1. Introduction
Rickettsia from the spotted fever group are important emerging vector borne infections of humans worldwide. These are obligate intracellular Gram-negative bacteria transmitted by ticks [
1]. The different SFG rickettsioses share several clinical features such as fever, headache, and rash, but may vary in disease severity [
1], and may have unique features (e.g. local lymphadenopathy [
1]), or different frequencies of occurrence (e.g. skin eschar (tache noire) formation at the tick bite site [
2]).
Molecular typing of infectious agents is an important tool for better understanding of ecological niches and identifying circulating species and their virulence [
3], as well as for epidemiologic and clinical use. In general, along with the development of the molecular typing and sequence analysis techniques the number of newly described SFG rickettsia species has increased [
4,
5].
Sequence analysis of PCR-amplified fragments of SFG targeting a number of genes has become a reliable and accepted method for the identification of rickettsia species. These genes encode for citrate synthase (
gltA),
Rickettsia-specific outer membrane protein (
rOmpA), 17kDa lipoprotein precursor antigen gene (
17kDa), as well as ribosomal
16S rRNA gene, among others [
3,
6,
7,
8,
9]. The
rOmpA encoding gene is present among all SFG members, as immune serum to
rOmpA reacts with all the SFG members. This gene is considered a good candidate for phylogenetic analysis, as it has been shown to exhibit high degrees of interspecies variation [
6]. Citrate synthase is a component of nearly all living cells and is a part of a central metabolic pathway, the citric acid cycle, which plays a key role in energy production and in providing important biosynthetic precursors [
7].
Previous studies from Israel found that the Israeli SFG
Rickettsia strain is
R. conorii israelensis, with substantial antigenic and genetic diversity existing within this strain [
10]. Recently however, other SFG rickettsia species have been molecularly identified in samples from questing ticks in Israel, including:
Rickettsia conorii caspia [
11],
Rickettsia massiliae [
2],
Rickettsia africae,
Candidatus Rickettsia barbariae [
3,
12], and others [
3,
13].
At the national rickettsiosis reference laboratory in Israel, nested PCR assays are the standard molecular diagnostic method for testing clinical samples. PCR methods for detection of
R. conorii in those samples include a nested assay for the
