Abstract
Serious outbreaks of foodborne disease have been caused by Listeria monocytogenes found in retail delicatessens and the severity of disease is significant, with high hospitalization and mortality rates. Little is understood about the formidable public health threat of L. monocytogenes in all four niches, humans, animals, food, and environment, in Egypt. This study analyzed the presence of L. monocytogenes collected from the four environmental niches and bioinformatics analysis was implemented to analyze and compare the data. PCR was used to detect virulence genes encoded by pathogenicity island (LIPI-1). prfA amino acid substation that causes constitutive expression of virulence was common in 77.7% of isolates. BLAST analysis did not match other isolates in the NCBI database, suggesting this may be a characteristic of the region associated with these isolates. A second group included the NH1 isolate originating in China, and BLAST analysis showed this prfA allele was shared with isolates from other global locations, such as Europe and North America. Identification of possible links and transmission pathways between the four niches helps to decrease the risk of disease in humans, to take more specific control measures in the context of disease prevention, to limit economic losses associated with food recalls, and highlights the need for treatment options.
1. Introduction
Listeria monocytogenes is a facultative intracellular pathogen, which may be classified as asapronoses (or saprozoonoses) with its environmental reservoir [1], where it may grow effectively outside of the host and to infect a relatively wide range of hosts (polyhostality) [1]. L. monocytogenes is present in natural ecosystems and is widely disseminated in different environmental niches [2]. It has been isolated from humans, and more than 50 species of wild and domestic animals, including mammals, birds, fish, crustaceans, and ticks, in addition to environmental sources, such as animal silage, soil, plants, sewage, stream water, and processing environments [3,4,5,6]. Although the genus Listeria includes many species, due to the pathogenicity of L. monocytogenes in human hosts and its ability to flourish in hostile environments, earlier research implicating genome sequencing were to a great extent concentrated on L. monocytogenes [7,8,9,10,11]. L. monocytogenes is implicated with human listeriosis and in the present decennium, numerous serious outbreaks of foodborne listeriosis have been recorded in different countries and continents [11,12,13,14,15,16], causing serious manifestations in the form of septicemia and meningitis, principally in the immunocompromised and old populaces in addition to pregnant women, who may bring forth stillborn babies or seriously contaminated infants [4,9,17,18,19,20,21,22,23,24,25,26].
The variance between L. monocytogenes strains in their virulence potentiality, that is consistent with the flagellar antigen groups, was exhibited in vitro and in vivo. Minimally, three distinct genetic lineages, and identified alphanumerically, were distinctive [27,28]: lineage I: which comprises serotypes causing pandemic listeriosis, lineage II: which embraces serotypes responsible for occasional listeriosis, and lineage III: which includes serotypes isolated from incidents of listeriosis on rare occasions. When the Listeria that were isolated from human, animal, and food were serotyped, 95% of the isolates were identified to be 1/2a, 1/2b and 4b [20,27,28,29]. The expression of several crucial virulence factors [8] are implicated in the pathogenesis of L. monocytogenes, which is harmonized and controlled by the regulatory prfA gene [8,30]. It has been hypothetically proposed that the shift of Listeria species from a facultative pathogen to saprobe [7] was due to losing its virulence genes, of which the prfA cluster is taken as an example, during natural selection of the Listeria species, giving the impression to believe that Listeria inclines to develop by preferably losing its virulence more readily than its possession of virulence traits.
Analysis of L. monocytogenes isolated from the farm milieu and its farm animals showed that some L. monocytogenes, which are correlated with human infection, circulate within the biosphere and the agroecosystems, contributing to the propagation of these pathogens throughout the food chain [9], thus posing a major health issue [9]. Ready-to-eat (RTE) foods, which include processed foods, are of special interest, as they are exposed to the processing milieu prior to packaging and molded for human consumption in the absence of any control measures capable of inactivating the bacterium, such as cooking or being stored at refrigerated or chilled temperatures, which allows growth of L. monocytogenes to high numbers. L. monocytogenes prevalence in the food milieu may not really be relative to its frequency in the food commodities [9,31].
In order to improve understanding of the pathogen’s ecology, the present endeavor was aimed to investigate L. monocytogenes contingency in human and animal clinical cases, retail food, and environment to establish their potential virulence and antibiotic resistance in Egypt. Consequently, this might permit to distinguish potential connections and conveyance routes amid these niches, the diversity in virulence, resistance, and additional features of clinical significance, allowing L. monocytogenes to become an intimidating epidemiological hazard and consequently, facilitate the prevention of humans and animals infection, to implement a specified and detailed course of action in the state of affairs in preventive medicine and to minimize financial losses accompanying food recalls.
2. Results
The current study examined the frequency of L. monocytogenes in different retail food and in animals or human clinical cases of abortion and septicemia in the Greater Cairo Area, based on the prevalence as determined in previous national studies on milk samples [32,33,34]. L. monocytogenes were found in 20/1607 (1.3%) samples analyzed (including 1406 retail food samples, 136 veterinaries, and 65 clinical samples). The results recorded in Table 1, and Supplementary Tables S1 and S2 disclose the frequency of L. monocytogenes in the different samples: Milk by-products, kariesh cheese (1/120); Chicken, broilers internal organs (3/120) and layers internal organs (3/120); Table eggs (1/100); Meat, meat by-products (hamburger 1/50); Ducks internal organs (1/60); Silage (3/90); Fish, frozen fish (1/100), fish filet (1/58), herring (1/66); Brain tissue, rabbit (1/30); fetal livers (goats 1/15); Septicemia (ewes 1/24 and women 1/65).

Table 1.
Virulence and antibiotic resistance profiles that showed variability among the L. monocytogenes isolates in this study.
The 20 isolated L. monocytogenes along with seven additional isolates from previous surveillance of milk [32,33,34] were examined for differences in serotype, genotypic virulence, and phenotypic virulence, biofilm formation, and antibiotic sensitivity. Table 1 is a condensed version of all results (Supplementary Tables S1 and S2) presenting phenotypic and genotypic differences between the isolates, with Figure 1 presenting a heat map of the isolates to visualize these differences. Nine of the isolates were determined to be serotype 1, while 18 isolates were serotype 4. All 27 isolates demonstrated the presence of genetic elements harbored on the Listeria pathogenicity island (LIPI-1; prfA, plcA, plcB, and actA), although two isolates demonstrated the absence of listeriolysin O, encoded by the hlyA gene (broilers 1/3 and the goat fetal liver). All isolates showed the presence of the minor virulence factor gene, iap, encoding the extracellular protein p60. All isolates exhibited causation of keratoconjunctivitis (Anton’s eye test), cytotoxicity of Vero cells, and successful infection and lethality toward mice and chick embryos, indicating that all were virulent isolates of L. monocytogenes. The flagellin-encoding flaA gene, which contributes to effective invasion during infection [8], was absent in only four of the 27 isolates. PCR screening of four key internalins encoded by inlA, inlB, inlC, and inlJ indicated the presence of all four genes in 12 isolates, including the isolates from veterinary and clinical samples, while it was absent in two of the isolates (1/3 from each of the broilers and layers). The frequency of the individual internalin genes inlA, inlB, inlC, and inlJ were 74.1% (20), 81.5% (22), 70.4% (19), and 66.7% (18), respectively. Notably, the isolates (4) from cow, she-camel, and buffalo milk, and the two isolates that caused septicemia, were positive for the presence of all 11 genes examined.
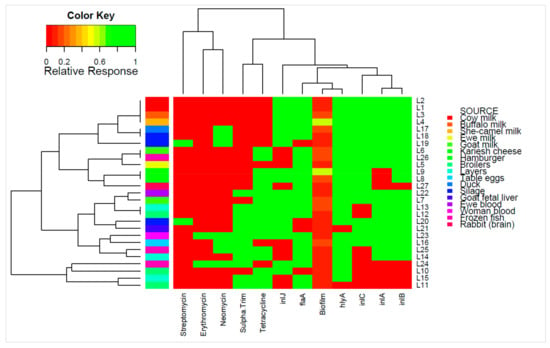
Figure 1.
Individual isolates showing hierarchical clustering of isolates and factors. Binary factors (such as antibiotics or genes) indicating presence as green (relative response 1) or absence as red (relative response 0). Clustering is based on Wald-like test (D2) and for binary data.
