogunsanya, P.; Valimunzigha, K.; Katsongo, K.; Ndemere, H.; Kumar, P.L. First Report of Maize Chlorotic Mottle Virus Infecting Maize in the Democratic Republic of the Congo. Plant Dis. 2014, 98, 1448. [Google Scholar] [CrossRef] [PubMed]
- Mahuku, G.; Lockhart, B.E.; Wanjala, B.; Jones, M.W.; Kimunye, J.N.; Stewart, L.R.; Cassone, B.J.; Sevgan, S.; Nyasani, J.O.; Kusia, E.; et al. Maize Lethal Necrosis (MLN), an Emerging Threat to Maize-Based Food Security in Sub-Saharan Africa. Phytopathology 2015, 105, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Mahuku, G.; Wangai, A.; Sadessa, K.; Teklewold, A.; Wegary, D.; Ayalneh, D.; Adams, I.; Smith, J.; Bottomley, E.; Bryce, S.; et al. First Report of Maize Chlorotic Mottle Virus and Maize Lethal Necrosis on Maize in Ethiopia. Plant Dis. 2015, 99, 1870. [Google Scholar] [CrossRef]
- Makone, S.M.; Menge, D.; Basweti, E. Impact of Maize Lethal Necrosis Disease on Maize Yield: A Case of Kisii, Kenya. Int. J. Agric. Ext. 2014, 2, 211–218. [Google Scholar]
- Adams, I.P.; Miano, D.W.; Kinyua, Z.M.; Wangai, A.; Kimani, E.; Phiri, N.; Reeder, R.; Harju, V.; Glover, R.; Hany, U.; et al. Use of Next-Generation Sequencing for the Identification and Characterization of Maize Chlorotic Mottle Virus and Sugarcane Mosaic Virus Causing Maize Lethal Necrosis in Kenya. Plant Pathol. 2013, 62, 741–749. [Google Scholar] [CrossRef]
- Lommel, S.A.; Kendall, T.L.; Xiong, Z.; Nutter, R.C. Identification of the Maize Chlorotic Mottle Virus Capsid Protein Cistron and Characterization of Its Subgenomic Messenger RNA. Virology 1991, 181, 382–385. [Google Scholar] [CrossRef]
- Gell, G.; Sebestyén, E.; Balázs, E. Recombination Analysis of Maize Dwarf Mosaic Virus (MDMV) in the Sugarcane Mosaic Virus (SCMV) Subgroup of Potyviruses. Virus Genes 2015, 50, 79–86. [Google Scholar] [CrossRef]
- Uyemoto, J.K. Biology and Control of Maize Chlorotic Mottle Virus. Plant Dis. 1983, 67, 7. [Google Scholar] [CrossRef]
- Scholthof, K.B.G. The Disease Triangle: Pathogens, the Environment and Society. Nat. Rev. Microbiol. 2007, 5, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.Q.; Meinke, L.J.; Wright, R.J.; Wilkinson, D.R.; Campbell, J.E. Maize Chlorotic Mottle Virus in Hawaiian-Grown Maize: Vector Relations, Host Range and Associated Viruses. Crop Prot. 1992, 11, 248–254. [Google Scholar] [CrossRef]
- Brault, V.; Uzest, M.; Monsion, B.; Jacquot, E.; Blanc, S. Aphids as Transport Devices for Plant Viruses. C. R. Biol. 2010, 333, 524–538. [Google Scholar] [CrossRef] [PubMed]
- Nault, L.R.; Styer, W.E.; Coffey, M.E.; Gordon, D.T.; Negi, L.S.; Niblett, C.L. Transmission of Maize Chlorotic Mottle Virus by Chrysomelid Beetles. Phytopathology 1978, 68, 1071–1074. [Google Scholar] [CrossRef]
- Schreinemachers, P.; Balasubramaniam, S.; Boopathi, N.M.; Ha, C.V.; Kenyon, L.; Praneetvatakul, S.; Sirijinda, A.; Le, N.T.; Srinivasan, R.; Wu, M.H. Farmers’ Perceptions and Management of Plant Viruses in Vegetables and Legumes in Tropical and Subtropical Asia. Crop Prot. 2015, 75, 115–123. [Google Scholar] [CrossRef]
- Coetzee, B.; Freeborough, M.J.; Maree, H.J.; Celton, J.M.; Rees, D.J.G.; Burger, J.T. Deep Sequencing Analysis of Viruses Infecting Grapevines: Virome of a Vineyard. Virology 2010, 400, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Agriculture. Agricultural Maps. Available online: https://www.kilimo.go.tz/index.php/en/maps (accessed on 15 September 2017).