17kDa-protein encoding gene. This PCR-assay allows identification of samples as rickettsia members as well as the classification of these positive samples as either SFG or typhi group (TG) rickettsiosis. The sequence used in this diagnostic assay is highly conserved among rickettsiosis species. Similarly, since members of the
Rickettsia are closely related phylogenetically and have a high degree of
16S-RNA nucleotide sequence similarity [
14], the
16S-RNA was also not applicable for the differentiation between rickettsia strains. Here, in this study, three positive patients were further analyzed by sequencing of unique regions from two conserved rickettsial genes. These analyses revealed a new
Rickettsia isolate showing no absolute identity with any known
Rickettsia species present in NCBI database, which caused disease in three independent patients.
2. Results
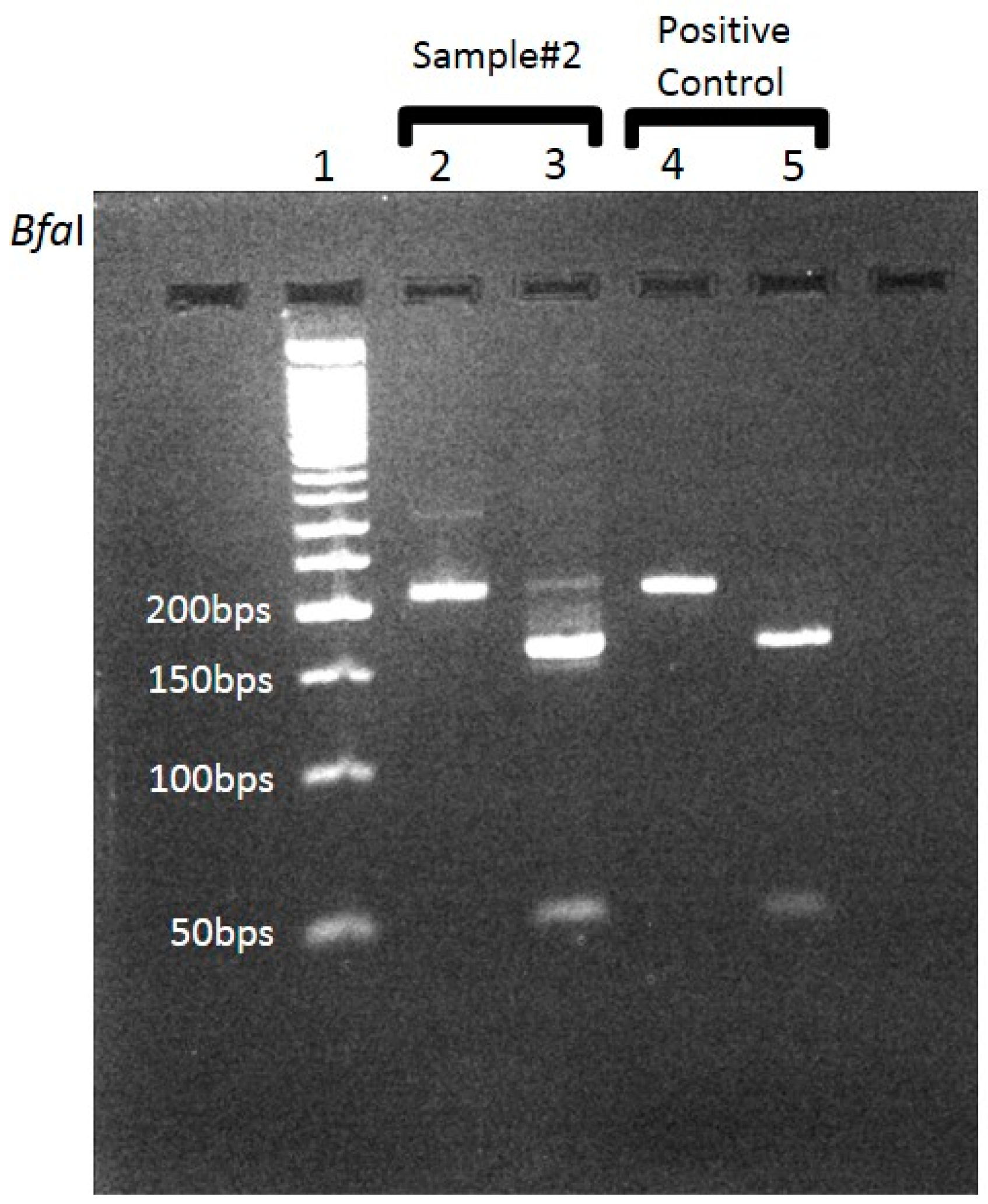
Whole blood samples and skin biopsy specimens from rash lesions taken (during admission) from three different patients suspected to have SFG rickettsiosis were received in our lab for diagnostic PCR assay. These samples were received from three different medical centers, located in distinct geographic regions in Israel, on different occasions. All samples were positive for SFG according to the differential PCR, based on the highly conserved
17kDa-protein encoding gene (
Figure 1) region, using appropriate controls. Following the attribution to the SFG group, these samples were further analyzed separately for determination of the SFG species by an additional PCR. We chose to focus on both
gltA and
rOmpA genomic regions. In order to ascertain the validity of this assay, it was previously validated by our group on an array of samples (data not shown). These included human and tick samples, found positive for SFG by the established diagnostic PCR assay. In addition, this assay was tested on DNA purified from known SFG strains (from our strain collection).
BLAST analysis of the
rOmpA diagnostic region (a 586 basepair region) showed no absolute identity with any known
Rickettsia species present in NCBI database. Species with high similarity to this isolate having 92.23%–96.89% homology were:
Rickettsia caspia (GenBank: U83437.1),
Rickettsia slovaca (GenBank: JX683121.1),
Rickettsia sibirica (GenBank: U83455.1), and
R. conorii israelensis (GenBank: U83441.1) (
Table 1). Phylogenetic tree analysis was performed using the EMBL–EBI Simple Phylogeny tool. The results did not conclusively allow the assignment of the new strain into an existing sub-group (
Figure S1.).
