Genome-Wide Analysis of Cyclophilin Proteins in 21 Oomycetes
Abstract
1. Introduction
2. Results and Discussion
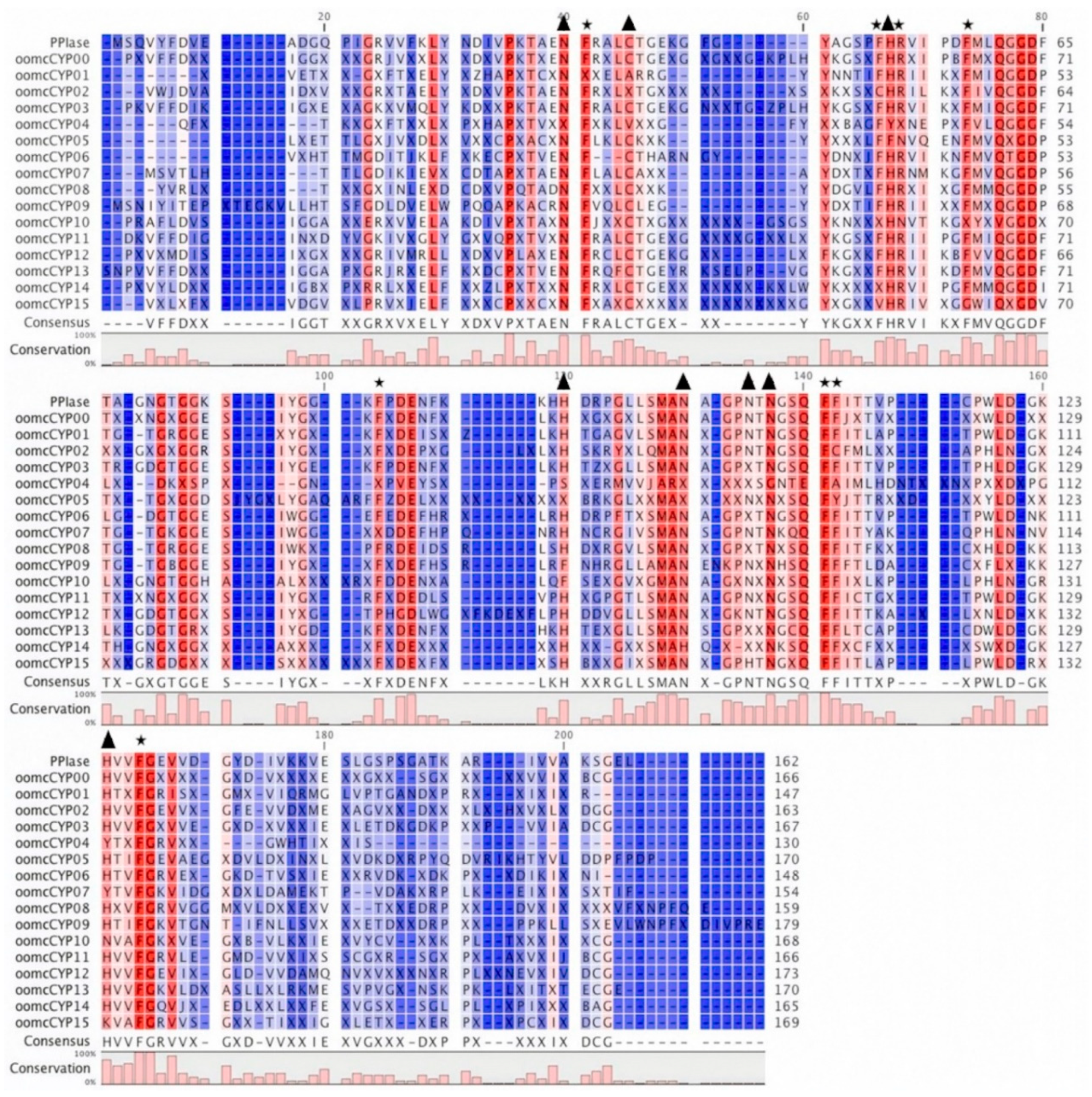
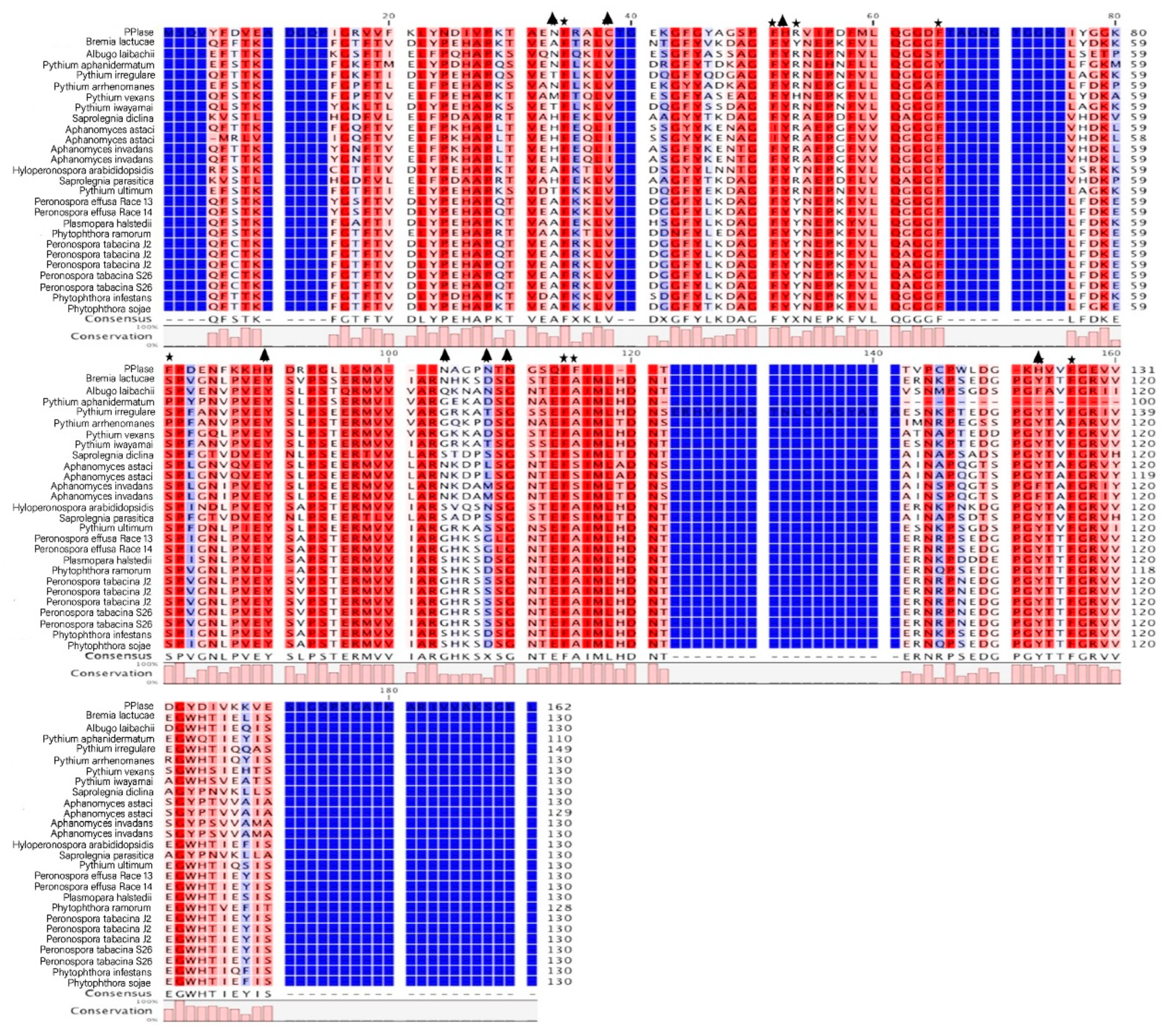
2.1. Structure Analysis
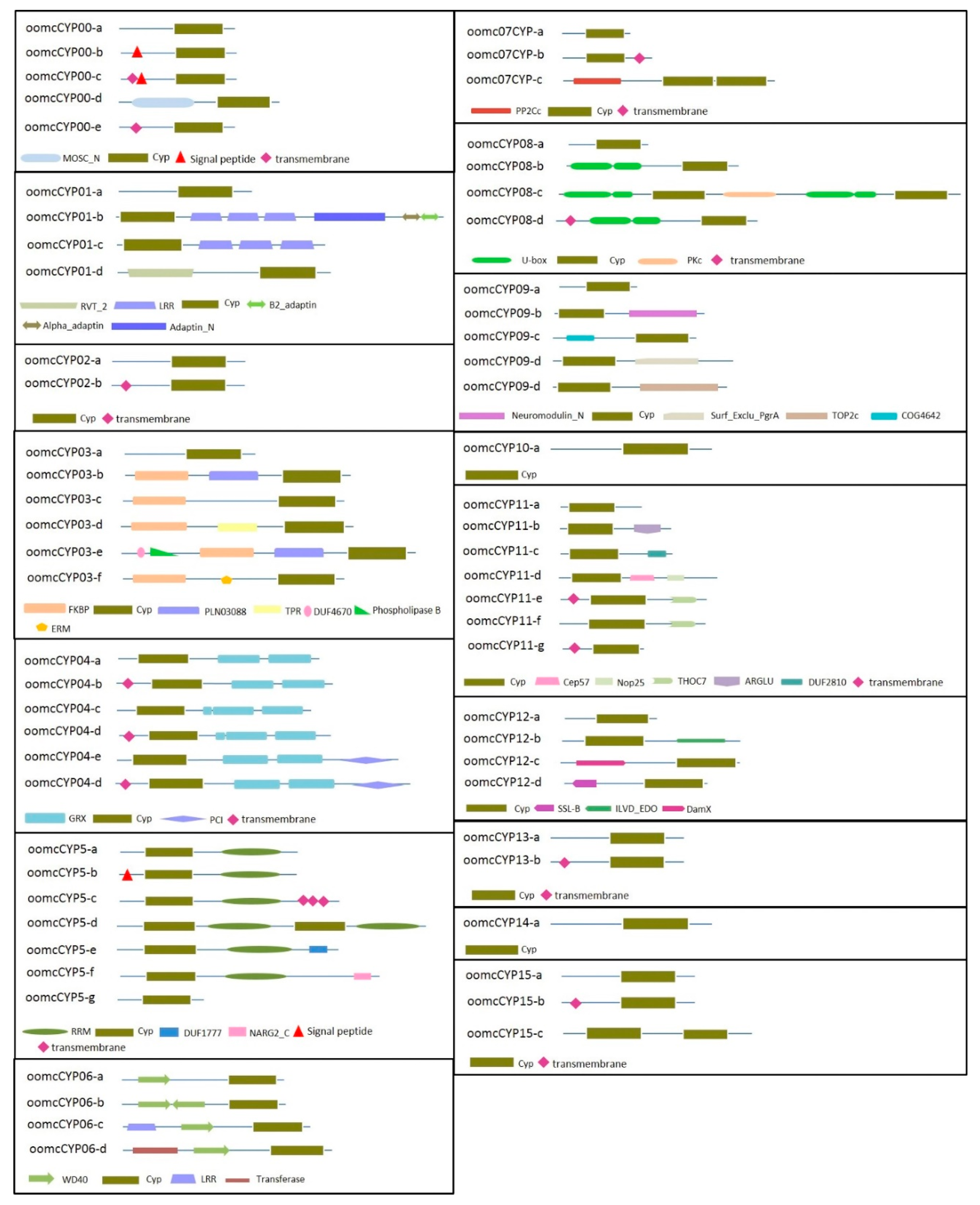
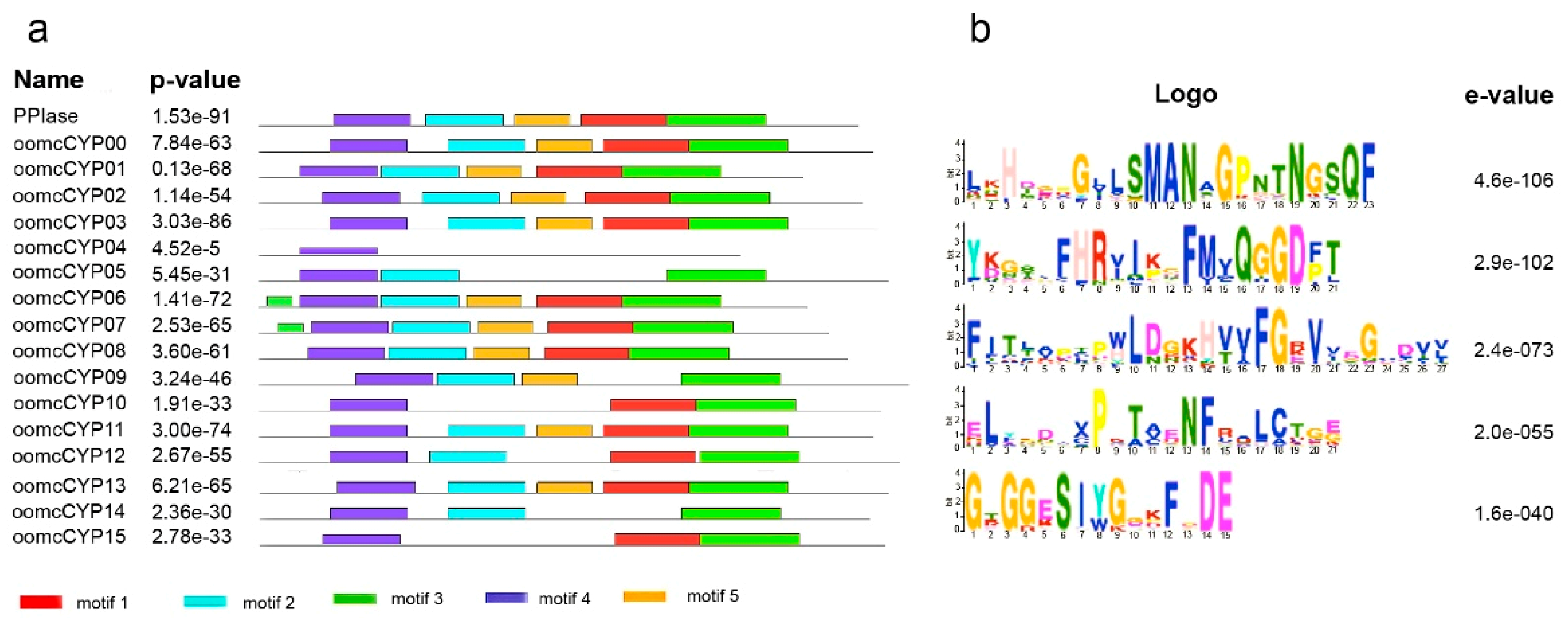
2.2. Multidomain Analysis
2.3. Phylogenetics of CYPs in Oomycetes
2.4. Expression of CYPs in Different Life Stages of P. infestans and P. halstedii
3. Methods
3.1. Identification of CYPs from Oomycete Species
3.2. Multiple Sequence Alignment and Phylogenetic Analysis
3.3. Expression Analysis of Phytophthora infestans and Plasmopara halstedii
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Chou, I.T.; Gasser, C.S. Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol. Biol. 1997, 35, 873–892. [Google Scholar] [CrossRef] [PubMed]
- Coaker, G.; Falick, A.; Staskawicz, B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 2005, 308, 548–550. [Google Scholar] [CrossRef] [PubMed]
- Dimou, M.; Venieraki, A.; Katinakis, P. Microbial cyclophilins: Specialized functions in virulence and beyond. World J. Microbiol. Biotechnol. 2017, 33, 164. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.H.; Fakhrana, I.N.; Abd Rasid, O.; Idris, A.; Ho, C.L.; Shaharuddin, N.A.; Parveez, G.K.A. Molecular cloning and expression analysis of Ganoderma boninense cyclophilins at different growth and infection stages. Physiol. Mol. Plant Pathol. 2017, 99, 31–40. [Google Scholar] [CrossRef]
- Dawar, F.U.; Wu, J.; Zhao, L.; Khattak, M.N.; Mei, J.; Lin, L. Updates in understanding the role of cyclophilin A in leukocyte chemotaxis. J. Leukoc. Biol. 2017, 101, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.Y.; Castel, S.E.; Ernst, E.; Kim, H.S.; Martienssen, R.A. The conserved RNA binding cyclophilin, Rct1, regulates small RNA biogenesis and splicing independent of heterochromatin assembly. Cell Rep. 2017, 19, 2477–2489. [Google Scholar] [CrossRef]