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive Metagenomic Visualization in a Web Browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- Nutter, R.C.; Scheets, K.; Panganiban, L.C.; Lommel, S.A. The Complete Nucleotide Sequence of the Maize Chlorotic Mottle Virus Genome. Nucleic Acids Res. 1989, 17, 3163–3177. [Google Scholar] [CrossRef]
- Stenger, D.C.; French, R. Complete Nucleotide Sequence of a Maize Chlorotic Mottle Virus Isolate from Nebraska. Arch. Virol. 2008, 153, 995–997. [Google Scholar] [CrossRef]
- Adams, M.J.; Antoniw, J.F.; Fauquet, C.M. Molecular Criteria for Genus and Species Discrimination within the Family Potyviridae. Arch. Virol. 2005, 150, 459–479. [Google Scholar] [CrossRef]
- Zhao, M.; Ho, H.; Wu, Y.; He, Y.; Li, M. Western Flower Thrips (Frankliniella Occidentalis) Transmits Maize Chlorotic Mottle Virus. J. Phytopathol. 2014, 162, 532–536. [Google Scholar] [CrossRef]
- Cabanas, D.; Watanabe, S.; Higashi, C.H.V.; Bressan, A. Dissecting the Mode of Maize Chlorotic Mottle Virus Transmission (Tombusviridae: Machlomovirus) by Frankliniella Williamsi (Thysanoptera: Thripidae). J. Econ. Entomol. 2013, 106, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Shukla, D.D. Taxonomy of Potyviruses Infecting Maize, Sorghum, and Sugarcane in Australia and the United States as Determined by Reactivities of Polyclonal Antibodies Directed towards Virus-Specific N-Termini of Coat Proteins. Phytopathology 1989, 79, 223–229. [Google Scholar] [CrossRef]
- Shepherd, D.N.; Martin, D.P.; Van Der Walt, E.; Dent, K.; Varsani, A.; Rybicki, E.P. Maize Streak Virus: An Old and Complex “emerging” Pathogen. Mol. Plant Pathol. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- Chen, S.; Jiang, G.; Wu, J.; Liu, Y.; Qian, Y.; Zhou, X. Characterization of a Novel Polerovirus Infecting Maize in China. Viruses 2016, 8, 120. [Google Scholar] [CrossRef]
- Massawe, D.P.; Stewart, L.R.; Kamatenesi, J.; Asiimwe, T.; Redinbaugh, M.G. Complete Sequence and Diversity of a Maize-Associated Polerovirus in East Africa. Virus Genes 2018, 54, 432–437. [Google Scholar] [CrossRef]
- Wamaitha, M.J.; Nigam, D.; Maina, S.; Stomeo, F.; Wangai, A.; Njuguna, J.N.; Holton, T.A.; Wanjala, B.W.; Wamalwa, M.; Lucas, T.; et al. Metagenomic Analysis of Viruses Associated with Maize Lethal Necrosis in Kenya. Virol. J. 2018, 15, 90. [Google Scholar] [CrossRef]
- Hammond, J.; Lecoq, H.; Raccah, B. Epidemiological Risks from Mixed Virus Infections and Transgenic Plants Expressing Viral Genes. Adv. Virus Res. 1999, 54, 189–314. [Google Scholar] [CrossRef]
- Chávez-Calvillo, G.; Contreras-Paredes, C.A.; Mora-Macias, J.; Noa-Carrazana, J.C.; Serrano-Rubio, A.A.; Dinkova, T.D.; Carrillo-Tripp, M.; Silva-Rosales, L. Antagonism or Synergism between Papaya Ringspot Virus and Papaya Mosaic Virus in Carica Papaya Is Determined by Their Order of Infection. Virology 2016, 489, 179–191. [Google Scholar] [CrossRef]
- Mascia, T.; Gallitelli, D. Synergies and Antagonisms in Virus Interactions. Plant Sci. 2016, 252, 176–192. [Google Scholar] [CrossRef]
- Xia, Z.; Zhao, Z.; Chen, L.; Li, M.; Zhou, T.; Deng, C.; Zhou, Q.; Fan, Z. Synergistic Infection of Two Viruses MCMV and SCMV Increases the Accumulations of Both MCMV and MCMV-Derived SiRNAs in Maize. Sci. Rep. 2016, 6, 20520. [Google Scholar] [CrossRef] [PubMed]
