The Impact of SsPI-1 Deletion on Streptococcus suis Virulence
Abstract
1. Introduction
2. Results
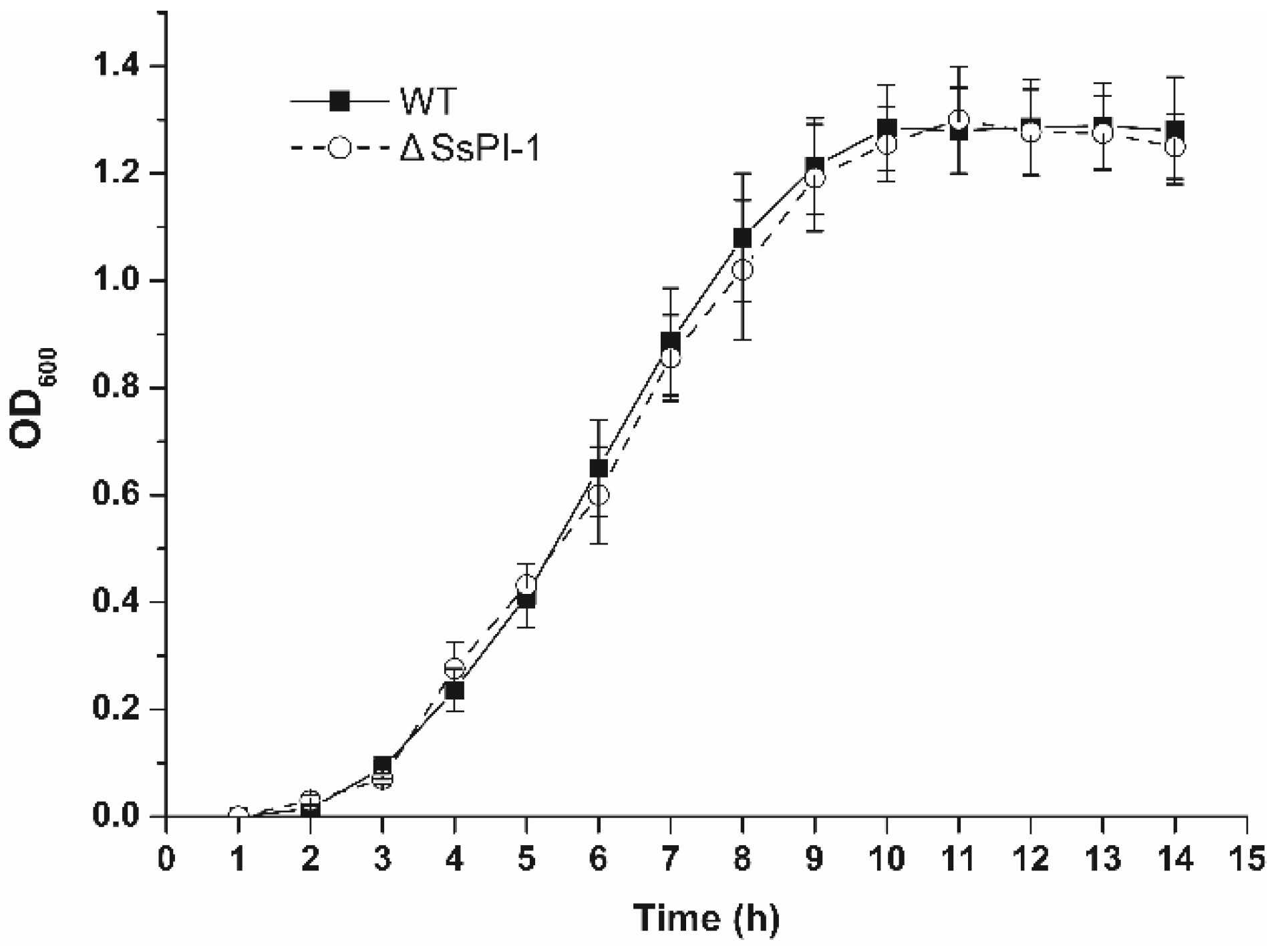
2.1. Elimination of SsPI-1 Does Not Affect the General Biological Characteristics of S. suis
2.2. Deletion of SsPI-1 Significantly Impairs the Adhesion Ability of S. suis
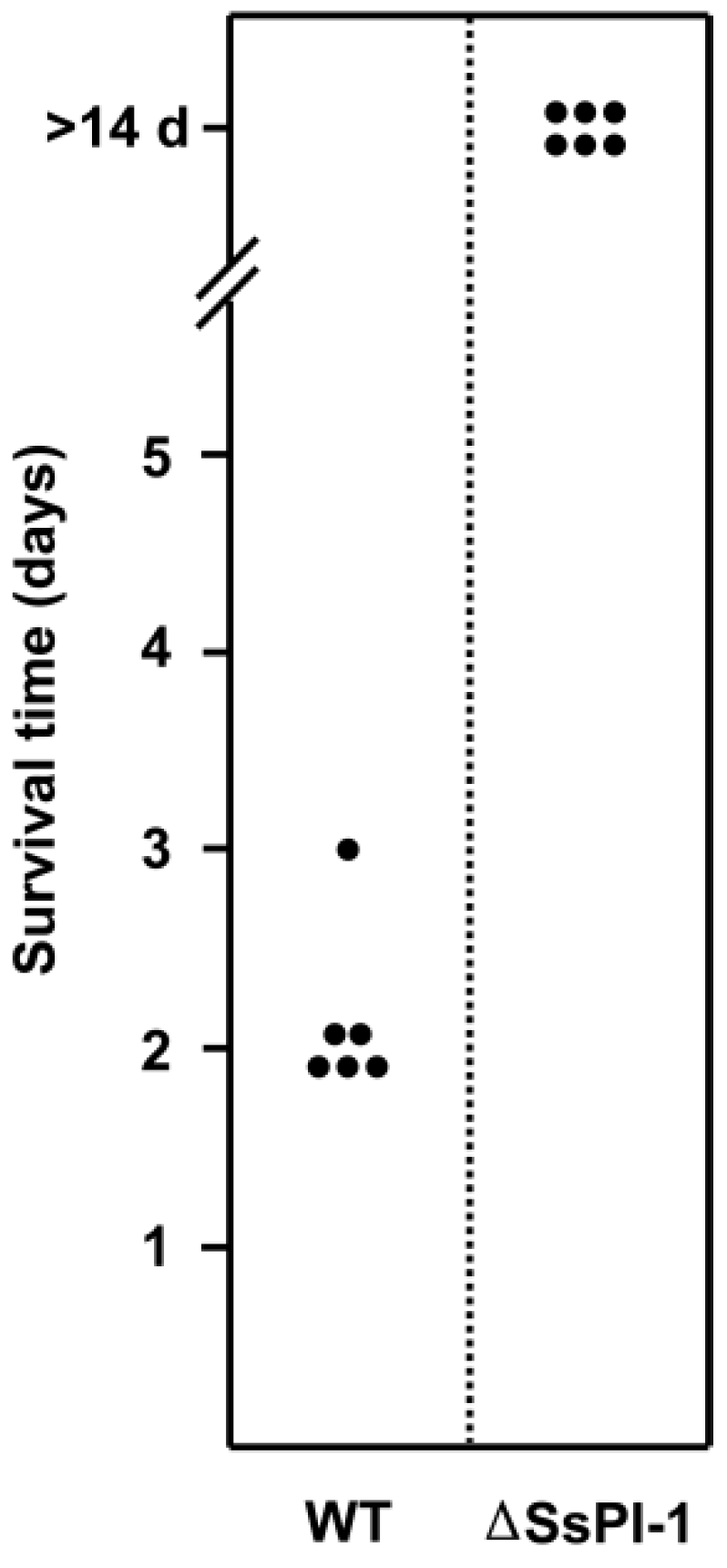
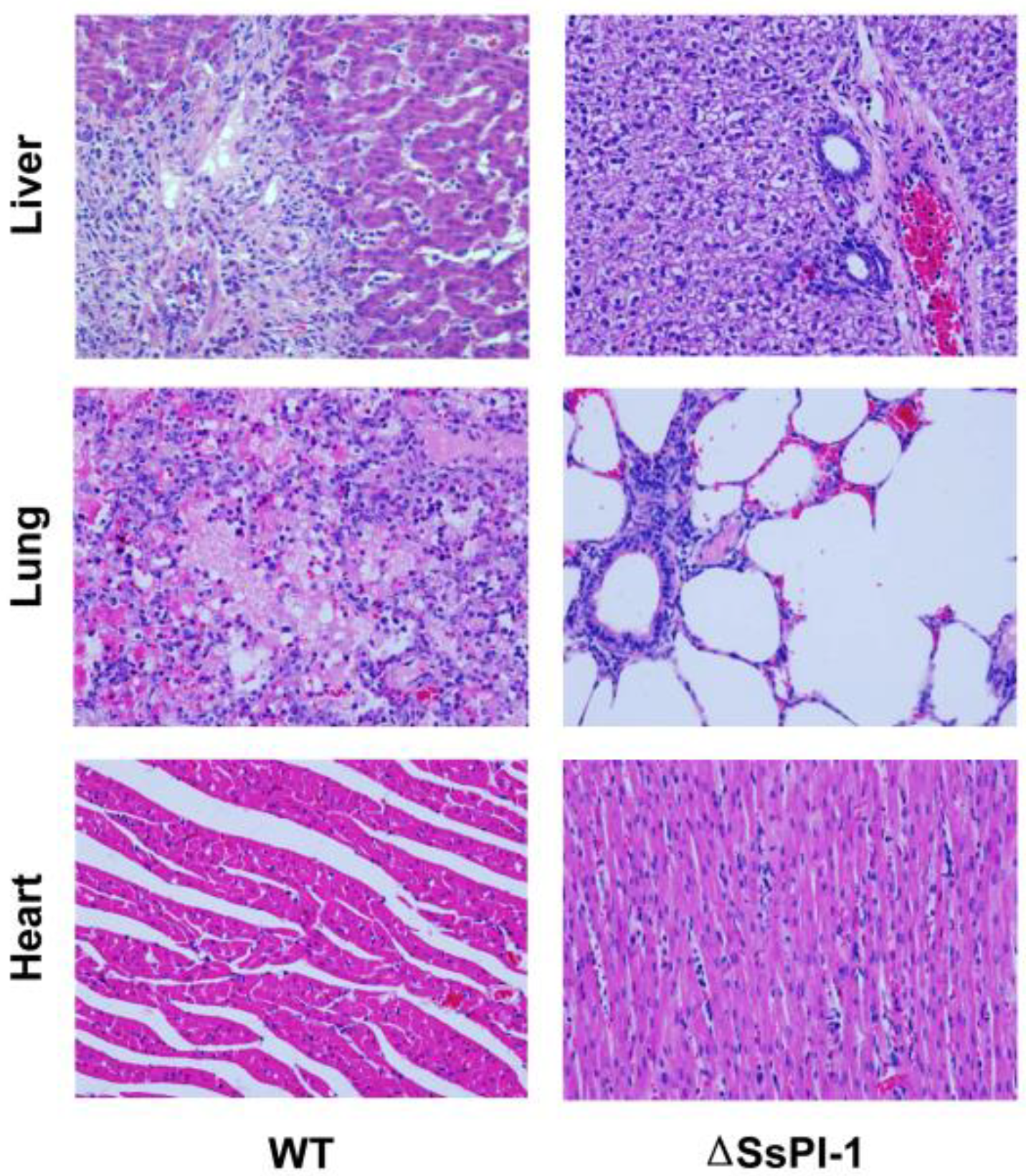
2.3. SsPI-1 Is Essential for the Full Virulence of S. suis
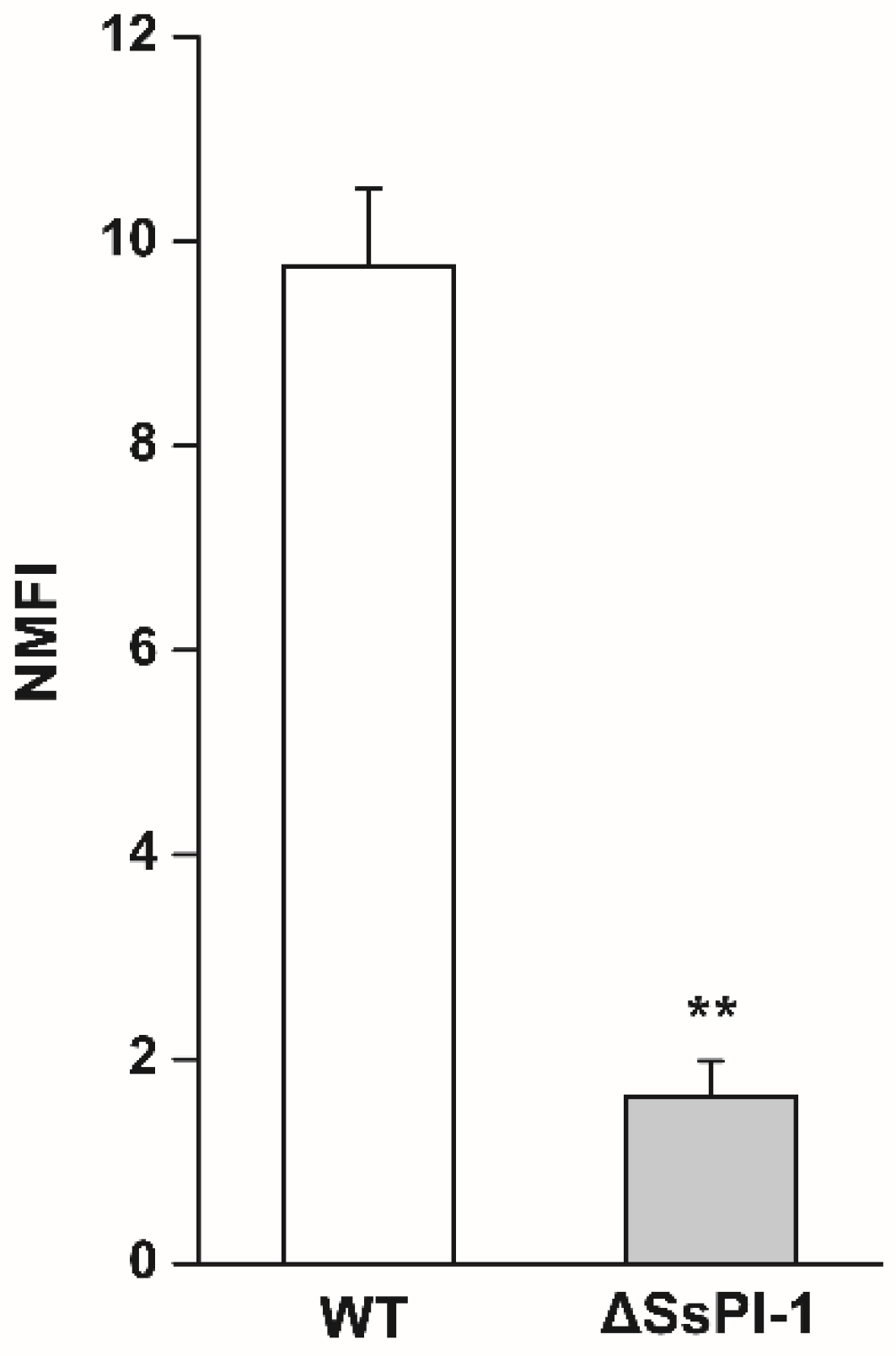
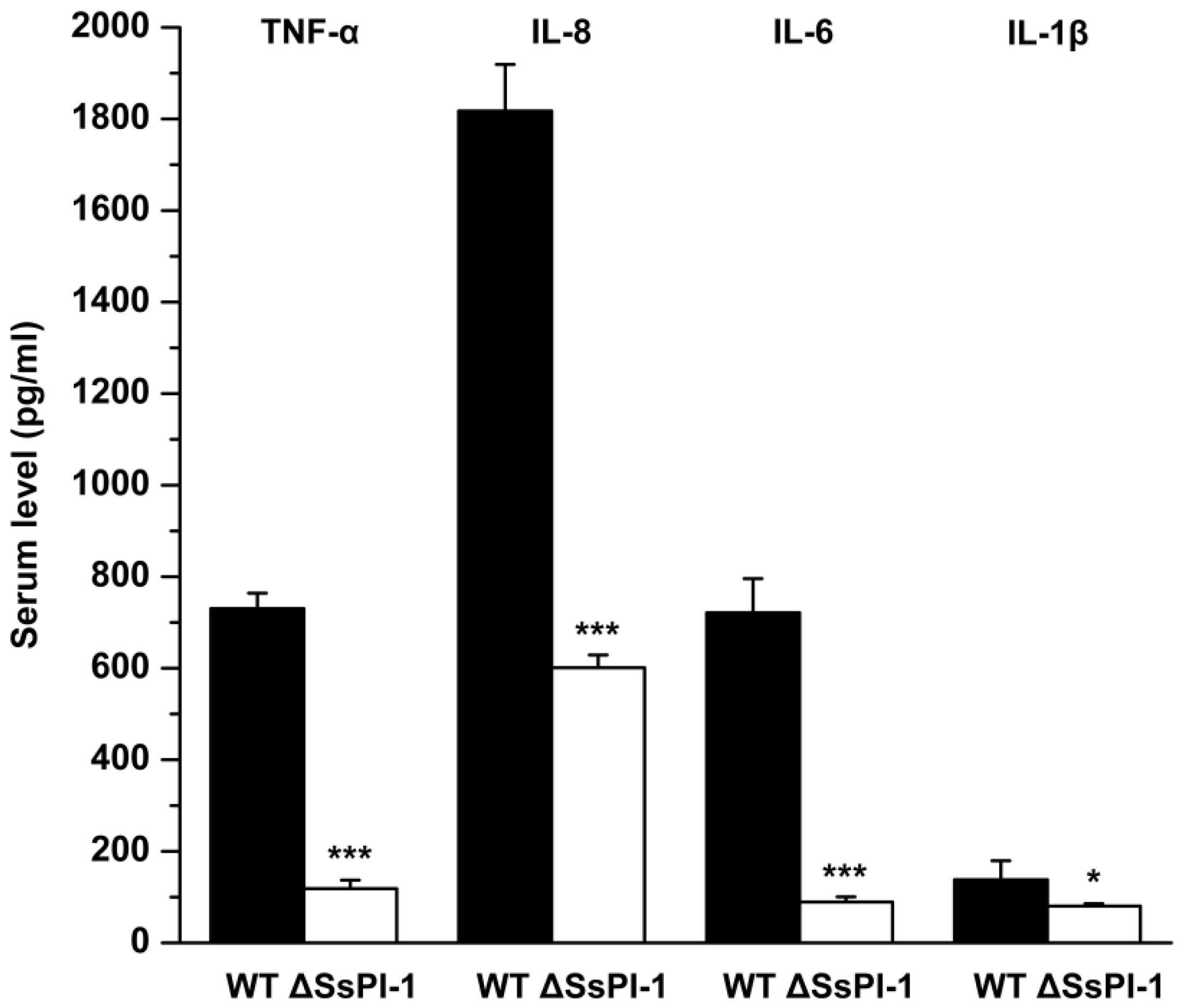
2.4. Elimination of SsPI-1 Markedly Decreased Proinflammatory Cytokine Production In Vivo
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Cell Adhesion Analysis
4.3. Experimental Infections of Piglets
4.4. Enzyme-Linked Immunosorbent Assay (ELISA)
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent—An update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef] [PubMed]
- Lun, Z.R.; Wang, Q.P.; Chen, X.G.; Li, A.X.; Zhu, X.Q. Streptococcus suis: An emerging zoonotic pathogen. Lancet Infect. Dis. 2007, 7, 201–209. [Google Scholar] [CrossRef]