- Horowitz, D.S.; Lee, E.J.; Mabon, S.A.; Misteli, T. A cyclophilin functions in pre-mRNA splicing. EMBO J. 2002, 21, 470–480. [Google Scholar] [CrossRef]
- Dubourg, B.; Kamphausen, T.; Weiwad, M.; Jahreis, G.; Feunteun, J.; Fischer, G.; Modjtahedi, N. The human nuclear SRcyp is a cell cycle-regulated cyclophilin. J. Biol. Chem. 2004, 279, 22322–22330. [Google Scholar] [CrossRef]
- Riggs, D.L.; Cox, M.B.; Tardif, H.L.; Hessling, M.; Buchner, J.; Smith, D.F. Noncatalytic role of the FKBP52 peptidyl-prolyl isomerase domain in the regulation of steroid hormone signaling. Mol. Cell. Biol. 2007, 27, 8658–8669. [Google Scholar] [CrossRef]
- Hatziioannou, T.; Goff, S.P. Infection of nondividing cells by Rous sarcoma virus. J. Virol. 2001, 75, 9526–9531. [Google Scholar] [CrossRef]
- Bannon, J.H.; O’Donovan, D.S.; Kennelly, S.M.E.; Mc Gee, M.M. The peptidyl prolyl isomerase cyclophilin A localizes at the centrosome and the midbody and is required for cytokinesis. Cell Cycle 2012, 11, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Pemberton, T.J.; Kay, J.E. The cyclophilin repertoire of the fission yeast Schizosaccharomyces pombe. Yeast 2005, 22, 927–945. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Heitman, J. The cyclophilins. Genome Biol. 2005, 6, 226. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Viaud, M.; Brunet-Simon, A.; Brygoo, Y.; Pradier, J.M.; Levis, C. Cyclophilin A and calcineurin functions investigated by gene inactivation, cyclosporin A inhibition and cDNA arrays approaches in the phytopathogenic fungus Botrytis cinerea. Mol. Microbiol. 2003, 50, 1451–1465. [Google Scholar] [CrossRef] [PubMed]