- Pita, J.S.; Sangaré, A.; Fodong, V.N.; Otim-Nape, G.W.; Ogwal, S.; Fauquet, C.M. Recombination, Pseudorecombination and Synergism of Geminiviruses Are Determinant Keys to the Epidemic of Severe Cassava Mosaic Disease in Uganda. J. Gen. Virol. 2001, 82, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Rey, M.E.C.; Ndunguru, J.; Berrie, L.C.; Paximadis, M.; Berry, S.; Cossa, N.; Nuaila, V.N.; Mabasa, K.G.; Abraham, N.; Rybicki, E.P.; et al. Diversity of Dicotyledenous-Infecting Geminiviruses and Their Associated DNA Molecules in Southern Africa, Including the South-West Indian Ocean Islands. Viruses 2012, 4, 1753–1791. [Google Scholar] [CrossRef] [PubMed]
- Gowda, M.; Das, B.; Makumbi, D.; Babu, R.; Semagn, K.; Mahuku, G.; Olsen, M.S.; Bright, J.M.; Beyene, Y.; Prasanna, B.M. Genome-Wide Association and Genomic Prediction of Resistance to Maize Lethal Necrosis Disease in Tropical Maize Germplasm. TAG. Theor. Appl. Genet. Theor. Angew. Genet. 2015, 128, 1957–1968. [Google Scholar] [CrossRef] [PubMed]
- Semagn, K.; Beyene, Y.; Babu, R.; Nair, S.; Gowda, M.; Das, B.; Tarekegne, A.; Mugo, S.; Mahuku, G.; Worku, M.; et al. Quantitative Trait Loci Mapping and Molecular Breeding for Developing Stress Resilient Maize for Sub-Saharan Africa. Crop Sci. 2015, 55, 1449–1459. [Google Scholar] [CrossRef]
- Beyene, Y.; Gowda, M.; Suresh, L.M.; Mugo, S.; Olsen, M.; Oikeh, S.O.; Juma, C.; Tarekegne, A.; Prasanna, B.M. Genetic Analysis of Tropical Maize Inbred Lines for Resistance to Maize Lethal Necrosis Disease. Euphytica 2017, 213, 224. [Google Scholar] [CrossRef]
- Elena, S.F.; Sanjuán, R. Adaptive Value of High Mutation Rates of RNA Viruses: Separating Causes from Consequences. J. Virol. 2005, 79, 11555–11558. [Google Scholar] [CrossRef]
- Li, Y.; Liu, R.; Zhou, T.; Fan, Z. Genetic Diversity and Population Structure of Sugarcane Mosaic Virus. Virus Res. 2013, 171, 242–246. [Google Scholar] [CrossRef]
- Steinhauer, D.A.; Domingo, E.; Holland, J.J. Lack of Evidence for Proofreading Mechanisms Associated with an RNA Virus Polymerase. Gene 1992, 122, 281–288. [Google Scholar] [CrossRef]
- Mutiga, S.K.; Gohole, L.S.; Auma, E.O. Effects of Integrating Companion Cropping and Nitrogen Application on the Performance and Infestation of Collards by Brevicoryne Brassicae. Entomol. Exp. Appl. 2010, 134, 234–244. [Google Scholar] [CrossRef]
- Maule, A.J.; Wang, D. Seed Transmission of Plant Viruses: A Lesson in Biological Complexity. Trends Microbiol. 1996, 4, 153–158. [Google Scholar] [CrossRef]
- Jensen, S.G.; Wysong, D.; Ball, E.; Higley, P. Seed Transmission of Maize Chlorotic Mottle Virus. Plant Dis. 1991, 75, 497. [Google Scholar] [CrossRef]
- Hassanali, A.; Herren, H.; Khan, Z.R.; Pickett, J.A.; Woodcock, C.M. Integrated Pest Management: The Push-Pull Approach for Controlling Insect Pests and Weeds of Cereals, and Its Potential for Other Agricultural Systems Including Animal Husbandry. Philos. Trans. R. Soc. Lond. Ser. B Biological Sci. 2008, 363, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Masek, T.; Vopalensky, V.; Suchomelova, P.; Pospisek, M. Denaturing RNA Electrophoresis in TAE Agarose Gels. Anal. Biochem. 2005, 336, 46–50. [Google Scholar] [CrossRef]
- Ortega, A. Insect Pests of Maize A Guide for Field Identification; CIMMYT: Mexico City, Mexico, 1987; pp. 9–38. [Google Scholar]
- Andrews, S. Babraham Bioinformatics—FastQC A Quality Control tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 August 2016).