- Feng, Y.J.; Zhang, H.M.; Ma, Y.; Gao, G.F. Uncovering newly emerging variants of Streptococcus suis, an important zoonotic agent. Trends Microbiol. 2010, 18, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.F.; Nghia, H.D.; Taylor, W.; Schultsz, C. Streptococcus suis: An emerging human pathogen. Clin. Infect. Dis. 2009, 48, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Segura, M. Streptococcus suis: An emerging human threat. J. Infect. Dis. 2009, 199, 4–6. [Google Scholar] [CrossRef]
- Tang, J.; Wang, C.; Feng, Y.; Yang, W.; Song, H.; Chen, Z.; Yu, H.; Pan, X.; Zhou, X.; Wang, H.; et al. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 2006, 3, e151. [Google Scholar]
- Gottschalk, M.; Segura, M.; Xu, J. Streptococcus suis infections in humans: The Chinese experience and the situation in North America. Anim. Health Res. Rev. 2007, 8, 29–45. [Google Scholar] [CrossRef]
- Lefebure, T.; Stanhope, M.J. Evolution of the core and pan-genome of Streptococcus: Positive selection, recombination, and genome composition. Genome Biol. 2007, 8, R71. [Google Scholar] [CrossRef]
- Feng, Y.J.; Zhang, H.M.; Wu, Z.W.; Wang, S.H.; Cao, M.; Hu, D.; Wang, C.J. Streptococcus suis infection: An emerging/reemerging challenge of bacterial infectious diseases? Virulence 2014, 5, 477–497. [Google Scholar] [CrossRef]