Biofilm formation was determined by Christensen’s test tube method (CT) and microtitre plate assay (MPA). The CT allows for qualitative analysis of attachment to glass surfaces and subsequent biofilm formation, which in our investigation showed moderate to strong biofilm formation by the isolates. The MPA for quantitative analysis of attachment and biofilm formation on plastic surfaces during static conditions displayed strong to very strong biofilm formation ranging from 0.13 to 0.56 of staining, where the reference strains recorded an O.D. of 0.16 (Supplementary Table S1).
The phenotypic antibiotic sensitivity results of the 27 L. monocytogenes isolates showed that all were sensitive to all penicillins tested: ampicillin, amoxicillin/clavulanic, amoxicillin, penicillin G, cloxacillin, and oxacillin, the second generation fluroquinolones: ofloxacin, enrofloxacin, and ciprofloxacin, the aminoglycosides: amikacin, gentamicin, kanamycin, and spiramycin, the glucopeptide, vancomycin, and rifamycin. All isolates were resistant to the fluroquinolones: flumequine and perfloxacin, the phenicol: chloramphenicol, the cephalosporins: cefotaxime and cephalothin, the lincosamides: lincomycin and clindamycin, and the polypeptide: bacitracin. The isolates showed 25.9% (7), 7.4% (2), 62.9% (17), 44.4% (12), and 7.4% (2) resistance to neomycin, streptomycin, tetracycline, sulphamethozole-trimethoprim, and erythromycin, respectively. The range of the multiple antibiotic resistance index (MARindex) was from 0.28 to 0.43, with the greatest MARindex attributed to the isolate from a case of human septicemia, which showed resistance to 12 antibiotics, including four of the five antibiotics showing variance within the isolates.
The prfA gene encodes a transcriptional activator of the determinants of pathogenicity in L. monocytogenes. The prfA gene was sequenced to determine possible differences in virulence and pathogenicity. There were seven nucleotide differences detected between codons 87 and 208 which caused five changes in the amino acid sequences compared to wild-type (Figure 2). Two of the changes were synonymous substitutions in the third position of codons for S127 and T170 in an isolate for cow milk and buffalo milk, respectively. The other five mutations were missense mutations. One isolate from cow milk showed a mutation of E101K and one isolate showed a mutation of G161D. The E101K and G161D mutations were not identified in the non-redundant protein database at NCBI by BLAST nor the literature. Of the 21 isolates demonstrating the mutation of G145S, only one was absent of additional mutations. The G145S mutation was present with K130I in four isolates, in one isolate which also harbored a G161D mutation, and in all 15 isolates with S184P mutations.
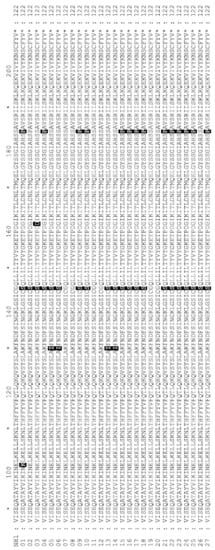
Figure 2.
Protein alignment of prfA protein from L. monocytogenesisolates. Alignment was made to prfA from Listeria monocytogenesstrain NH1 (Genebank: GCA_002969195.1). Differences to NH1 are highlighted and can be found in Table 1.
Examination of correlations between the phenotypic and genotypic characterizations of the 27 isolates of L. monocytogenes was performed (Figure 3). There was a significant positive correlation of the MARindex with resistance to neomycin, sulphamethozole-trimethoprim, and erythromycin (p < 0.05). The strongest correlation of the MARindex was with the antibiotic combination of sulphamethozole-trimethoprim, suggesting this resistance is most important in multi-antibiotic resistance of L. monocytogenes. The presence of the S184P mutation in the pfrA gene was positively and strongly correlated with the G145S mutation, as expected based on initial analysis (p < 0.05). There was a significant negative correlation of the S184P and K130I mutations in pfrA gene (p < 0.05). Correlations of genes and phenotypes that are assumed to be independent of one-another included a strong negative correlation of streptomycin resistance with the flagellin encoding, flaA, gene and the pfrA gene with the G145S mutation (p < 0.05). There was also a significant negative correlation between the internalin encoded by the inlC gene and the antibiotic combination of sulphamethozole-trimethoprim. PCA analysis (Figure 4) supports a number of these correlations, however, MANOVA analysis indicated no statistical differences in the levels of individual test and there were no strong differences in the test based on source or serotype (p > 0.56).
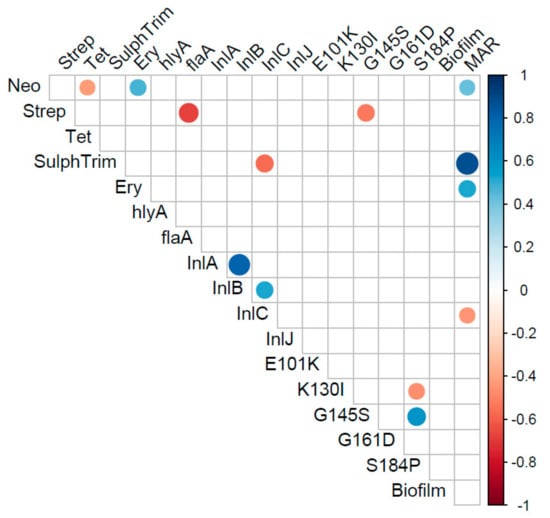
Figure 3.
Correlation matrix of virulence and antibiotic resistance profiles that were different among the recovered Listeria monocytogenes. Only correlations that were significant (p < 0.05) are represented in the matrix.
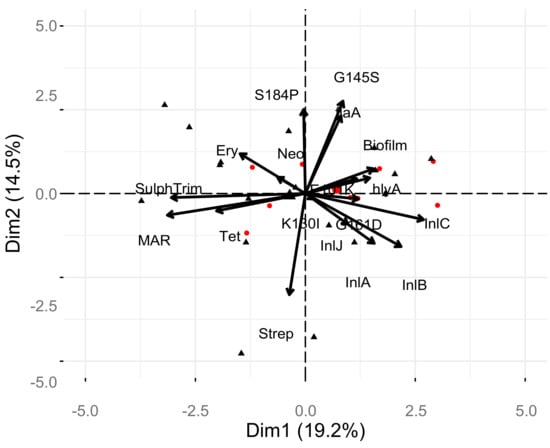
Figure 4.
Principle component analysis of factors and relationship with serotype and individual isolates.
prfA Phylogenetic Analysis
The 27 isolates were clustered into seven unique prfA alleles. The largest group included 14 isolates, which included isolates of each of the sample types, including the human isolate. BLAST analysis of this allele did not match other isolates in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi), suggesting this may be a characteristic of the region associated with these isolates (Figure 5). The second largest group included the NH1 isolate originating in China, and BLAST analysis showed this allele was shared with isolates from other global locations, such as Europe and North America (data not shown). No clear association of a single sample type and specific allele was apparent, with multiple alleles identified among most animal sources included.
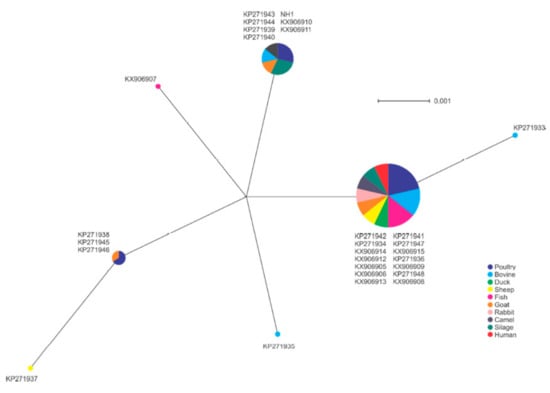
Figure 5.
Phylogenetic analysis of prfA gene sequences among isolates in this study. Node size indicates proportion of isolates sharing a specific genotype.
3. Discussion
The assessment of the actual prevalence of the diverse serotypes causing listeriosis and analyzing the environmental distribution and their bionomics are principal steps to elucidate any potential linkage allying environmental reservoirs of L. monocytogenes to human disease. In a review published by Walland et al. [35], the paper outlines listeriosis in animals and exchanges views on/about the scarcity in our knowledge concerning the reservoirs harboring L. monocytogenes and its cycling between animal and human hosts. This insufficient understanding is caused by gaps in the available literature on the genetic subtypes and its attribution to the L. monocytogenes bionomics, virulence, risk factors, in vivo diagnostics, and listerial pathogeny in animals. The ecology of L. monocytogenes serotypes in the reservoir of agricultural and animals’ environments is not known in Egypt.