- Patel, R.K.; Jain, M. NGS QC Toolkit: A Toolkit for Quality Control of next Generation Sequencing Data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-Glance Quality Assessment of Illumina Second-Generation Sequencing Data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Shiryev, S.A.; Papadopoulos, J.S.; Schäffer, A.A.; Agarwala, R. Improved BLAST Searches Using Longer Words for Protein Seeding. Bioinformatics 2007, 23, 2949–2951. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- NCBI. BankIt. Available online: https://www.ncbi.nlm.nih.gov/WebSub/ (accessed on 17 June 2017).

| Region | Villages | Agro-Ecological Zones (AEZs) a | Sampled Farms (n) | * Maize Plants with MLN Symptoms (%) | Leaf Samples Collected (n) | Samples with MCMV (n) by RT-PCR | Samples Selected for NGS | Samples with SCMV (n) by NGS | Co-infection with SCMV and MCMV (n) by NGS |
|---|---|---|---|---|---|---|---|---|---|
| Kilimanjaro | Lyamungu Kati | N4 | 8 | 20.6 ± 2.4A | 44 | 44 | 6 | 6 | 6 |
| Mandaka Mnono | E2 | 5 | 24.0 ± 2.9A | 31 | 31 | 9 | 0 | 0 | |
| Sub-total 1 | 13 | 22.0 ± 1.9A | 75 | 75 | 15 | 6 | |||
| Arusha | Ngaramtoni | N5 | 14 | 19.1 ± 1.6A | 58 | 57 | 8 | 6 | 6 |
| Madira-Sing’isi | N5 | 3 | 16.0 ± 3.4AB | 35 | 35 | 8 | 8 | 8 | |
| Tengeru | N5 | 6 | 4.7 ± 2.6B | - | - | - | - | - | |
| Mlangarini | N5 | 3 | 2.8 ± 4.2B | 20 | 16 | 6 | 6 | 6 | |
| Sub-total 2 | 26 | 14.0 ± 1.6B | 113 | 108 | 22 | 20 | |||
| Manyara | Ayasanda | E2 | 1 | 10.0 ± 5.2AB | - | - | - | - | - |
| Nyunguu | E2 | 2 | 9.9 ± 4.2AB | 35 | 33 | 11 | 4 | 4 | |
| Sub-total 3 | 3 | 10.0 ± 3.3B | 35 | 33 | 11 | 4 | 4 | ||
| Total | 41 | 223 | 216 (97%) | 48 | 30 | 30 (62%) |
| Virus | Isolate | Region Collected | Accession Number | Read Mapped | % Read Mapped | Average Depth of Sequence | % Genome Coverage | Genome Length (nt *) |
|---|---|---|---|---|---|---|---|---|
| MCMV | MA5-Tz | Arusha | MF467384 | 578,660 | 65.7 | 15,057 | 99.9 | 4432 |
| MA7-Tz | Arusha | MF467383 | 408,433 | 82.8 | 10,668 | 99.4 | 4410 | |
| NA11-Tz | Arusha | MF467374 | 433,159 | 69.5 | 11,176 | 99.7 | 4422 | |
| LK14-Tz | Kilimanjaro | MF467392 | 731053 | 80.4 | 18,797 | 99.9 | 4431 | |
| NA16-Tz | Arusha | MF467375 | 429,822 | 37.3 | 10,795 | 99.9 | 4431 | |
| NM19-Tz | Manyara | MF467382 | 466,171 | 38.9 | 11,955 | 99.8 | 4428 | |
| MK21-Tz | Kilimanjaro | MF467385 | 480,071 | 50.1 | 11,738 | 99.5 | 4416 | |
| MK23-Tz | Kilimanjaro | MF467376 | 710,295 | 52.9 | 16,979 | 99.9 | 4431 | |
| NM27-Tz | Manyara | MF467379 | 548,930 | 64.9 | 13,866 | 99.8 | 4427 | |
| NM28-Tz | Manyara | MF467390 | 442,863 | 40.2 | 11,291 | 99.8 | 4429 | |
| LK12-Tz | Kilimanjaro | MF467387 | 453,118 | 73.7 | 11,767 | 99.75 | 4425 | |
| MK34-Tz | Kilimanjaro | MF467386 | 498,579 | 49.2 | 12,741 | 99.8 | 4428 | |
| MK24-Tz | Kilimanjaro | MF467388 | 663,071 | 73.4 | 16,529 | 99.9 | 4431 | |
| MK47-Tz | Kilimanjaro | MF467391 | 38,431 | 2.04 | 928 | 99.7 | 4423 | |
| NM17-Tz | Manyara | MF467377 | 496,273 | 59.3 | 12,672 | 99.5 | 4415 | |