- Campos, B.M.; Sforca, M.L.; Ambrosio, A.L.B.; Domingues, M.N.; Brasil de Souza, T.d.A.C.; Barbosa, J.A.R.G.; Leme, A.F.P.; Perez, C.A.; Whittaker, S.B.-M.; Murakami, M.T.; et al. A redox 2-Cys mechanism regulates the catalytic activity of divergent cyclophilins. Plant Physiol. 2013, 162, 1311–1323. [Google Scholar] [CrossRef]
- Potenza, M.; Galat, A.; Minning, T.A.; Ruiz, A.M.; Duran, R.; Tarleton, R.L.; Marín, M.; Fichera, L.E.; Búa, J. Analysis of the Trypanosoma cruzi cyclophilin gene family and identification of Cyclosporin A binding proteins. Parasitology 2006, 132, 867–882. [Google Scholar] [CrossRef]
- Gan, P.H.; Shan, W.; Blackman, L.M.; Hardham, A.R. Characterization of cyclophilin-encoding genes in Phytophthora. Mol. Genet. Genom. 2009, 281, 565–578. [Google Scholar] [CrossRef]
- Liu, J.; Jesse, D.; Farmer, J.; Lane, W.S.; Friedman, J.; Weissman, I.; Schreiber, S.L. Calcineurin is a common target of Cyclophilin-Cyclosporin A and FKBP-FK506 Complexes. Cell 1991, 66, 807–815. [Google Scholar] [CrossRef]
- Viaud, M.C.; Balhadère, P.V.; Talbot, N.J. A Magnaporthe grisea cyclophilin acts as a virulence determinant during plant infection. Plant Cell 2002, 14, 917–930. [Google Scholar] [CrossRef]
- Chen, M.M.; Jiang, M.; Shang, J.; Lan, X.; Yang, F.; Huang, J.; Nuss, D.L.; Chen, B. CYP1, a hypovirus-regulated cyclophilin, is required for virulence in the chestnut blight fungus. Mol. Plant Pathol. 2011, 12, 239–246. [Google Scholar] [CrossRef]
- Kong, G.; Zhao, Y.; Jing, M.; Huang, J.; Yang, J.; Xia, Y.; Kong, L.; Ye, W.; Xiong, Q.; Qiao, Y.; et al. The activation of Phytophthora effector Avr3b by plant cyclophilin is required for the Nudix hydrolase activity of Avr3b. PLoS Pathog. 2015, 11, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Wang, Y. Nudix Effectors: A common weapon in the arsenal of plant pathogens. PLoS Pathog. 