The general low prevalence of L. monocytogenes of less than one percent detected within this study is lower than previous studies, which reflects increased awareness, vigilance, and good practices in food preparation in the Egyptian community in the area of study [3,9,11,36,37,38,39,40,41]. A recent study by Leong et al. [42] of small food businesses in Ireland showed a similar decrease over the course of a three-year study. Leong et al. [42] found a comparable food and environmental occurrence of L. monocytogenes at 4.2% and 3.8%, respectively. The highest occurrence was within meat at 7.5% and the lowest in seafood at 1.8%. A similar trend was observed in the current study with occurrence of 2.4% in meat and 1.3% in seafood. The occurrence of L. monocytogenes in raw meat, milk, and cheese in this study was lower than that found in other countries [4,11,16,39,40,41,42,43,44,45]. The global L. monocytogenes population diversity in the implicated food vehicles could be attributed to several factors: (i) improvements in detection methodologies, (ii) packing facility, (iii) increases in populations of lactic acid bacteria, yeasts, and molds that might have an impact on the microenvironment supporting growth of L. monocytogenes, (iv) period and temperature degree of the storage condition, (v) handling by the consumers, (vi) unhygienic conditions, (vii) uncontrolled temperature, (viii) glove/hand issues, (ix) environmental (hygienic conditions of the farms, hygienic conditions of the slaughter houses, rodents, workers, the slicer, trash handling, and cleanup operations) contamination and subsequent cross contamination to other products, (x) transport, (xi) in the processing facilities, and (xii) during handling at the outlets. In the cheese industry, where ripening and storage are critical stages of modernization of the process, several additional parameters must be taken into consideration, including [40]: (i) time needed to ripen, (ii) temperature during storage in the store place, (iii) degree of the primary bacterial load, (iv) post processing contamination, (v) physio-chemical conditions of the cheese process, and (vi) packaging.
Globally, the prevalence rates in fish show great variability [3,25,44,46,47,48,49]: The prevalence of L. monocytogenes in fish are presumably from contaminated waters or during manipulation and processing with contaminated environment and/or equipment [47,50]. The L. monocytogenes isolated from our frozen fish samples indicates that L. monocytogenes has the ability to endure freezing of food, thereby acting as a reservoir to participate in future human listerial outbreaks [9,25,51,52,53].
The presence of the genes of LPI-1, including prfA, plcA, plcB, and actA detected in all isolates highlights the potential for virulence by the isolates, which was further confirmed by infection and mortality of mice, chick embryos, and Vero cells [54]. The lack of detection of the listerolysin O encoding hlyA gene of LIPI-1, required for intracellular pathogenesis, in two of the 27 isolates may suggest a decreased sequence homology, indicating evolutionary plasticity of the hlyA gene [55]. A similar lack of detection of this gene was observed by Al-Nabulsi et al. [56] and Ndahi et al. [57]. The 27 L. monocytogenes isolates exhibited hemolysis on sheep blood agar plates and with the Staphylococcus aureus in the CAMP test. Yet, we detected two hemolytic L. monocytogenes isolates (recovered from broiler and fetal liver), while at the same time, we were not able to detect the listeriolysin hylA gene in these two isolates, which was previously encountered with serotype 4, as manifested in our results [58], indicating that hemolysis by L. monocytogenes could occur by other unrecovered hemolysins. Therefore, en route to explain our recorded phenomenon, Cotter et al. [59] identified a second haemolysin Listeriolysin S (LLS) existing in a subdivision of strains of lineage I, which is the evolutionary lineage of L. monocytogenes that assists in the majority of unpremeditated listerial epidemic outbreaks. Thus, the presence of LIPI-1 in all of the isolates indicates a potential health risk with the associated contaminated food products.
Studies of the functions of flagellar-related proteins in L. monocytogenes are in the preliminary stage. Although the characteristic tumbling motility of L. monocytogenes was observed in all of our 27 isolates, the flaA gene was not detected from four of these motile isolates, stipulating that there are alternative mechanisms controlling the flagella and the secretion of other flagella-related proteins involved in the synthesis of the L. monocytogenes flagellum required for its motility and the secretion of supplementary flagellar-related proteins [60]. Attachment and formation of biofilm allows the colonization of surfaces, including food processing environments [61]. The presence of LIPI-1 genes, hly and prfA, is required for efficient biofilm formation by L. monocytogenes [62]; however, the current study was unable to detect a correlation between the strength of attachment and biofilm formation and detection of the hlyA gene or amino acid substitutions in prfA. There was also no positive correlation with the absence of the flagellin-encoding flaA gene and biofilm formation. The isolates lacking the flaA gene did not show an increase in biofilm formation; however, several genes play a role in biofilm formation which were not screened [63]. Likewise, the presence of strong biofilm formation in two isolates might be due to disruption or overexpression of genes with a role in biofilm formation [63]. Further exploration of differences between environmental and clinical isolates in relation to attachment and biofilm formation will lead to greater understanding of the reservoirs of L. monocytogenes.
An important element of the LIPI-1 virulence determinant is the prfA gene. The pfrA gene encodes the key regulator (the PfrA protein) involved in the activation of pathogenicity determinants in L. monocytogenes. The high representation of the G145S substitution in pfrA suggests that the majority of isolates would have putative constitutive overexpression of virulence factors [64]. However, this overexpression in virulence can lead to decreased fitness outside the host and decrease environmental survival [65]. Additional mutations of K130I, G161D, or S184P were present in PrfA in isolates with the G145S mutation. Substitution of K130 with glutamine (K130Q) caused complete loss of prfA activity [66]; however, the isoleucine (K130I) substitution may not have disrupted the putative functional pocket in which it is found, or the G145S substitution causing constitutive activity was not abolished by K130I. The detection of the S184P mutation within 14 of the 21 isolates with the G145S mutation is interesting as the S184 forms direct hydrogen bonds with the nucleotides within the major groove of the DNA binding site [67]. An alanine substitution of S184 (S184A) decreases DNA binding and virulence in mouse model infections [67], however the S184P mutation did not lead to changes in infection of mice or chick embryos in the current study. The G161D mutation was in one isolate and it most likely does not influence the DNA binding domain of prfA, however, G161 is conserved with the G180 of srv, a PrfA-like Group A Streptococcus regulator of virulence, within the putative DNA-binding domain [68]. The detected negative correlations between the G145S mutation and streptomycin resistance was driven by two isolates from silage that were streptomycin-resistant and had no detected mutations within the PrfA protein, suggesting that this was an artifact of low isolate number. The understanding of the benefit and cost of these mutations of PrfA on the regulation of LIPI-1 mediated virulence in the host and environment will inform models of potential risk of environmental strains of L. monocytogenes.
In addition to the LIPI-1, the internalin family of proteins are necessary for attachment and host cell invasion in non-phagocytic cells. Internalins are associated with or anchored to the cell wall or may be secreted and together mediate diverse functions in virulence. Currently, only the surface proteins inlA and inlB are linked with internalization per se [69]. inlA is necessary for the recognition and invasion of epithelial cells and is essential in invasion of enterocytes and to crossing the placental barrier, while inlB is important for liver and spleen colonization but does not appear necessary for crossing the intestinal epithelium [69]. The small secreted internalin inlC may contribute to inlA-mediated internalization and inlJ contributes to virulence after intravenous infections; however, inlC and inlJ are not known to affect internalization, intracellular proliferation, nor cell-to-cell spread [69]. The isolates showed differences in carriage of the internalins encoded by the inlA, inlB, inlC, and inlJ genes. There were 12 isolates with all four of these internalins, while seven isolates lacked inlA but did carry inlB. There were no detected differences in the virulence of the isolates based on infection and mortality of mice, chick embryo, and Vero cells, suggesting that the presence of other determinants present could facilitate an invasive pathology, or the isolates contained a sequence variant of inlA or inlB not detected in the assay. Interestingly, there was a significant negative correlation of the detection of inlC and phenotypic resistance to the antibiotic combination of sulphamethozole-trimethoprim, suggesting that strains with inlC may currently have decreased incidences of mechanisms yielding resistance to suplfamethozole-trimethoprim. However, a greater number of isolates and identification of the specific mechanisms of antibiotic resistance will be needed to further support this potential correlation. The absence of genes encoding the inlC or inlJ in only four of the isolates suggests a L. monocytogenes population with potentially diverse markers associated with infectivity. L. monocytogenes isolates from imported frozen fish lacked the four main internalin genes (inlA, inlB, inlC, and inlJ) in contrast to the previous studies, where internalin genes were present in almost all previously examined L. monocytogenes isolates from all 4 niches—humans, animals, food, and environment [46,47,48,70,71,72,73,74,75,76,77,78].