- Wu, Z.; Li, M.; Wang, C.; Li, J.; Lu, N.; Zhang, R.; Jiang, Y.; Yang, R.; Liu, C.; Liao, H.; et al. Probing genomic diversity and evolution of Streptococcus suis serotype 2 by NimbleGen tiling arrays. BMC Genom. 2011, 12, 219. [Google Scholar] [CrossRef]
- Chen, C.; Tang, J.; Dong, W.; Wang, C.; Feng, Y.; Wang, J.; Zheng, F.; Pan, X.; Liu, D.; Li, M.; et al. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS ONE 2007, 2, e315. [Google Scholar] [CrossRef] [PubMed]
- Hacker, J.; Carniel, E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001, 2, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Dobrindt, U.; Hochhut, B.; Hentschel, U.; Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004, 2, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Yang, M.; Hu, P.; Wu, J.; Chen, B.; Hua, Y.; Yu, J.; Chen, H.; Xiao, J.; Jin, M. Comparative genomic analysis of Streptococcus suis reveals significant genomic diversity among different serotypes. BMC Genom. 2011, 12, 523. [Google Scholar] [CrossRef]
- Li, M.; Shen, X.; Yan, J.; Han, H.; Zheng, B.; Liu, D.; Cheng, H.; Zhao, Y.; Rao, X.; Wang, C.; et al. GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol. Microbiol. 2011, 79, 1670–1683. [Google Scholar] [CrossRef]
- Yao, X.Y.; Chen, T.; Shen, X.D.; Zhao, Y.; Wang, M.; Rao, X.C.; Yin, S.P.; Wang, J.; Gong, Y.L.; Lu, S.G.; et al. The chromosomal SezAT toxin-antitoxin system promotes the maintenance of the SsPI-1 pathogenicity island in epidemic Streptococcus suis. Mol. Microbiol. 2015, 98, 243–257. [Google Scholar] [CrossRef]
- Shen, X.; Zhong, Q.; Zhao, Y.; Yin, S.; Chen, T.; Hu, F.; Li, M. Proteome analysis of the two-component SalK/SalR system in Epidemic Streptococcus suis serotype 2. Curr. Microbiol. 2013, 67, 118–122. [Google Scholar] [CrossRef]
- Li, M.; Wang, C.; Feng, Y.; Pan, X.; Cheng, G.; Wang, J.; Ge, J.; Zheng, F.; Cao, M.; Dong, Y.; et al. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS ONE 2008, 3, e2080. [Google Scholar] [CrossRef]
- Xu, J.; Fu, S.; Liu, M.; Xu, Q.; Bei, W.; Chen, H.; Tan, C. The two-component system NisK/NisR contributes to the virulence of Streptococcus suis serotype 2. Microbiol. Res. 2014, 169, 541–546. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, G.; Li, S.; Wang, M.; Song, J.; Wang, J.; Tang, J.; Li, M.; Hu, F. Role of a type IV-like secretion system of Streptococcus suis 2 in the development of Streptococcal toxic shock syndrome. J. Infect. Dis. 2011, 204, 274–281. [Google Scholar] [CrossRef]
- Yao, X.Y.; Li, M.; Wang, J.; Wang, C.J.; Hu, D.; Zheng, F.; Pan, X.Z.; Tan, Y.L.; Zhao, Y.; Hu, L.W.; et al. Isolation and characterization of a native avirulent strain of Streptococcus suis serotype 2: A perspective for vaccine development. Sci. Rep. 2015, 5, 9835. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Zheng, H.; Zhang, J.; Jing, H.; Wang, L.; Xiong, Y.; Wang, W.; Zhou, Z.; Sun, Q.; Luo, X.; et al. Clinical, experimental, and genomic differences between intermediately pathogenic, highly pathogenic, and epidemic Streptococcus suis. J. Infect. Dis. 2009, 199, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, J.; Yu, J.; Xu, Z.; Liu, L.; Song, Y.; Sun, X.; Zhang, A.; Jin, M. HP1330 contributes to Streptococcus suis virulence by inducing Toll-like receptor 2-and ERK1/2-dependent pro-inflammatory responses and influencing in vivo S. suis loads. Front. Immunol. 2017, 8, 869. [Google Scholar] [CrossRef] [PubMed]