2016, 12, e1005704. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y. Phytophthora sojae effectors orchestrate warfare with host immunity. Curr. Opin. Microbiol. 2018, 46, 7–13. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, C.G.P.; Fitzpatrick, D.A. Phylogenomic reconstruction of the oomycete phylogeny derived from 37 genomes. Msphere 2017, 2, e00095-17. [Google Scholar] [CrossRef] [PubMed]
- Seidl, M.F.; Van den Ackerveken, G.; Govers, F.; Snel, B. A domain-centric analysis of oomycete plant pathogen genomes reveals unique protein organization. Plant Physiol. 2011, 155, 628–644. [Google Scholar] [CrossRef] [PubMed]
- Lane, W.S.; Liu, J.; Farmer, J.D.; Weissman, I.; Schreiber, S.L.; Friedman, J. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 2004, 66, 807–815. [Google Scholar]
- Lillig, C.H.; Berndt, C.; Holmgren, A. Glutaredoxin systems. BBA Gen. Subj. 2008, 1780, 1304–1317. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Guo, M.; Jeong, B.R.; Tian, F.; Elthon, T.E.; Cerny, R.L.; Staiger, D.; Alfano, J.R. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 2007, 447, 284–288. [Google Scholar] [CrossRef]
- Jain, B.P.; Pandey, S. WD40 repeat proteins: Signaling scaffold with diverse functions. Protein J. 2018, 37, 391–406. [Google Scholar] [CrossRef]
- He, Q.; McLellan, H.; Boevink, P.C.; Sadanandom, A.; Xie, C.; Birch, P.R.J.; Tian, Z. U-box E3 ubiquitin ligase PUB17 acts in the nucleus to promote specific immune pathways triggered by Phytophthora infestans. J. Exp. Bot. 2015, 66, 3189–3199. [Google Scholar] [CrossRef]