L. monocytogenes exhibiting resistance to antibiotics commonly used for the treatment of listeriosis, including ampicillin and other penicillins, and gentamicin resulting in treatment failure, are of major concern [79]. While antibiotic resistance of L. monocytogenes isolates from food, the environment, and human have been reported during the present decade [49], differences between the phenotypic antibiotic resistance of isolates from different ecological niches appear to vary by antimicrobial usage in humans and animals of different geographical location [3,46,50,80]. The isolates in this study showed sensitivity to antibiotics within the penicillin class, second generation fluoroquinolones, glucopeptide class, and rifampicin, while showing complete resistance to phenicols, polypeptides, and lincosamides. The isolates also showed the characteristic intrinsic resistance to cephalosporins and first generation fluoroquinolones [81]. The isolates were variable in their resistance to the macrolide, erythromycin, the aminoglycosides, neomycin and streptomycin, tetracycline, and the combination folic acid pathway inhibitors, sulphamethozole-trimethoprim, indicating acquired resistance from mutation or horizontal gene transfer. The prevalence of 44% (12/27) of isolates showing sulphamethozole-trimethoprim resistance is concerning as there are very few reports of this resistance. Importantly, this is a treatment option utilized in cases of penicillin resistance and so presents a significant risk for human treatment of listeriosis. Bertsch et al. [82] detected the presence of the trimethoprim resistance genes drfA, drfD, and drfG within isolates demonstrating resistance to trimethoprim. Horizontal gene transfer of tetracycline resistance has been identified previously in L. monocytogenes in relation to the tet(M) gene on a Tn916 family conjugative transposons [82] and plasmid [83] and tet(S) bearing conjugative plasmid [84]. Plasmid-mediated resistance to erythromycin and streptomycin has also been documented in L. monocytogenes [84,85]. The MARindex indicates the relative level of resistance to antibiotics. The MARindex was driven by the antibiotic resistance, which was variable in the isolates. The highest MARindex was identified in the isolate from clinical septicemia with the variable resistance to neomycin, tetracycline, erythromycin, and sulphamethozole-trimethoprim. The majority of higher MARindex were associated with isolates from chickens (layers or broilers), silage, and frozen fish, which was driven by the presence of resistance to tetracycline and sulphamethozole-trimethoprim. L. monocytogenes strains resistant to one or more antibiotics have been recovered from environmental, food, and from sporadic cases of human listeriosis [3,27,28,46,49,50,51,80]. Importantly, the common-use drug treatment combination of ampicillin-gentamicin was not disrupted in any of the isolates, indicating high potential success of treatment. However, the high resistance to sulphamethozole-trimethoprim is concerning.
4. Materials and Methods
The present investigation was carried out in accordance with recommendations published by the National Institutes of Health (NRC, 2010) in the updated Guide for the Care and Use of Laboratory Animals. The procedures were approved in accordance with the United Kingdom (UK) Animals (scientific procedures) Act of 1986 prior to experimentation by the Cairo University Ethical Committee.
4.1. Food and Clinical Samples
Prevalence studies were carried out in the Greater Cairo Area (GCA), which includes the cities in the Cairo, Giza, and in the Qalyubia Governorates, with a total population estimated at 20,500,000 (as of 2012); area: 1709 km2; density: 10,400/km2 to detect the presence of L. monocytogenes in retail food and from animals or human clinical cases showing septicemia and abortion. A total of 1607 samples were collected from different environmental niches and tested for the presence of L. monocytogenes following International Organization for Standards 11290-1 (NF EN ISO 11290-1, 1996) during the year 2016: Pasteurized milk (n = 77); Milk by-products, kariesh cheese (n = 120) and yogurt (n = 70); Chicken, chicken filet (n=100), broilers internal organs (n = 120) and layers internal organs (n = 120); Table eggs (n = 100); Meat, retail meat (n = 100) and meat by-products (hamburger n = 50 and basturma n = 50); Duck internal organs (n=60); Silage (n = 90); Frozen seafood, fish (n = 100), fish filet (n = 58), herring (n = 66) and shrimp (n = 50); Brain tissue, cattle (n = 25), sheep (n = 20) and rabbits (n = 30); Abortion, uterine discharge (cows, n = 20 and ewes n = 10) and fetal livers (cows n = 15 and goats n = 15); Septicemia (cows n = 18, buffaloes n = 14, ewes n = 24, goats n = 20 and women n = 65). Samples consisting of 100 g from different retail stores were kept on ice during transportation and analysis was initiated within 4 h.
4.2. Isolation, Identification, and Serotyping of L. monocytogenes
Isolation of Listeria was performed as in Osman et al. [32,34]. Briefly, subsamples of 25 g or 25 mL were aseptically transferred into sterile stomacher closure bags containing 500 mL of half-strength Fraser enrichment broth with CCFA supplement (pre-enrichment broth) and homogenized for 1 min. Samples in pre-treatment broth were incubated at 30 °C for 48 h. The pre-enrichment culture was diluted 1:100 into 10 mL of full strength Fraser enrichment broth with CCFA supplement (enrichment broth) and incubated at 35 °C for 48 h. A loopful of the enrichment culture was streaked onto PALCAM Listeria agar plates and incubated at 37 °C for 24 to 48 h for presumptive isolation and differentiation for Listeria species. Colonies were transferred onto tryptic soy yeast extract agar (TSYEA) and incubated at 30 °C for 24 h. Strains were identified using the API Listeria system and the Oxoid Microbact Listeria 12L system. As previously described [32,34], isolates were examined for morphological and biochemical characteristics by Gram stain, tumbling motility at 20–25 °C, catalase test, Methyl Red (MR), and Voges-Prosakuer tests, hemolysis determined by 5% sheep blood agar, carbohydrate utilization, CAMP test, and phosphatidylinositol-specific phospholipase C (PI-PLC) assay. The enrichment and isolation resulted in 20 identified L. monocytogenes. In addition, seven previously isolated L. monocytogenes from raw milk samples collected from ewes, goats, buffaloes, and cows were included in this study [32,34]. Serotype was assayed following the manufacturer’s instructions of commercially available antisera against serovars 1 and 4 (Behringwerke AG).
4.3. Antibiogram Profile
The 27 isolated L. monocytogenes were tested for their resistance to 28 antibiotics belonging to 11 drug classes by the Kirby–Bauer disk diffusion antibiotic sensitivity testing using the breakpoints for Staphylococci [86] and interpreted according to the CLSI standards [87] or manufacturer’s instructions.
4.4. Pathogenicity and Biofilm Formation
The pathogenicity of the 27 L. monocytogenes isolates was accessed, as previously implemented by Osman et al. [32,34], by: Anton’s eye test for purulent keratoconjunctivitis, the infection and mortality of experimental mice and chick embryos, cytotoxicity of Vero cells, and biofilm formation by Christensen’s tube method and microtitre plate assays.