| MA6-Tz | Arusha | MF467389 | 397,723 | 75.7 | 10,425 | 99.5 | 4416 | |
| NA41-Tz | Arusha | MF467380 | 448,926 | 58.5 | 11,521 | 99.9 | 4432 | |
| NA44-Tz | Arusha | MF467378 | 657,532 | 60.3 | 16,422 | 99.6 | 4418 | |
| MDA43-Tz | Arusha | MF467381 | 452,173 | 38.5 | 11,294 | 99.9 | 4431 | |
| SCMV | MK23-Tz | Kilimanjaro | MF467394 | 27,976 | 2.1 | 309 | 99.4 | 9522 |
| MDA43-Tz | Arusha | MF467400 | 14,677 | 1.3 | 168 | 100 | 9575 | |
| MK46-Tz | Kilimanjaro | MF467395 | 11,321 | 0.8 | 131 | 99.3 | 9511 | |
| NA15-Tz | Arusha | MF467402 | 17,531 | 2.0 | 209 | 99.0 | 9484 | |
| NA41-Tz | Arusha | MF467399 | 18,657 | 2.4 | 211 | 99.1 | 9491 | |
| NM27-Tz | Manyara | MF467398 | 13,241 | 1.6 | 152 | 99.0 | 9482 | |
| NA11-Tz | Arusha | MF467393 | 9473 | 1.5 | 113 | 99.4 | 9520 | |
| MA35-Tz | Arusha | MF467403 | 9292 | 1.8 | 115 | 99.5 | 9527 | |
| NM17-Tz | Manyara | MF467397 | 9743 | 1.5 | 115 | 99.4 | 9520 | |
| MDA25-Tz | Arusha | MF467401 | 14,343 | 1.9 | 164 | 99.1 | 9492 | |
| MA5-Tz | Arusha | MF467404 | 11,621 | 1.3 | 137 | 99.1 | 9494 | |
| LK13-Tz | Kilimanjaro | MF467396 | 10,510 | 1.4 | 125 | 99.1 | 9487 | |
| MSV | NA15-Tz | Arusha | MH667487 | 8937 | 0.4 | 377 | 100 | 2689 |
| MDA26-Tz | Arusha | MH667488 | 992 | 0.1 | 37 | 100 | 2689 |
| Region | Villages | Agro-Ecological Zones (AEZs) | Interviewed Farmers (n) | Farmers had Recognized MLN in Their Farms (%) | Farmers Observed Known Insect-Vectors of MLN (%) | Farmers Reported Complete Maize Yield Loss Due to MLN in 2014 (n) |
|---|---|---|---|---|---|---|
| Kilimanjaro | Lyamungu Kati | N4 | 29 | 59 | 48 | 29 |
| Mandaka Mnono | E2 | 24 | 67 | 17 | 23 | |
| Sub-total 1 | 53 | 52 | ||||
| Arusha | Ngaramtoni | N5 | 27 | 78 | 30 | 25 |
| Mlangarini | N5 | 30 | 50 | 27 | 19 | |
| Sub-total 2 | 57 | 44 | ||||
| Manyara | Ayasanda | E2 | 27 | 67 | 22 | 25 |
| Sub-total 3 | 27 | 25 | ||||
| Total | - | 137 | 121 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiruwa, F.H.; Mutiga, S.; Njuguna, J.; Machuka, E.; Senay, S.; Feyissa, T.; Ndakidemi, P.A.; Stomeo, F. Status and Epidemiology of Maize Lethal Necrotic Disease in Northern Tanzania. Pathogens 2020, 9, 4. https://doi.org/10.3390/pathogens9010004
Kiruwa FH, Mutiga S, Njuguna J, Machuka E, Senay S, Feyissa T, Ndakidemi PA, Stomeo F. Status and Epidemiology of Maize Lethal Necrotic Disease in Northern Tanzania. Pathogens. 2020; 9(1):4. https://doi.org/10.3390/pathogens9010004
Chicago/Turabian StyleKiruwa, Fatma Hussein, Samuel Mutiga, Joyce Njuguna, Eunice Machuka, Senait Senay, Tileye Feyissa, Patrick Alois Ndakidemi, and Francesca Stomeo. 2020. "Status and Epidemiology of Maize Lethal Necrotic Disease in Northern Tanzania" Pathogens 9, no. 1: 4. https://doi.org/10.3390/pathogens9010004
APA StyleKiruwa, F. H., Mutiga, S., Njuguna, J., Machuka, E., Senay, S., Feyissa, T., Ndakidemi, P. A., & Stomeo, F. (2020). Status and Epidemiology of Maize Lethal Necrotic Disease in Northern Tanzania. Pathogens, 9(1), 4. https://doi.org/10.3390/pathogens9010004