- Barik, S. Dual-family peptidylprolyl isomerases (immunophilins) of select monocellular organisms. Biomolecules 2018, 8, 148. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.; Musiyenko, A.; Kumar, R.; Barik, S. A novel class of dual-family immunophilins. J. Biol. Chem. 2005, 280, 24308–24314. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S.; Yada, M.; Matsumoto, M.; Ishida, N.; Nakayama, K.I. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 2001, 276, 33111–33120. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ballinger, C.A.; Wu, Y.; Dai, Q.; Cyr, D.M.; Hohfeld, J.; Patterson, C. CHIP is a U-box-dependent E3 ubiquitin ligase: Identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 2001, 276, 42938–42944. [Google Scholar] [CrossRef]
- Maris, C.; Dominguez, C.; Allain, F.H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef]
- Leeper, T.; Zhang, S.; Van Voorhis, W.C.; Myler, P.J.; Varani, G. Comparative analysis of glutaredoxin domains from bacterial opportunistic pathogens. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 1141–1147. [Google Scholar] [CrossRef]
- Xu, C.; Min, J. Structure and function of WD40 domain proteins. Protein Cell 2011, 2, 202–214. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rafiqi, M.; Gan, P.H.; Hardham, A.R.; Jones, D.A.; Ellis, J.G. Effectors of biotrophic fungi and oomycetes: Pathogenicity factors and triggers of host resistance. New Phytol. 2009, 183, 993–1000. [Google Scholar] [CrossRef]
- Fawke, S.; Doumane, M.; Schornack, S. Oomycete interactions with plants: Infection strategies and resistance principles. Microbiol. Mol. Biol. Rev. 2015, 79, 263–280. [Google Scholar] [CrossRef]
- Mi, H.; Kops, O.; Zimmermann, E.; Jaschke, A.; Tropschug, M. A nuclear RNA-binding cyclophilin in human T cells. FEBS Lett. 1996, 398, 201–205. [Google Scholar] [CrossRef]
- Romano, P.G.; Horton, P.; Gray, J.E. The Arabidopsis cyclophilin gene family. Plant Physiol. 2004, 134, 1268–1282. [Google Scholar] [CrossRef] [PubMed]