4.5. Molecular Confirmation of L. monocytogenes and Detection of Virulence Genes
The 27 isolates, 20 isolates from recent sampling, and the 7 resuscitated from frozen glycerol stocks (40% glycerol stored at −70 °C), freshly grown on TSYEA were used for DNA extraction. DNA extraction and PCR confirmation of species were performed as previously described [33]. Colonies of isolates were boiled in 400 µL of TE (10 mMTris-HCl, 1 mM EDTA, pH 8.0) for 10 min and centrifuged at 14,000 rpm for 10 min. The supernatant was immediately used for PCR reactions. The genus was confirmed by PCR as previously described utilizing primers (Supplementary Table S3) specific for the 16S rRNA gene of Listeria. PCR reaction were performed in 50 µL containing 2 µL of DNA extract, 1.5 mM MgCl2, 250 µM dNTPs, 1x Ampli PCR buffer, 0.5 µM of each primer, and 1.25 U of AmpliTaq DNA polymerase, and filled with molecular grade water to volume. The reaction was overlaid with mineral oil and tubes were placed in a thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.). The conditions for amplification were initially denatured for 94 °C for 4 min, followed by 25 cycles of 1 min at 94 °C, 60 °C for 1 min, and 72 °C for 1 min, ending with a final extension for 5 min at 72 °C. L. monocytogenes strain (ATCC 19115) and an E. coli strain (ATCC 25922) were included as positive and negative controls, respectively. Intragenic regions of 11 virulence genes (prfA, hlyA, inlA, inlB, inlC, inlJ, plcA, plcB, Iap, actA, and flaA) were examined by PCR. The primers are included in Supplementary Table S3. The L. monocytogenes virulence genes form the LIPI-1 pathogenicity island (plcA, plcB, prfA, and actA), the genes encoding internalin proteins (inlA, inlB, inlC and inlJ), an adhesion protein (lap), and a flagellin protein (flaA). Bacterial isolates and reference strains were subjected to PCR assay. The PCR conditions were optimized to 1 mM of each primer, 0.65 UTaq DNA Polymerase, 0.2 mM dNTPs, 1X PCR buffer, 1.5 mM MgCl2, and 2 µL of template in 25 μl final volume. Amplification was performed in a thermal cycler with an initial denaturation step of 95 °C for 5 min, followed by 35 amplification cycles of 15 sec at 95 °C (denaturation), 30 sec at 55 °C (annealing), and 90 sec at 72 °C (primer extension), followed by a final extension step of 72 °C for 10 min. The PCR products were determined by separation by electrophoresis in 1.5% agarose and TAE (Tris-acetate EDTA) buffer. Gels were visualized with a UV transilluminator after staining with ethidium bromide.
Partial prfA gene PCR products were purified with a GeneJET PCR Purification Kit, sequenced using BigDye Terminator V3.1 Cycle Sequencing kit with forward and reverse primers following the manufacturer’s instructions, and the resulting reactions were detected by an ABI 3730 xl DNA analyzer. The sequences were trimmed at the 5’ and 3’ end to remove quality less than a confidence of 30 and were aligned to full length prfA and merged into a single sequence representing the PCR product. The sequences were aligned using Mega7 [88] software and the Muscle alignment [89] with default settings. Single nucleotide changes and the subsequent amino acid change, if applicable, were visualized in Mega7. The gene sequences of prfA gene fragments determined in this study were deposited in the GenBank database under accession numbers: KP271933, KP271934, KP271935, KP271936, KP271937, KP271938, KP271939, KP271940, KP271941, KP271942, KP271943, KP271944, KP271945, KP271946, KP271947, KP271948, KX906905, KX906906, KX906907, KX906908, KX906909, KX906910, KX906911, KX906912, KX906913, KX906914, and KX906915.
4.6. Statistical Analysis
Analysis was performed only on factors that showed differences. Antibiotic resistance phenotypic profiles, gene presence, and biochemical results were converted into numerical coding. Sensitivity to an antibiotic was represented as 0 and resistance was represented as 1. Presence or absence of a specific gene (e.g., hlyA) were represented as 1 and 0, respectively. Statistical analysis was performed using the open statistical program R [90]. Heatmap representations with dendrograms and partitions were plotted using the function ’heatmap.3’ in the ‘GMD’ package [91]. Correlations for variables were calculated using the ‘cor’ function and ‘cor.test’ function to determine significance. Significant correlations were visualized utilizing the ‘corrplot’ function from the ‘corrplot’ R package [92]. False Discovery Rate function was used to correct p-values for multiple comparisons [93]. The R packages ‘FactoMineR’ [94] and ‘factoextra’ [95] were used to perform and visualize principal component analysis (PCA). Multivariant statistical analysis was performed using functions in the R package ‘vegan’ [96]. Binomial similarity matrices were calculated for the isolate profiles using ‘vegdist’ function as input for per mutational multivariate analysis of variance (ANOVA) (MANOVA) analyses using the ‘adonis’ function.
4.7. Phylogenetic Analysis of the prfA gene
Partial prfA gene PCR products were purified with GeneJET PCR Purification Kit, sequenced using BigDye Terminator V3.1 Cycle Sequencing kit with forward and reverse primers following manufacturer’s instructions, and the resulting reactions were detected by ABI 3730 xl DNA analyzer. The sequences were trimmed at the 5’ and 3’ end to remove quality less than a confidence of 30 and were aligned to full length prfA and merged into a single sequence representing the PCR product. The sequences were aligned using Mega7 [88] software and the Muscle alignment [89] with default settings. Single nucleotide changes and the subsequent amino acid change, if applicable, were visualized in Mega7.
The prfA nucleotide sequence of each isolate was trimmed, in frame, to 360 bp in length. A translation alignment was performed using Geneious software [97], followed by phylogenetic Neighbor Net analysis using SplitsTree software [98,99].
5. Conclusions
While additional isolates would be preferable in a study such as this, the low incidence of isolation is an indication of good practices in food preparation that can be further improved. Additional isolates may have strengthened the detected significant correlations while also identifying additional correlations. To upgrade our perception of the pathogen’s ecology, it will be empirical to execute additional coherent experiments and high-resolution population genomic analysis of L. monocytogenes isolated from the four niches—humans, animals, food, and environment. This will allow us to trace the evolutionary origins and to prognosticate if there have been any L. monocytogenes host switches which are considered to be a serious hazard to food security. The importance of such studies allows us to speculate the fact that animals could be reservoirs and hotbeds for the blooming of novel virulent L. monocytogenes infecting humans, facilitating the assessment of their potential public health significance in addition to the implementation of control steps to prevent diseases to mitigate infection of humans and animals, and to minimize financial deficit accompanying food recalls and animal welfare. Moreover, genetic identification related to the host-pathogen associations, being central to host adaptation, prompts the pharmaceutical companies to advance new medications to control listeriosis.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/9/1/5/s1, Table S1: athogenicity profiles of Listeria monocytogenes recovered from different samples, Table S1: cont. Pathogenicity profiles of Listeria monocytogenes recovered from different samples, Table S2: Antibiogram for L. monocytogenes isolates, Table S3: PCR Primers and references for 16S rRNA of Listeria and virulence genes. Data S1: Fasta formatted alignments of sequences obtained from MEGA7.
Author Contributions
K.M.O. and A.S. supervised the research work, designed the experimental work, literature search, data collection, and prepared the manuscript. A.O. was responsible for the sample collection and carried out the experiments. A.D.K. conducted the bioinformatics analysis and interpretation. E.M.F. carried out the phylogenetic analysis of prfA gene sequences among isolates in this study. K.M.O., A.S., A.D.K., and E.M.F. were involved in the preparation of the manuscript. All authors read and approved the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by The Science and Technology Development Fund (STDF) of the Ministry of Higher Education, Egypt, through project number 25782. The funders approved the study design, methods of data collection and analysis, decision to publish, and preparation of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Adgamov, R.; Zaytseva, E.; Thiberge, J.; Brisse, S.; Ermolaeva, S. Genetically related Listeria Monocytogenes strains isolated from lethal human cases and wild animals. In Genetic Diversity in Microorganisms; Caliskan, M., Ed.; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef]
- Linke, K.; Rückerl, I.; Brugger, K.; Karpiskova, R.; Walland, J.; Muri-Klinger, S.; Tichy, A.; Wagner, M.; Stessl, B. Reservoirs of Listeria species in three environmental ecosystems. Appl. Environ. Microbiol. 2014, 80, 5583–5592. [Google Scholar] [CrossRef] [PubMed]
- Jami, M.; Ghanbari, M.; Zunabovic, M.; Domig, K.J.; Kneifel, W. Listeria monocytogenes in aquatic food products—A review. Comp. Rev. Food Sci. Food Saf. 2014, 13, 798–813. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSAJ 2017, 15, e5077. [Google Scholar]
- Nastasijevica, I.; Milanov, D.; Velebit, B.; Djordjevic, V.; Swift, C.; Painset, A.; Lakicevic, B. Tracking of Listeria monocytogenes in meat establishment using Whole Genome Sequencing as a food safety management tool: A proof of concept. Int. J. Food Microbiol. 2017, 257, 157–164. [Google Scholar] [CrossRef]
- Editors of Encyclopædia Britannica, Listeriosis Pathology, Last Updated: 29 November 2018.