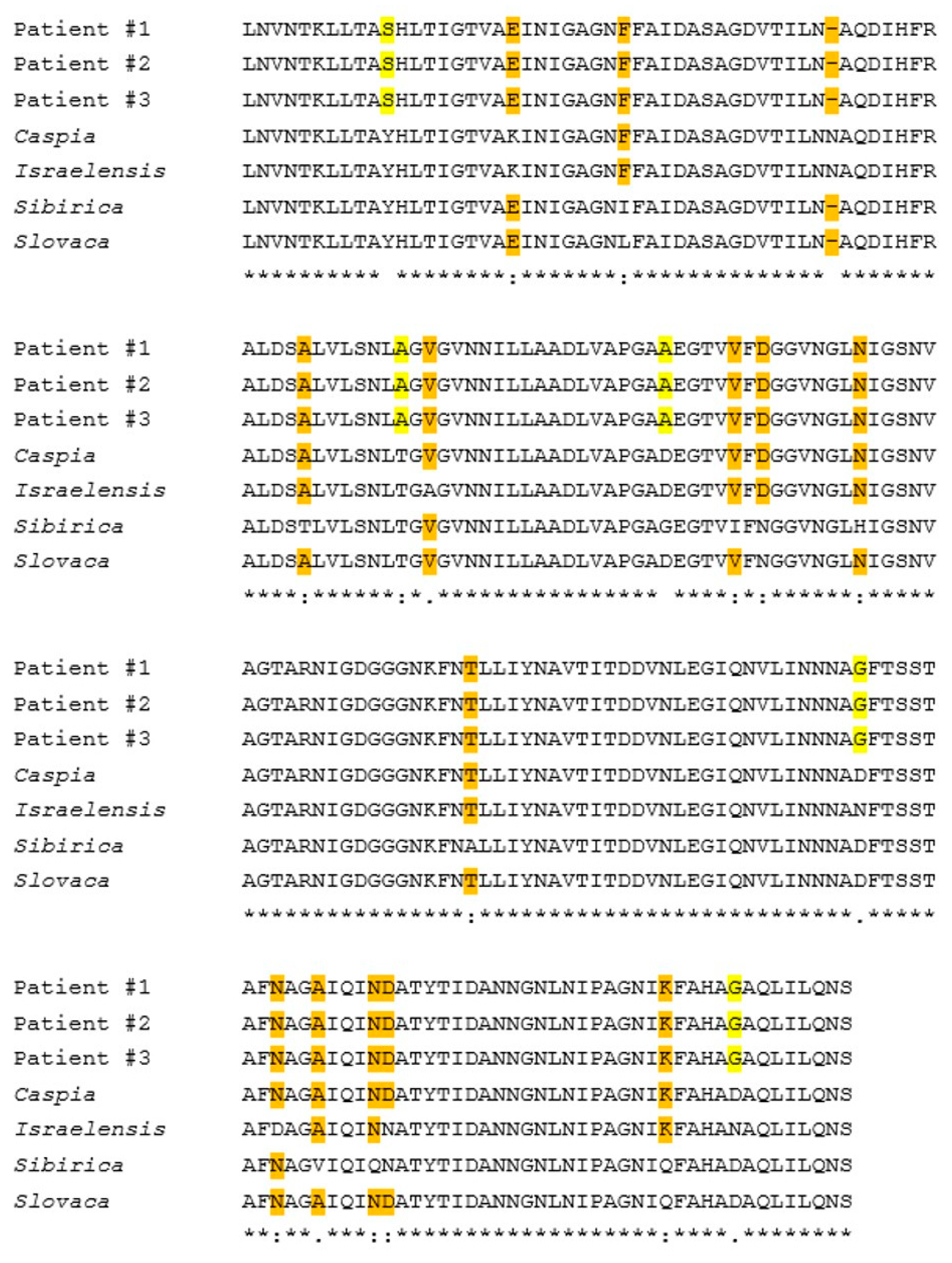
Multiple sequence alignment of the three samples provides evidence of the perfect match between these samples in the sequenced segments; meaning these samples were identical. When compared to documented
Rickettsia species we discovered 21 SNP mutations differentiating these samples from the above mentioned species (in the sequenced region) (
Figure S2). Of these 21 SNPs, 19 led to changes at the predicted protein translation as shown in
Figure 2. The remaining two SNPs were translationally-synonymous. Out of 194 amino-acids (AA) translation, five AA differed from the closest species: two of non-conservative groups; two are of semi-conservative groups; and one is conservative. The BLAST analysis and multiple sequence alignment analysis of the
gltA gene sequences of these three samples demonstrated the same identity seen between these three samples, as shown for the
rOmpA gene, despite containing less species-specific SNPs compared to other species (
Figure S3). Overall, these comparisons have revealed a new sequence segments for both
rOmpA and
gltA genes, which had no 100% identity to any other SFG species (
Figure 2).