- Links, M.G.; Holub, E.; Jiang, R.H.; Sharpe, A.G.; Hegedus, D.; Beynon, E.; Sillito, D.; Clarke, W.E.; Uzuhashi, S.; Borhan, M.H. De novo sequence assembly of Albugo candida reveals a small genome relative to other biotrophic oomycetes. BMC Genom. 2011, 12, 503. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, K.; Klosterman, S.J.; Derevnina, L.; Martin, F.; Bertier, L.D.; Koike, S.; Reyes-Chin-Wo, S.; Mou, B.; Michelmore, R. Comparative genomics of downy mildews reveals potential adaptations to biotrophy. BMC Genom. 2018, 19, 851. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Fabritius, A.L.; Cvitanich, C.; Judelson, H.S. Stage-specific gene expression during sexual development in Phytophthora infestans. Mol. Microbiol. 2002, 45, 1057–1066. [Google Scholar] [CrossRef]
- Erben, E.D.; Daum, S.; Téllez-Iñón, M.T. The Trypanosoma cruzi PIN1 gene encodes a parvulin peptidyl-prolyl cis/trans isomerase able to replace the essential ESS1 in Saccharomyces cerevisiae. Mol. Biochem. Parasitol. 2007, 153, 186–193. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef]
- Derevnina, L.; Chin-Wo-Reyes, S.; Martin, F.; Wood, K.; Froenicke, L.; Spring, O.; Michelmore, R. Genome sequence and architecture of the tobacco downy mildew pathogen Peronospora tabacina. Mol. Plant Microbe Interact. 2015, 28, 1198–1215. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families’ database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, P.; Piao, Y.; Shon, H.S.; Ryu, K.H. Comparing the normalization methods for the differential analysis of Illumina high-throughput RNA-Seq data. BMC Bioinform. 2015, 16, 1–9. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, 1–9. [Google Scholar] [CrossRef]