- Tan, M.F.; Siow, C.C.; Dutta, A.A.; Mutha, N.V.R.; Wee, W.Y.; Heydari, H.; Tan, S.Y.; Ang, M.Y.; Wong, G.J.; Choo, S.W. Development of ListeriaBase and comparative analysis of Listeria monocytogenes. BMC Genom. 2015, 16, 755. [Google Scholar] [CrossRef]
- Orsi, R.H.; Wiedmann, M. Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl. Microbiol. Biotechnol. 2016, 100, 5273–5287. [Google Scholar] [CrossRef]
- Buchanana, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- Hingston, P.; Chen, J.; Dhillon, B.K.; Laing, C.; Bertelli, C.; Gannon, V.; Tasara, T.; Allen, K.; Brinkman, F.S.; Truelstrup Hansen, L.; et al. Genotypes Associated with Listeria monocytogenes isolates displaying impaired or enhanced tolerances to cold, salt, acid, or desiccation stress. Front. Microbiol. 2017, 8, 369. [Google Scholar] [CrossRef]
- Jennison, A.V.; Masson, J.J.; Fang, N.X.; Graham, R.M.; Bradbury, M.I.; Fegan, N.; Gobius, K.S.; Graham, T.M.; Guglielmino, C.J.; Brown, J.L.; et al. Analysis of the Listeria monocytogenes population structure among isolates from 1931 to 2015 in Australia. Front. Microbiol. 2017, 8, 603. [Google Scholar] [CrossRef]
- Chenal-Francisque, V.; Lopez, J.; Cantinelli, T.; Caro, V.; Tran, C.; Leclercq, A.; Lecuit, M.; Brisse, S. Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 2011, 17, 1110–1112. [Google Scholar] [CrossRef]
- Ariza-Miguel, J.; Fernández-Natal, M.I.; Soriano, F.; Hernández, M.; Stessl, B.; Rodríguez-Lázaro, D. Molecular epidemiology of invasive listeriosis due to Listeria monocytogenes in a Spanish hospital over a nine-year study period, 2006–2014. BioMed Res. Int. 2015, 2015, 191409. [Google Scholar] [CrossRef] [PubMed]
- Voronina, O.L.; Ryzhova, N.; Marina, K.; Andrey, S.; Ekaterina, A.; Irina, E.; Denis, K.; Ermolaeva, S.; Alexander, L.G. Diversity and pathogenic potential of Listeria monocytogenes isolated from environmental sources in the Russian Federation. Int. J. Modern Eng. Res. (IJMER) 2015, 5, 5–15. [Google Scholar]
- Marini, E.; Magi, G.; Vincenzi, C.; Manso, E.; Facinelli, B. Ongoing outbreak of invasive listeriosis due to serotype 1/2a Listeria monocytogenes, Ancona province, Italy, January 2015 to February 2016. Euro. Surveill. 2016, 21, 17. [Google Scholar] [CrossRef] [PubMed]
- EFSA Biohaz Panel (EFSA Panel on Biological Hazards); Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Salvador, P.F.E.; Girones, R.; Herman, L.; Koutsoumanis, K. Scientific Opinion on the Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSAJ 2018, 16, e05134. [Google Scholar]
- Wadhwa, D.R.; Smith, M.A. Pregnancy-related listeriosis. Birth Defects Res. 2017, 109, 324–335. [Google Scholar] [CrossRef]
- Madjunkov, M.; Chaudhry, S.; Ito, S. Listeriosis during pregnancy. Arch. Gynecol. Obstet. 2017, 296, 143–152. [Google Scholar] [CrossRef]
- Acciari, V.A.; Iannetti, L.; Gattuso, A.; Sonnessa, M.; Scavia, G.; Montagna, C.; Addante, N.; Torresi, M.; Zocchi, L.; Scattolini, S.; et al. Tracing sources of Listeria contamination in traditional Italian cheese associated with a US outbreak: Investigations in Italy. Epidemiol. Infect. 2016, 144, 2719–2727. [Google Scholar] [CrossRef]
- Althaus, D.; Hofer, E.; Corti, S.; Julmi, A.; Stephan, R. Bacteriological Survey of Ready-to-Eat Lettuce, Fresh-Cut Fruit, and Sprouts Collected from the Swiss Market. J. Food Prot. 2012, 75, 1338–1341. [Google Scholar] [CrossRef]
- Heiman, K.E.; Garalde, V.B.; Gronostaj, M.; Jackson, K.A.; Beam, S.; Joseph, L.; Saupe, A.; Ricotta, E.; Waechter, H.; Wellman, A.; et al. Multistate outbreak of listeriosis caused by imported cheese and evidence of cross-contamination of other cheeses, USA, 2012. Epidemiol. Infect. 2016, 144, 2698–2708. [Google Scholar] [CrossRef]
- Henriques, A.R.; Cristino, J.M.; Fraqueza, M.J. Genetic characterization of Listeria monocytogenes Isolates from industrial and retail ready-to-eat meat-based foods and their relationship with clinical strains from human listeriosis in Portugal. J. Food Prot. 2017, 80, 551–560. [Google Scholar] [CrossRef]
- Henriques, A.R.; Gama, L.T.; Fraqueza, M.J. Tracking Listeria monocytogenes contamination and virulence-associated characteristics in the ready-to-eat meat-based food products industry according to the hygiene level. Int. J. Food Microbiol. 2017, 242, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Gooneratne, R.; Hussain, M.A. Listeria monocytogenes in Fresh Produce: Outbreaks, Prevalence and Contamination Levels. Foods (Basel, Switzerland) 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control; European Food Safety Authority. Multi-country outbreak of Listeria monocytogenes sequence type 8 infections linked to consumption of salmon products –25 October 2018. Stockholm and Parma. ECDC/EFSA J. 2018, 15, 1496E. [Google Scholar]
- WHO. Listeriosis Fact sheet February; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Chen, J.Q.; Regan, P.; Laksanalamai, P.; Healey, S.; Hu, Z. Prevalence and methodologies for detection, characterization and subtyping of Listeria monocytogenes and L. ivanovii in foods and environmental sources. Food Sci. Human Wellness 2017, 6, 97–120. [Google Scholar] [CrossRef]
- Usman, U.B.; Kwaga, J.K.; Kabir, J.; Olonitola, O.S.; Radu, S.; Bande, F. Molecular characterization and phylogenetic analysis of Listeria monocytogenes isolated from milk and milk products in Kaduna, Nigeria. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 4313827. [Google Scholar] [CrossRef]
- Raheem, D. Outbreaks of listeriosis associated with deli meats and cheese: an overview. AIMS Microbiol. 2016, 2, 230–250. [Google Scholar] [CrossRef]
- Vasanthakrishnan, R.B.; de Las Heras, A.; Scortti, M.; Deshayes, C.; Colegrave, N.; Vázquez-Boland, J.A. PrfA regulation offsets the cost of Listeria virulence outside the host. Environ. Microbiol. 2015, 17, 4566–4579. [Google Scholar] [CrossRef]
- D’Amico, D.J.; Donnelly, C.W. Microbiological quality of raw milk used for small-scale artisan cheese production in Vermont: Effect of farm characteristics and practices. J. Dairy Sci. 2010, 93, 134e147. [Google Scholar] [CrossRef]
- Osman, K.M.; Zolnikov, T.R.; Samir, A.; Orabi, A. Prevalence, pathogenic capability, virulence genes, biofilm formation, and antibiotic resistance of Listeria in goat and sheep milk confirms need of hygienic milking conditions. Pathog. Glob. Health 2014, 108, 21–29. [Google Scholar] [CrossRef]
- Osman, K.M.; Samir, A.; Abo-Shama, U.H.; Mohamed, E.H.; Orabi, A.; Zolnikov, T. Determination of virulence and antibiotic resistance pattern of biofilm producing Listeria species isolated from retail raw milk. BMC Microbiol. 2016, 16, 1–13. [Google Scholar] [CrossRef]
- Osman, K.M.; Samir, A.; Orabi, A.; Zolnikov, T.R. Confirmed low prevalence of Listeria mastitis in she-camel milk delivers a safe, alternative milk for human consumption. Acta Trop. 2014, 130, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Walland, J.; Lauper, J.; Frey, J.; Imhof, R.; Stephan, R.; Seuberlich, T.; Oevermann, A. Listeria monocytogenes infection in ruminants: Is there a link to the environment, food and human health? A review. Schweiz. Arch. Tierheilkd. 2015, 157, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Lomonaco, S.; Nucera, D.; Filipello, V. The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect. Genet. Evol. 2015, 35, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Şanlıbaba, P.; Tezel, B.U.; Çakmak, G.A. Prevalence and Antibiotic Resistance of Listeria monocytogenes Isolated from Ready-to-Eat Foods in Turkey. J. Food Quality 2018, 2018, 7693782. [Google Scholar] [CrossRef]
- CDC. Centers for Disease Control and Prevention, Listeria (Listeriosis) | Listeria | National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Foodborne, Waterborne, and Environmental Diseases (DFWED) Page last reviewed: December 18, 2018.