3. Discussion
The
R. conorii group, the etiologic agent of Mediterranean spotted fever, includes several distinct subspecies: the
Malish,
Indian tick typhus rickettsia (ITTR),
Israeli spotted fever rickettsia (ISFR), and
Astrakhan fever rickettsia (AFR). These strains are closely related by genotype (four MLST genotypes with pairwise similarity in nucleotide sequence that varies from 98.2% to 100%), but differ serotypically and cause diseases with distinct clinical features [
15].
For example,
R. conorii israelensis, which is endemic to Israel [
16], has a higher ICU-admission and case-fatality rates than the
R. conorii Malish strain (29% vs. 13%; 36% vs. 22%) [
17]. In addition, the infections of the different strains often manifest clinically different according to strain (for example,
R. israelensis usually manifests as a fever disease with maculopapular rashes and may result in multi-organ failure and acute encephalitis [
18];
R. slovaca is implicated in development of tick-borne lymphadenopathy (TIBOLA) or dermacentor-borne necrosis erythema and lymphadenopathy (DEBONEL) [
19];
Rickettsia sibirica causes lymphadenopathy and lymphangitis [
20]; while
Rickesttsia caspia infections are characterized by febrile illness and a generalized maculopapular rash [
15]). These clinical distinctions according to strain constitute the motivation and significance of our finding.
In the current study, we have discovered a new rickettsia isolate from clinical samples of three independent patients with SFG rickettsiosis, based on sequence analysis of three genes that are highly conserved within the group. We identified novel SNP mutations in the rOmpA and gltA genes, common to these samples. The sequenced fragments of this isolate showed no identity to any other known species documented in the NCBI database. Within a sequence of 194 predicted amino acids, five have no similarity to any of the species that show the highest identity (caspia, conorii israelensis, slovaca, sibirica). These SNP mutations were identified in the rOmpA and gltA genes.
Rickettsial pathogens produce two conserved immunodominant outer membrane proteins, rOmpA and rOmpB, which are thought to be essential for pathogenesis. Mutations in these genes may affect virulence, as previously demonstrated by comparing virulent
R. rickettsii strains; in which rOmpA is present with the avirulent strain
Iowa, in which a premature stop codon prevents its production [
21]. Together, these clinical samples (from patients admitted to three different Hospitals in Israel, from distinct geographic locations, and temporally separated by one to two years from each other) establish the basis for the identification of a previously unknown new isolate that has clinical significance. Further research should focus on the endemic prevalence and distribution of this new isolate in Israel. This may be carried out by epidemiologic mapping of Israeli tick reservoir and routine molecular studying of all clinical cases of SFG in Israel.
This new isolate may also cause a distinct type of clinical presentation and outcome, possibly explaining the varying outcomes observed in patients with SFG rickettsiosis, not always explained strictly by host factors. Therefore, this new SFG rickettsia should be familiar to both physicians and researchers, since there are clinical and epidemiological implications on initial diagnosis, treatment and outcomes.
4. Materials and Methods
PCR amplification
Genomic DNA was extracted from clinical samples using the QIAamp DNA Mini Kit (Qiagen cat.51304) according to manufacturer’s instructions.
Diagnostic nested PCR and differentiating digestion were carried out as previously described [
22].
Further PCR reactions were performed separately for each sample, using the oligonucleotide primers listed in
Table 2. Most of the primers were designed according to conserved regions after alignment of about 15 SFG rickettsia species.
Five microliters (µL) of the extracted DNA were added to a 50 µL reaction mixture, containing: 5 µL of 5 picomol/µL of each primer, 10 µL of MyTaqTM 5x Buffer (Bioline cat.BIO-21111), 0.4 µL enzyme MyTaqTM (Bioline cat.BIO-21111) and 24.6 µL DDW.
The following conditions were used for amplification: initial denaturation for 3 min at 94 °C, followed by 40 cycles of annealing for 30 sec at 55 °C, extension for 1 min and 30 sec at 72 °C, denaturation for 30 sec at 94 °C. After these cycles, a final cycle of annealing for 30 sec at 55 °C, then 5 min at 72 °C to allow complete extension of the PCR products was performed.
