Abstract
Protein phosphorylation/dephosphorylation is an important regulatory mechanism that controls many key physiological processes. Numerous pathogens successfully use kinases and phosphatases to internalize, replicate, and survive, modifying the host′s phosphorylation profile or signal transduction pathways. Multiple phosphatases and kinases from diverse bacterial pathogens have been implicated in human infections before. In this work, we have identified and characterized the dual specificity protein/lipid phosphatase LmDUSP1 as a novel virulence factor governing Leishmania mexicana infection. The LmDUSP1-encoding gene (LmxM.22.0250 in L. mexicana) has been acquired from bacteria via horizontal gene transfer. Importantly, its orthologues have been associated with virulence in several bacterial species, such as Mycobacterium tuberculosis and Listeria monocytogenes. Leishmania mexicana with ablated LmxM.22.0250 demonstrated severely attenuated virulence in the experimental infection of primary mouse macrophages, suggesting that this gene facilitates Leishmania pathogenicity in vertebrates. Despite significant upregulation of LmxM.22.0250 expression in metacyclic promastigotes, its ablation did not affect the ability of mutant cells to differentiate into virulent stages in insects. It remains to be further investigated which specific biochemical pathways involve LmDUSP1 and how this facilitates the parasite′s survival in the host. One of the interesting possibilities is that LmDUSP1 may target host′s substrate(s), thereby affecting its signal transduction pathways.
1. Introduction
The genus Leishmania (Kinetoplastea, Trypanosomatidae) contains parasitic flagellates causing leishmaniasis. This disease is clinically pleomorphic, varying from fairly harmless self-healing skin lesions to fatal visceral organ failure [1]. Leishmania spp. are dixenous parasites, i.e. they have two different hosts in their life cycle [2,3]. The flagellated extracellular promastigotes develop in the gut of insect vectors, while the intracellular non-flagellated amastigotes occupy the phagolysosomes of vertebrates′ macrophages [4,5]. Genes and gene products, governing differentiation or infection maintenance, are putative virulence factors. At present, they are identified primarily by the next-generation sequencing (NGS)-based approaches [3,6] and analyzed using functional genomics [7,8,9,10,11].
One of the genes previously identified in our analyses is LmxM.22.0250. Its orthologues were upregulated in metacyclic promastigotes of Leishmania mexicana M379 and in a virulent strain (as compared to its avirulent kin) of Leishmania major LV561 [12]. Confirming this and highlighting its potential importance, LmxM.22.0250 orthologue was one of only four genes consistently upregulated in both fly-derived and axenically differentiated metacyclic promastigotes of L. major [13]. This gene is present in most trypanosomatid lineages, genomes of which are available from the TriTrypDB [14]. Remarkably, it appears to be selectively lost from the genomes of Trypanosoma and Blechomonas spp. [15], while its distribution within Leishmaniinae is not uniform. Orthologues of LmxM.22.0250 are pseudogenized in some Leishmania species or strains [16].
LmxM.22.0250 encodes a dual specificity phosphatase (DUSP), which we named LmDUSP1. In general, such enzymes can recognize and work upon more than one molecular substrate [17]. Specifically, LmDUSP1 can dephosphorylate both proteins and lipids [16,18]. It contains a conserved P-loop signature sequence HCXXGKDR, governing substrate binding and its subsequent dephosphorylation [19]. This enzyme can remove phosphates from phospho-tyrosine (P-Tyr) and phospho-serine (P-Ser) at the rates of 12 and 0.6 nmoles/mg min, respectively, indicating that it has a higher preference towards P-Tyr. In addition, it can also dephosphorylate different mono-phosphorylated inositols at the rate of about 10 nmoles/mg min [18]. Of note, all these biochemical activities have been measured in vitro, and it remains to be established what role LmDUSP1 plays under physiological conditions. Another lipid phosphatase (encoded by LbrM.25.21.80) may contribute to the parasite virulence, as it was shown to be among the 100 most abundantly expressed genes in Leishmania braziliensis skin lesions of humans with cutaneous leishmaniasis [20].