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSAJ 2018, 16, e05500. [Google Scholar]
- Gerard, A.; El-Hajjaji, S.; Niyonzima, E.; Daube, G.; Sindic, M. Prevalence and survival of Listeria monocytogenes in various types of cheese—A review. Int. J. Dairy Technol. 2018, 71, 825–843. [Google Scholar] [CrossRef]
- Li, W.; Bai, L.; Fu, P.; Han, H.; Liu, J.; Guo, Y. The Epidemiology of Listeria monocytogenes in China. Foodborne Pathog. Dis. 2018, 15, 459–466. [Google Scholar] [CrossRef]
- Leong, D.; NicAogáin, K.; Luque-Sastre, L.; McManamon, O.; Hunt, K.; Alvarez-Ordóñez, A.; Scollard, J.; Schmalenberger, A.; Fanning, S.; O’Byrne, C.; et al. A 3-year multi-food study of the presence and persistence of Listeria monocytogenes in 54 small food businesses in Ireland. Int. J. Food Microbiol. 2017, 249, 18–26. [Google Scholar] [CrossRef]
- Kurpas, M.; Wieczorek, K.; Osek, J. Ready-to-eat meat products as a source of Listeria monocytogenes. J. Vet. Res. 2018, 61, 49–55. [Google Scholar] [CrossRef]
- Rodrigues, C.S.; de Sá, C.V.G.C.; de Melo, C.B. An overview of Listeria monocytogenes contamination in ready to eat meat, dairy and fishery foods. Ciência Rural 2017, 47, e20160721. [Google Scholar] [CrossRef]
- Indrawattana, N.; Nibaddhasobon, T.; Sookrung, N.; Chongsa-Nguan, M.; Tungtrongchitr, A.; Makino, S.; Tungyong, W.; Chaicumpa, W. Prevalence of Listeria monocytogenes in raw meats marketed in Bangkok and characterization of the isolates by phenotypic and molecular methods. J. Heal. Popul. Nutr. 2011, 29, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Cheng, J.; Wu, Q.; Zhang, J.; Chen, Y.; Xue, L.; Lei, T.; Zeng, H.; Wu, S.; Ye, Q.; et al. Occurrence, antibiotic resistance, and population diversity of Listeria monocytogenes Isolated from fresh aquatic products in China. Front. Microbiol. 2018, 9, 2215. [Google Scholar] [CrossRef] [PubMed]
- Jamali, H.; Paydar, M.; Ismail, S.; Looi, C.Y.; Wong, W.F.; Radmehr, B.; Abedini, A. Prevalence, antimicrobial susceptibility and virulotyping of Listeria species and Listeria monocytogenes isolated from open-air fish markets. BMC Microbiol. 2015, 15, 144. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, K.; Osek, J. Prevalence, genetic diversity and antimicrobial resistance of Listeria monocytogenes isolated from fresh and smoked fish in Poland. Food Microbiol. 2017, 64, 164–171. [Google Scholar] [CrossRef]
- Obaidat, M.M.; Bani Salman, A.E.; Lafi, S.Q.; Al-Abboodi, A.R. Characterization of Listeria monocytogenesfrom three countries and antibiotic resistance differencesamong countries and Listeria monocytogenes serogroups. Lett. Appl. Microbiol. 2015, 60, 609–614. [Google Scholar] [CrossRef]
- Skowron, K.; Kwiecińska-Piróg, J.; Grudlewska, K.; Świeca, A.; Paluszak, Z.; Bauza-Kaszewska, J.; Wałecka-Zacharska, E.; Gospodarek-Komkowska, E.; Gospodarek-Komkowska, E. The occurrence, transmission, virulence and antibiotic resistance of Listeria monocytogenes in fish processing plant. Int. J. Food Microbiol. 2018, 282, 71–83. [Google Scholar] [CrossRef]
- Schjørring, S.; Gillesberg Lassen, S.; Jensen, T.; Moura, A.; Kjeldgaard, J.S.; Müller, L.; Thielke, S.; Leclercq, A.; Maury, M.M.; Tourdjman, M.; et al. Cross-border outbreak of listeriosis caused by cold-smoked salmon, revealed by integrated surveillance and whole genome sequencing (WGS), Denmark and France, 2015 to 2017. Eur. Surveill. 2017, 22, pii:17-00762. [Google Scholar] [CrossRef]
- Gillesberg Lassen, S.; Ethelberg, S.; Björkman, J.T.; Jensen, T.; Sørensen, G.; Kvistholm Jensen, A.; Müller, L.; Nielsen, E.M.; Mølbak, K. Two listeria outbreaks caused by smoked fish consumptionusingwhole-genome sequencing for outbreak investigations. Clin. Microbiol. Infect. 2016, 22, 620e624. [Google Scholar] [CrossRef]
- Bae, D.; Mezal, E.; Smiley, R.; Cheng, C.; Khan, A. The sub-species characterization and antimicrobialresistance of Listeria monocytogenes isolated from domesticand imported food products from 2004 to 2011. Food Res. Int. 2014, 64, 656–663. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.A.; Domínguez-Bernal, G.; González-Zorn, B.; Kreft, J.; Goebel, W. Pathogenicity islands and virulence evolution in Listeria. Microb. Infect. 2001, 3, 571–584. [Google Scholar] [CrossRef]
- Soni, D.K.; Dubey, S.K. Phylogenetic analysis of the Listeria monocytogenes based on sequencing of 16S rRNA and hlyA genes. Mol. Biol. Rep. 2014, 41, 8219–8229. [Google Scholar] [CrossRef] [PubMed]
- Al-Nabulsi, A.; Osaili, T.M.; Awad, A.A.; Olaimat, A.N.; Shaker, R.R.; Holley, R.A. Occurrence and antibiotic susceptibility of Listeria monocytogenes isolated from raw and processed meat products in Amman, Jordan. CyTA J. Food 2014, 13, 1–7. [Google Scholar]
- Ndahi, M.D.; Kwaga, J.K.; Bello, M.; Kabir, J.; Umoh, V.J.; Yakubu, S.E.; Nok, A.J. Prevalence and antimicrobial susceptibility of Listeria monocytogenes and methicillin-resistant Staphylococcus aureus strains from raw meat and meat products in Zaria, Nigeria. Lett. Appl. Microbiol. 2014, 58, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.M.; Tyler, S.D.; Ewan, E.P.; Ashton, F.E.; Wang, G.; Rozee, K.R. Detection of genes coding for listeriolysin and Listeria monocytogenes antigen A (ImaA) in Listeria spp. by the polymerase chain reaction. Microb. Pathog. 1992, 12, 79–86. [Google Scholar] [CrossRef]
- Cotter, P.D.; Draper, L.A.; Lawton, E.M.; Daly, K.M.; Groeger, D.S.; Casey, P.G.; Ross, R.P.; Hill, C. Listeriolysin S, a Novel Peptide Haemolysin Associated with a Subset of Lineage I Listeria monocytogenes. PLoS Pathog. 2008, 4, e1000144. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wang, H.; Ma, T.; Han, X.; Yang, Y.; Sun, J.; Chen, Z.; Yu, H.; Hang, Y.; Liu, F.; et al. Flagellar Basal Body Structural Proteins FlhB, FliM, and FliY Are Required for Flagellar-Associated Protein Expression in Listeria monocytogenes. Front. Microbiol. 2018, 9, 208. [Google Scholar] [CrossRef]
- Kumar, S.; Parvathi, A.; George, J.; Krohne, G.; Karunasagar, I.; Karunasagar, I. A study on the effects of some laboratory-derived genetic mutations on biofilm formation by Listeria monocytogenes. World J. Microbiol. Biotechnol. 2009, 25, 527–531. [Google Scholar] [CrossRef]
- Price, R.; Jayeola, V.; Niedermeyer, J.; Parsons, C.; Kathariou, S. The Listeria monocytogenes Key Virulence Determinants hly and prfA are involved in Biofilm Formation and Aggregation but not Colonization of Fresh Produce. Pathogens 2018, 7, 18. [Google Scholar] [CrossRef]
- Piercey, M.J.; Hingston, P.A.; Truelstrup Hansen, L. Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15 °C. Int. J. Food Microbiol. 2016, 223, 63–74. [Google Scholar] [CrossRef]
- Ripio, M.T.; Domínguez-Bernal, G.; Lara, M.; Suárez, M.; Vázquez-Boland, J.A. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 1997, 179, 1533–1540. [Google Scholar] [CrossRef] [PubMed]