- Lachance, C.; Gottschalk, M.; Gerber, P.; Lemire, P.; Xu, J.; Segura, M. Exacerbated type II interferon response drives hypervirulence and toxic shock by an emergent epidemic strain of Streptococcus suis. Infect. lmmun. 2013, 81, 1928–1939. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, J.; Lin, L.; Pan, S.; Fu, L.; Han, L.; Jin, M.; Zhou, R.; Zhang, A. Targeting TREM-1 signaling in the presence of antibiotics is effective against Streptococcal toxic-shock-like syndrome (STSLS) caused by Streptococcus suis. Front. Cell Infect. Microbiol. 2015, 5, 79. [Google Scholar] [CrossRef]
- Yin, S.P.; Li, M.; Rao, X.C.; Yao, X.Y.; Zhong, Q.; Wang, M.; Wang, J.; Peng, Y.Z.; Tang, J.Q.; Hu, F.Q.; et al. Subtilisin-like protease-1 secreted through type IV secretion system contributes to high virulence of Streptococcus suis 2. Sci. Rep. 2016, 6, 27369. [Google Scholar] [CrossRef]
- Jiang, X.; Yang, Y.; Zhou, J.; Zhu, L.; Gu, Y.; Zhang, X.; Li, X.; Fang, W. Roles of the putative type IV-like secretion system key component VirD4 and PrsA in pathogenesis of Streptococcus suis type 2. Front. Cell. Infect. Microbiol. 2016, 6, 172. [Google Scholar] [CrossRef]