It has been previously shown that LmDUSP1 has bacterial origin and is similar to several proteins identified as virulence factors in some pathogenic microorganisms [16,18]. The most notable examples are secreted phosphatases MptpB from Mycobacterium tuberculosis [21] and LipA from Listeria monocytogenes [22]. Two secreted tyrosine phosphatases of M. tuberculosis, MptpA and MptpB, play an important role in the interaction of the pathogen with the host cell [23]. While MptpA homologs are present in both pathogenic (M. tuberculosis complex) and non-pathogenic (e.g. Mycobacterium smegmatis) mycobacteria, MptpB is restricted to members of the former complex. Importantly, mycobacteria with inhibited MptpB or ablated mptpB gene are severely defective in their ability to survive in activated mouse macrophages and in guinea pigs [24,25]. It has been hypothesized that MptpB dephosphorylates the P-Tyr residues of the myelin basic protein, thus enabling the mycobacteria to survive within its host [23,26]. Similarly, the virulence of Listeria monocytogenes strains lacking the lmo1800 gene, which encodes a secreted tyrosine phosphatase LipA, is severely attenuated in vivo [22].
In this work, we investigated the role of LmDUSP1 in Leishmania virulence both in vitro and in vivo.
2. Results
2.1. LmxM.22.0250 Encodes a Dual Specificity Phosphatase LmDUSP1: in Silico Analyses
Phylogenetic analysis revealed clustering of the putative LmxM.22.0250 homologs into four clades. One of them included the gene under study along with the sequences identified in all other Leishmaniinae, whose genomes had been sequenced thus far, and those of the genus Strigomonas from the endosymbiont-harboring subfamily Strigomonadinae (Figure 1). Of note, all recognized subgenera of Leishmania (Leishmania, Viannia¸ Sauroleishmania, and Mundinia [27,28]) were included in the analysis, but the homologous sequences of the L. (Viannia) species were pseudogenized. The three other clades composed a super-clade, which was sister to the first one. Only one of these three clades contained sequences of Leishmania spp., which were exclusively from the members of the nominative subgenus L. (Leishmania). In addition, this clade included one gene from Leptomonas pyrrhocoris and one from a single member of Strigomonadinae, Angomonas deanei. The third and fourth clades contained genes of Strigomonadinae and monoxenous Leishmaniinae, and those of Strigomonas, Phytomonas spp., and Paratrypanosoma confusum, respectively. The inferred phylogeny of DUSPs and, in particular, the absence of this gene in the free-living eubodonid Bodo saltans and its presence in various clades, including the earliest branching P. confusum, suggests that the acquisition this gene occurred via lateral gene transfer from a bacterium in the last common ancestor of all trypanosomatids. Subsequently, it experienced at least three ancient duplications, which occurred not later than in the common ancestor of Leishmaniinae and Strigomonadinae, and multiple independent losses in different lineages of Trypanosomatidae. In trypanosomes and monoxenous parasites of fleas (Blechomonas spp.), these losses resulted in a complete ablation of the DUSP-encoding genes.
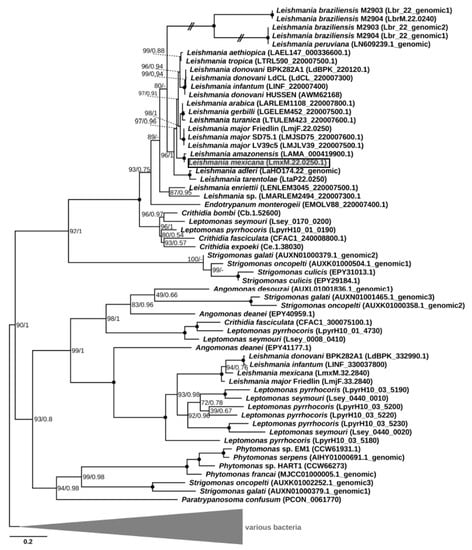
Figure 1.
Maximum-likelihood phylogenetic tree of LmxM.22.0250 and its homologs. Numbers at the nodes represent bootstrap percentage/posterior probability. Nodes with maximal support are marked with black circles. Dashes indicate different topology in the Bayesian tree. The scale bar shows the number of substitutions per site. Double-crossed branches are shown at 25% of their actual length. The gene of interest is boxed and shaded. The tree is rooted with bacterial sequences. In cases where a sequence was not annotated in the genome and could be identified only using TBLASTN, a sequence ID is in the following format: chromosome#_genomic.