PCR reactions were carried out in a T100TM thermal cycler (BIO-RAD, 1861096).
Sequencing reaction:
Sequencing using the same primers as used for amplification, was carried out by Hy Laboratories Ltd (hylabs) using an ABI 3730xl DNA Analyzer. Each sample was sequenced separately.
Data analysis:
The sequences used in this study were approved following a process of meticulous manual examination, looking into the quality of the specific peaks of the SNPs described. In addition, the fragments were separately sequenced on both directions, yielding 100% homologous results. Data was analyzed using the BioEdit Sequence Alignment Editor. BLAST was performed using NCBI BLAST (National Center for Biotechnology Information)–BLASTN for nucleotide BLAST or BLASTX for protein BLAST. Sequences were aligned using EMBL-EBI T-Coffee MSA tool.
Ethical statement:
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of The Sanz Medical Center, Lanaido Hospital, Netanya (Project identification codes 0002-19-LND).
Supplementary Materials
The following are available online at
https://www.mdpi.com/2076-0817/9/1/11/s1, Figure S1: Phylogenetic tree analysis of the new isolate along known rickettsial species according to: A.
rOmpA, B.
gltA, Figure S2: Multiple sequence alignment, for the
rOmpA nucleotide sequence of the three clinical samples (patients 1–3) and four known rickettsia species, Figure S3: Multiple sequence alignment, for the
gltA nucleotide sequence of the three clinical samples (patients 1–3) and four known rickettsia species.
Author Contributions
Data curation, D.K.; investigation, D.K., A.B.-D., and S.L.; methodology, D.K.; supervision, Y.A.-N.; writing—original draft, D.K. and Y.A.-N.; writing—review and editing, A.B.-D., R.C., I.G., and H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Faccini-Martínez, Á.A.; García-Álvarez, L.; Hidalgo, M.; Oteo, J.A. Syndromic classification of rickettsioses: An approach for clinical practice. Int. J. Infect. Dis. 2014, 28, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Harrus, S.; Perlman-Avrahami, A.; Mumcuoglu, K.Y.; Morick, D.; Baneth, G. Molecular detection of Rickettsia massiliae, Rickettsia sibirica mongolitimonae and Rickettsia conorii israelensis in ticks from Israel. Clin. Microbiol. Infect. 2011, 17, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Ereqat, S.; Nasereddin, A.; Al-Jawabreh, A.; Azmi, K.; Harrus, S.; Mumcuoglu, K.; Apanaskevich, D.; Abdeen, Z. Molecular Detection and Identification of Spotted Fever Group Rickettsiae in Ticks Collected from the West Bank, Palestinian Territories. PLoS Negl. Trop. Dis. 2016, 10, e0004348. [Google Scholar] [CrossRef] [PubMed]
- Weinert, L.A.; Werren, J.H.; Aebi, A.; Stone, G.N.; Jiggins, F.M. Evolution and diversity of Rickettsia bacteria. BMC Biol. 2009, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Tomassone, L.; Portillo, A.; Nováková, M.; De Sousa, R.; Oteo, J.A. Neglected aspects of tick-borne rickettsioses. Parasites Vectors 2018, 11, 263. [Google Scholar] [CrossRef] [PubMed]
- Fournier, P.E.; Roux, V.; Raoult, D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int. J. Syst. Evol. Microbiol. 1998, 48, 839–849. [Google Scholar] [CrossRef]
- Roux, V.; Rydkina, E.; Eremeeva, M.; Raoult, D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int. J. Syst. Evol. Microbiol. 1997, 47, 252–261. [Google Scholar] [CrossRef]
- Roux, V.; Raoult, D. Phylogenetic analysis of the genus Rickettsia by 16S rDNA sequencing. Res Microbiol. 1995, 146, 385–396. [Google Scholar] [CrossRef]