- Xu, Z.M.; Chen, B.; Zhang, Q.; Liu, L.; Zhang, A.D.; Yang, Y.J.; Huang, K.S.; Yan, S.X.; Yu, J.P.; Sun, X.M.; et al. Streptococcus suis 2 transcriptional regulator TstS stimulates cytokine production and bacteremia to promote Streptococcal toxic shock-like syndrome. Front. Microbiol. 2018, 9, 1309. [Google Scholar] [CrossRef]
- Zheng, J.X.; Li, Y.; Zhang, H.; Fan, H.J.; Lu, C.P. Identification and characterization of a novel hemolysis-related gene in Streptococcus suis serotype 2. PLoS ONE 2013, 8, e74674. [Google Scholar] [CrossRef]
- Bi, L.L.; Pian, Y.Y.; Chen, S.L.; Ren, Z.Q.; Liu, P.; Lv, Q.Y.; Zheng, Y.L.; Zhang, S.W.; Hao, H.J.; Yuan, Y.; et al. Toll-like receptor 4 confers inflammatory response to Suilysin. Front. Microbiol. 2015, 6, 644. [Google Scholar] [CrossRef]
- Lin, L.; Xu, L.; Lv, W.H.; Han, L.; Xiang, Y.Z.; Fu, L.; Jin, M.L.; Zhou, R.; Chen, H.C.; Zhang, A.D. An NLRP3 inflammasome-triggered cytokine storm contributes to Streptococcal toxic shock-like syndrome (STSLS). PLoS Pathog. 2019, 15, e1007795. [Google Scholar] [CrossRef] [PubMed]