- Xayarath, B.; Freitag, N.E. Optimizing the balance between host and environmental survival skills: lessons learned from Listeria monocytogenes. Future Microbiol. 2012, 7, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Xayarath, B.; Volz, K.W.; Smart, J.I.; Freitag, N.E. Probing the role of protein surface charge in the activation of PrfA, the central regulator of listeria monocytogenes pathogenesis. PLoS ONE 2011, 6, e23502. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, B.; Klarsfeld, A.; Ebright, R.; Cossart, P. A single substitution in the putative helix-turn-helix motif of the pleiotropic activator PrfA attenuates Listeria monocytogenes virulence. Mol. Microbiol. 1996, 20, 785–797. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, H.; Zhu, Y.; Gao, P. Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes. Protein Cell. 2017, 8, 308–312. [Google Scholar] [CrossRef]
- Bierne, H.; Sabet, C.; Personnic, N.; Cossart, P. Internalins: A complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 2007, 9, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.; Kim, S.; Lee, S.; Lee, H.; Ha, J.; Lee, J.; Choi, Y.; Choi, K.-H.; Yoon, Y. Prevalence and Genetic Characteristics of Meatborne Listeria monocytogenes Isolates from Livestock Farms in Korea. Korean J. Food Sci. Anim. Res. 2016, 36, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Owusu-Kwarteng, J.; Wuni, A.; Akabanda, F.; Jespersen, L. Prevalence and characteristics of Listeria monocytogenes isolates in raw milk, heated milkand nunu, a spontaneously fermented milk beverage, in Ghana. Beverages 2018, 4, 40. [Google Scholar] [CrossRef]
- Chen, J.Q.; Healey, S.; Regan, P.; Laksanalamai, P.; Hu, Z. PCR-based methodologies for detection and characterization of Listeria monocytogenes and Listeria ivanovii in foods and environmental sources. Food Sci. Human Wellness 2017, 6, 39–59. [Google Scholar] [CrossRef]
- Karthikeyan, R.; Gunasekaran, P.; Rajendhran, J. Molecular Serotyping and Pathogenic Potential of Listeria monocytogenes isolated from milk and milk products in Tamil Nadu, India. Foodborne Pathog. Dis. 2015, 12, 522–528. [Google Scholar] [CrossRef]
- Mammina, C.; Aleo, A.; Romani, C.; Pellissier, N.; Nicoletti, P.; Pecile, P.; Nastasi, A.; Pontello, M.M. Characterization of Listeria monocytogenes isolates from human listeriosis cases in Italy. J. Clin. Microbiol. 2009, 47, 2925–2930. [Google Scholar] [CrossRef] [PubMed]
- Sant Ana, A.S.; Igarashi, M.C.; Landgraf, M.; Destro, M.T.; Franco, B.D.G.M. Prevalence, populations and pheno- and genotypic characteristics of Listeria monocytogenes isolated from ready-to-eat vegetables marketed in São Paulo, Brazil. Int. J. Food Microbiol. 2012, 155, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jamali, H.; Radmehr, B. Frequency, virulence genes and antimicrobial resistance of Listeria spp. isolated from bovine clinical mastitis. Vet. J. 2013, 198, 541–542. [Google Scholar] [CrossRef] [PubMed]
- Jamali, H.; Thong, K.L. Genotypic characterization and antimicrobial resistance of Listeria monocytogenes from ready-to-eat foods. Food Control 2014, 44, 1–6. [Google Scholar] [CrossRef]
- GelbíčoVá, T.; KaRpíšKoVá, R. Outdoor environment as a source of Listeria monocytogenes in food chain. Czech J. Food Sci. 2012, 30, 83–88. [Google Scholar] [CrossRef]
- Newman, T. Listeria: What you need to know: Medical News Today. MediLexicon. Intl. 27 April 2017. Available online: https://www.medicalnewstoday.com/articles/180370.php (accessed on 12 November 2019).
- Wilson, A.; Gray, J.; Chandry, P.S.; Fox, E.M. Phenotypic and Genotypic Analysis of Antimicrobial Resistance among Listeria monocytogenes Isolated from Australian Food Production Chains. Genes 2018, 9, 80. [Google Scholar] [CrossRef]
- Krawczyk-Balska, A.; Markiewicz, Z. The intrinsic cephalosporin resistome of Listeria monocytogenes in the context of stress response, gene regulation, pathogenesis and therapeutics. J. Appl. Microbiol. 2016, 120, 251–265. [Google Scholar] [CrossRef]
- Bertsch, D.; Muelli, M.; Weller, M.; Uruty, A.; Lacroix, C.; Meile, L. Antimicrobial susceptibility and antibiotic resistance gene transfer analysis of foodborne, clinical, and environmental Listeria spp. isolates including Listeria monocytogenes. Microbiol. Open 2014, 3, 118–127. [Google Scholar] [CrossRef]
- Haubert, L.; Mendonça, M.; Lopes, G.V.; de Itapema Cardoso, M.R.; da Silva, W.P. Listeria monocytogenes isolates from food and food environment harbouring tetM and ermB resistance genes. Lett. Appl. Microbiol. 2016, 62, 23–29. [Google Scholar] [CrossRef]
- Li, L.; Olsen, R.H.; Shi, L.; Ye, L.; He, J.; Meng, H. Characterization of a plasmid carrying cat, ermB and tetS genes in a foodborne Listeria monocytogenes strain and uptake of the plasmid by cariogenic Streptococcus mutans. Int. J. Food Microbiol. 2016, 238, 68–71. [Google Scholar] [CrossRef]
- Poyart-Salmeron, C.; Carlier, C.; Trieu-Cuot, P.; Courvalin, P.; Courtieu, A.L. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet 1990, 335, 1422–1426. [Google Scholar] [CrossRef]
- Chen, M.; Cheng, J.; Zhang, J.; Chen, Y.; Zeng, H.; Xue, L.; Lei, T.; Pang, R.; Wu, S.; Wu, H.; et al. Isolation, potential virulence, and population diversity of listeria monocytogenes from meat and meat products in China. Front. Microbiol. 2019, 10, 946. [Google Scholar] [CrossRef] [PubMed]
- Clinical, and Laboratory Standards Institute [CLSI]. Performance Standards for Antimicrobial Susceptibility Testing: 24th Informational Supplement (M100S24); Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2014. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.r-project.org/ (accessed on 12 November 2019).
- Zhao, X.; Sandelin, A. GMD: Measuring the distance between histograms with applications on high-throughput sequencing reads. Bioinformatics 2012, 28, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Simko, V. The Corrplot Package. CRAN Repos. 2016. Available online: http://www.sthda.com/french/wiki/matrice-de-correlation-la-fonction-r-qui-fait-tout (accessed on 12 November 2019).
- Benjamini, Y.; Hochberg, Y.; Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Factoextra: Extract and visualize the Results of Multivariate Data Analyses. In R Packag Version 1; 2016; Available online: https://rpkgs.datanovia.com/factoextra/index.html. (accessed on 12 November 2019).
- Dixon, P. Vegan, a package for R funcitons for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Bryant, D.; Moulton, V. Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004, 21, 255–265. [Google Scholar] [CrossRef]
- Huson, D.H. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics 1998, 14, 68–73. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).