- Racine, J.S. RStudio: A platform-independent IDE for R and Sweave. J. Appl. Econom. 2012, 27, 167–172. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. gplots: Various R programming tools for plotting data. R Package Version 2009, 2, 1. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
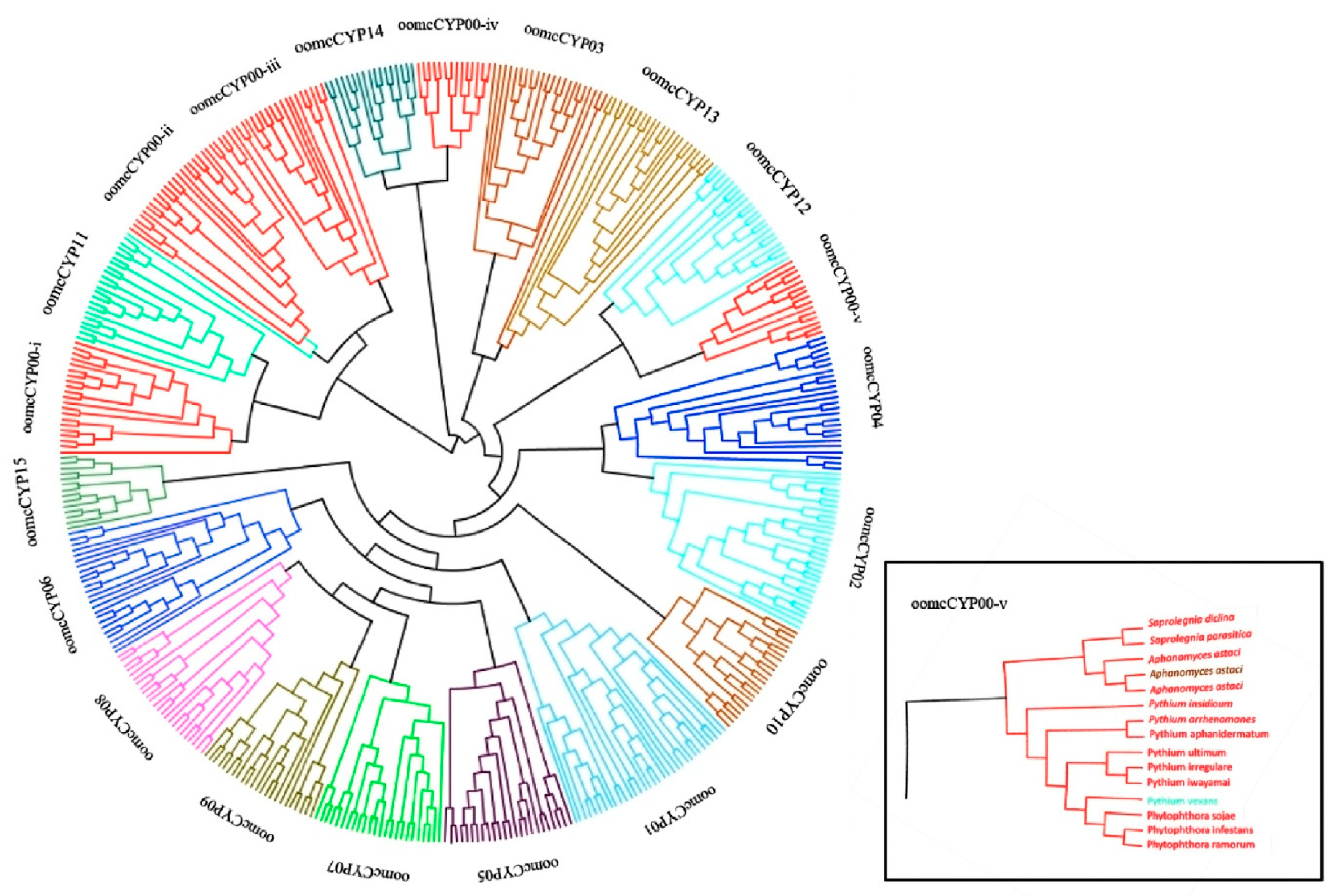
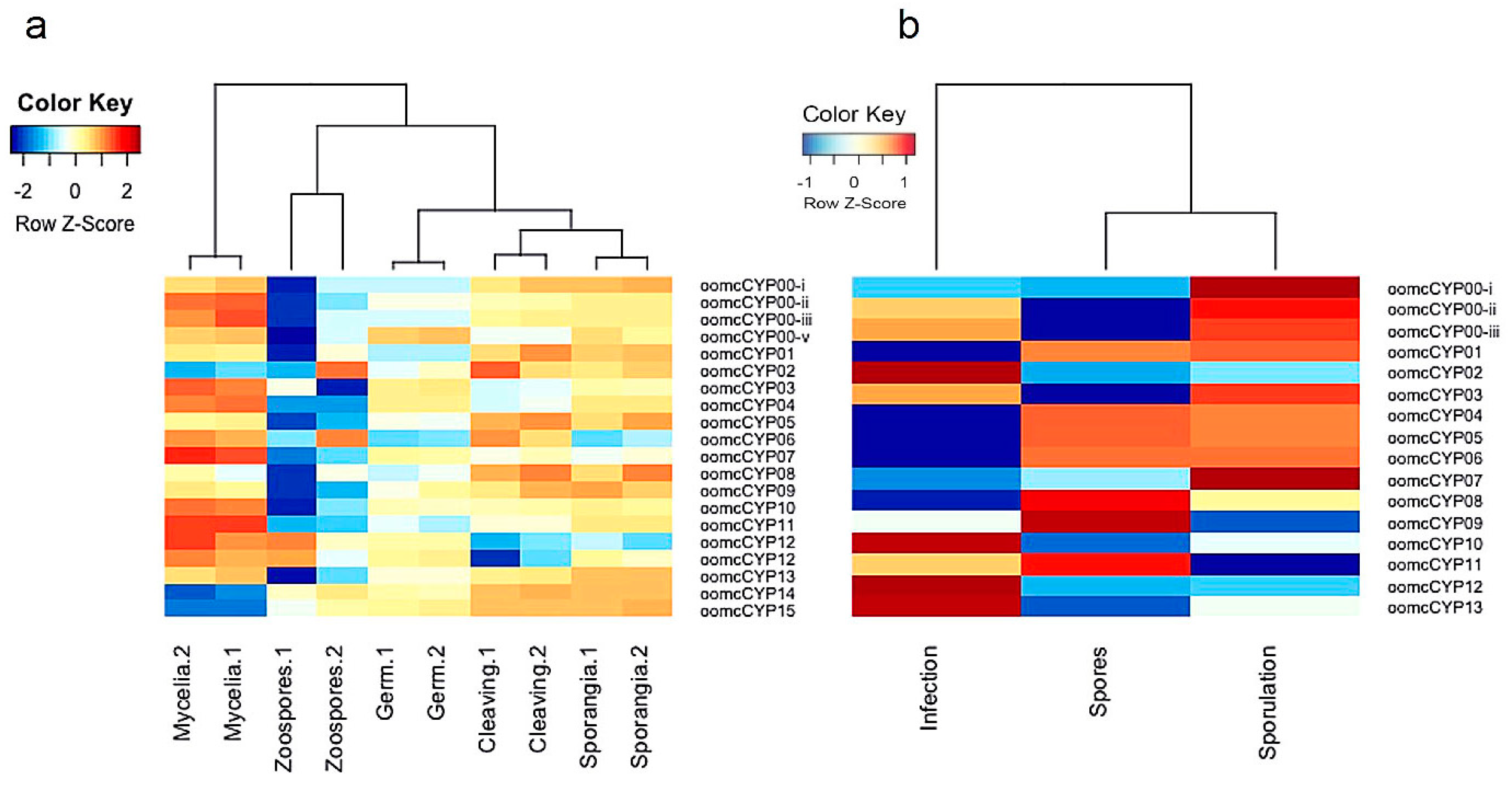
| Species Name | Obligate Biotroph? | CYP Proteins | CYP Domains Predicted Complete | oomcCYP00 | oomcCYP01 | oomcCYP02 | oomcCYP03 | oomcCYP04 | oomcCYP05 | oomcCYP06 | oomcCYP07 | oomcCYP08 | oomcCYP09 | oomcCYP10 | oomcCYP11 | oomcCYP12 | oomcCYP13 | oomcCYP14 | oomcCYP15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Albugo candida | + | 15 | 14 | 2 | 1 | 1 | 2 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 |
| Albugo laibachii | + | 15 | 15 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 0 | 0 | 0 | 0 |
| Aphanomyces astaci | - | 35 | 30 | 6 | 8 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 3 | 2 |
| Aphanomyces invadans | - | 33 | 30 | 7 | 1 | 6 | 2 | 2 | 1 | 3 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Bremia lactucae | + | 18 | 15 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| Hyaloperonospora arabidopsidis | + | 15 | 12 | 4 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| Peronospora effusa Race 13 | + | 16 | 14 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| Peronospora effusa Race 14 | + | 16 | 15 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| Peronospora tabacina J2 | + | 28 | 20 | 5 | 3 | 2 | 1 | 3 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 2 | 2 | 0 | 0 |
| Peron o spora tabacina S26 | + | 28 | 25 | 6 | 2 | 1 | 2 | 2 | 3 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 0 | 0 |