- Fournier, P.E.; Dumler, J.S.; Greub, G.; Zhang, J.; Wu, Y.; Raoult, D. Gene sequence-based criteria for identification of new rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J. Clin. Microbiol. 2003, 41, 5456–5465. [Google Scholar] [CrossRef]
- Walker, D.H.; Feng, H.M.; Saada, J.I.; Crocquet-Valdes, P.; Radulovic, S.; Popov, V.L.; Manor, E. Comparative antigenic analysis of spotted fever group rickettsiae from Israel and other closely related organisms. Am. J. Trop. Med. Hyg. 1995, 52, 569–576. [Google Scholar] [CrossRef]
- Kleinerman, G.; Baneth, G.; Mumcuoglu, K.Y.; van Straten, M.; Berlin, D.; Apanaskevich, D.A.; Harrus, S. Molecular detection of Rickettsia africae, Rickettsia aeschlimannii, and Rickettsia sibirica mongolitimonae in camels and Hyalomma spp. ticks from Israel. Vector Borne Zoonotic Dis. 2013, 13, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Waner, T.; Keysary, A.; Eremeeva, M.E.; Din, A.B.; Mumcuoglu, K.Y.; King, R.; Atiya-Nasagi, Y. Rickettsia africae and Candidatus Rickettsia barbariae in ticks in Israel. Am. J. Trop. Med. Hyg. 2014, 90, 920–922. [Google Scholar] [CrossRef] [PubMed]
- Keysary, A.; Eremeeva, M.E.; Leitner, M.; Din, A.B.; Wikswo, M.E.; Mumcuoglu, K.Y.; Inbar, M.; Wallach, A.D.; Shanas, U.; King, R.; et al. Spotted fever group rickettsiae in ticks collected from wild animals in Israel. Am. J. Trop. Med. Hyg. 2011, 85, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D.; Fournier, P.E.; Eremeeva, M.; Graves, S.; Kelly, P.J.; Oteo, J.A.; Sekeyova, Z.; Tamura, A.; Zhang, L. Naming of Rickettsiae and rickettsial diseases. Ann. N. Y. Acad. Sci. 2005, 1063, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Fournier, P.E.; Eremeeva, M.; Raoult, D. Proposal to create subspecies of Rickettsia conorii based on multi-locus sequence typing and an emended description of Rickettsia conorii. BMC Microbiol. 2005, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Mumcuoglu, K.Y.; Keysary, A.; Gilead, L. Mediterranean spotted fever in Israel: A tick-borne disease. Isr. Med. Assoc. J. 2002, 4, 44–49. [Google Scholar] [PubMed]
- De Sousa, R.; França, A.; Nòbrega, S.D.; Belo, A.; Amaro, M.; Abreu, T.; Poças, J.; Proença, P.; Vaz, J.; Torgal, J.; et al. Host- and microbe-related risk factors for and pathophysiology of fatal Rickettsia conorii infection in Portuguese patients. J. Infect. Dis. 2008, 198, 576–585. [Google Scholar] [CrossRef]
- Cohen, R.; Babushkin, F.; Shapiro, M.; Uda, M.; Atiya-Nasagi, Y.; Klein, D.; Finn, T. Two Cases of Israeli Spotted Fever with Purpura Fulminans, Sharon District, Israel. Emerg. Infect. Dis. 2018, 24, 835–840. [Google Scholar] [CrossRef]
- Blanda, V.; Torina, A.; La Russa, F.; D’Agostino, R.; Randazzo, K.; Scimeca, S.; Giudice, E.; Caracappa, S.; Cascio, A.; de la Fuente, J. A retrospective study of the characterization of Rickettsia species in ticks collected from humans. Ticks Tick Borne Dis. 2017, 8, 610–614. [Google Scholar] [CrossRef]
- Kuscu, F.; Orkun, O.; Ulu, A.; Kurtaran, B.; Komur, S.; Inal, A.S.; Erdogan, D.; Tasova, Y.; Aksu, H.S. Rickettsia sibirica mongolitimonae Infection, Turkey, 2016. Emerg. Infect. Dis. 2017, 23, 1214–1216. [Google Scholar] [CrossRef]
- Clark, T.R.; Noriea, N.F.; Bublitz, D.C.; Ellison, D.W.; Martens, C.; Lutter, E.I.; Hackstadt, T. Comparative genome sequencing of Rickettsia rickettsii strains that differ in virulence. Infect. Immun. 2015, 83, 1568–1576. [Google Scholar] [CrossRef] [PubMed]
- Leitner, M.; Yitzhaki, S.; Rzotkiewicz, S.; Keysary, A. Polymerase chain reaction-based diagnosis of Mediterranean spotted fever in serum and tissue samples. Am. J. Trop. Med. Hyg. 2002, 67, 166–169. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).