- Segura, M. Streptococcus suis vaccines: Candidate antigens and progress. Expert Rev. Vaccines 2015, 14, 1587–1608. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Ji, H.; Cao, M.; Wang, C.; Feng, Y.; Li, M.; Pan, X.; Wang, J.; Qin, Y.; Hu, F.; et al. Contribution of the Rgg transcription regulator to metabolism and virulence of Streptococcus suis serotype 2. Infect. Immun. 2011, 79, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Li, G.; Yao, X.-Y.; Lu, S.-G.; Wang, J.; Shen, X.-D.; Li, M. The Impact of SsPI-1 Deletion on Streptococcus suis Virulence. Pathogens 2019, 8, 287. https://doi.org/10.3390/pathogens8040287
Zhao Y, Li G, Yao X-Y, Lu S-G, Wang J, Shen X-D, Li M. The Impact of SsPI-1 Deletion on Streptococcus suis Virulence. Pathogens. 2019; 8(4):287. https://doi.org/10.3390/pathogens8040287
Chicago/Turabian StyleZhao, Yan, Gang Li, Xin-Yue Yao, Shu-Guang Lu, Jing Wang, Xiao-Dong Shen, and Ming Li. 2019. "The Impact of SsPI-1 Deletion on Streptococcus suis Virulence" Pathogens 8, no. 4: 287. https://doi.org/10.3390/pathogens8040287
APA StyleZhao, Y., Li, G., Yao, X.-Y., Lu, S.-G., Wang, J., Shen, X.-D., & Li, M. (2019). The Impact of SsPI-1 Deletion on Streptococcus suis Virulence. Pathogens, 8(4), 287. https://doi.org/10.3390/pathogens8040287