| Plasmopara halstedii | + | 16 | 15 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| Phytophthora infestans | - | 20 | 20 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| Phytophthora sojae | - | 19 | 19 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| Phytophthora ramorum | - | 20 | 19 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 2 | 1 |
| Pythium aphanidermatum | - | 20 | 17 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Pythium arrhenomanes | - | 22 | 18 | 6 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| Pythium insidiosum | - | 13 | 12 | 3 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 |
| Pythium irregulare | - | 20 | 20 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Pythium iwayam a i | - | 20 | 17 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Pythium ultimum | - | 20 | 18 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Pythium vexans | - | 20 | 18 | 4 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Saprolegnia diclina | - | 21 | 21 | 6 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Saprolegnia parasitica | - | 22 | 22 | 6 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Fletcher, K.; Han, R.; Michelmore, R.; Yang, R. Genome-Wide Analysis of Cyclophilin Proteins in 21 Oomycetes. Pathogens 2020, 9, 24. https://doi.org/10.3390/pathogens9010024
Zhang Y, Fletcher K, Han R, Michelmore R, Yang R. Genome-Wide Analysis of Cyclophilin Proteins in 21 Oomycetes. Pathogens. 2020; 9(1):24. https://doi.org/10.3390/pathogens9010024
Chicago/Turabian StyleZhang, Yan, Kyle Fletcher, Rongkui Han, Richard Michelmore, and Ruiwu Yang. 2020. "Genome-Wide Analysis of Cyclophilin Proteins in 21 Oomycetes" Pathogens 9, no. 1: 24. https://doi.org/10.3390/pathogens9010024
APA StyleZhang, Y., Fletcher, K., Han, R., Michelmore, R., & Yang, R. (2020). Genome-Wide Analysis of Cyclophilin Proteins in 21 Oomycetes. Pathogens, 9(1), 24. https://doi.org/10.3390/pathogens9010024