It is not clear whether the genes falling into different clades have the same function, since the corresponding proteins are characterized by rather low sequence identity, even within the same species. For example, the amino acid sequence identity between the two paralogs found in L. mexicana (LmxM.22.0250 and LmxM.32.2840) is just ~28%. In all the analyzed sequences except for the pseudogenes of L. (Viannia), a tyrosine phosphatase domain was clearly identifiable.
2.2. Conventional Genetic Ablation of LmxM.22.0250
To examine whether LmDUSP1 is involved in L. mexicana virulence, we first subsequently deleted two alleles of LmxM.22.0250 by replacing them with antibiotic resistance genes for Sat and Hyg (Figure S1A). Similarly to the case of LmxBTN1 reported previously [9], Southern blotting showed that LmxM.22.0250 ablation was not complete (Figure S1B, panel 5′ UTR). We concluded that at least one extra copy of this gene must be present elsewhere in the L. mexicana genome. Whether this is a naturally occurring phenomenon or a result of genetic manipulations remains to be examined further.
2.3. CRISPR-Cas9-mediated Genetic Ablation of LmxM.22.0250
In order to avoid the hurdles with LmxM.22.0250 conventional knock-out, we decided to use the CRISPR-Cas9 system, previously established in our lab [9], to ablate this gene in L. mexicana (Figure 2). The gRNA under control of the U6 promoter in forward orientation was stably expressed from the L. mexicana 18S ribosomal DNA locus. PCR (Figure 2B), Southern blotting (Figure 2C), and qRT-PCR (quantitative Reverse Transcription Polymerase Chain Reaction) data (Figure 2D) confirmed complete ablation of the LmxM.22.0250 gene. The resulting strain was named LmDUSP1 KO.
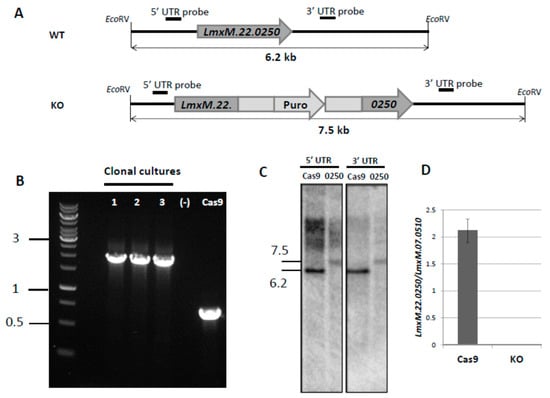
Figure 2.
Knock-out of LmxM.22.0250 with CRISPR-Cas9 approach. (A) Schematic depictions of the WT (Cas9) and recombined alleles after replacement with puromycin-resistant gene; annealing positions of the probes and expected fragment sizes are shown. (B) PCR analysis of LmDUSP1 KO clonal cultures, a wild type (Cas9), and a negative control; 1 kb DNA ladder is on the left. (C) Southern blotting results of the EcoRV digested L. mexicana genomic DNA of the wild type (Cas9, labelled Cas9) and LmxM.22.0250 ablated strains (clonal culture 1 from 2B, labelled 0250) with LmxM.22.0250 5’ UTR and LmxM.22.0250 3’ UTR probes. (D) Quantitative RT-PCR analysis of LmxM.22.0250 gene expression in the wild type (Cas9) and LmDUSP1 KO L. mexicana. Gene expression was normalized to LmxM.07.0510 [29] and presented as means and standard deviations of three independent biological replicates. Sizes in (B,C) are in kb.
2.4. Growth Kinetics of Leishmania Strains In Vitro
We first investigated the expression profile of LmxM.22.0250 in the wild type L. mexicana. In agreement with previous reports [12,13], we documented a significant upregulation of its expression in axenically differentiated metacyclic promastigotes (Figure 3A). We propose using this gene as a marker of metacyclogenesis in Leishmania (Leishmania) spp.
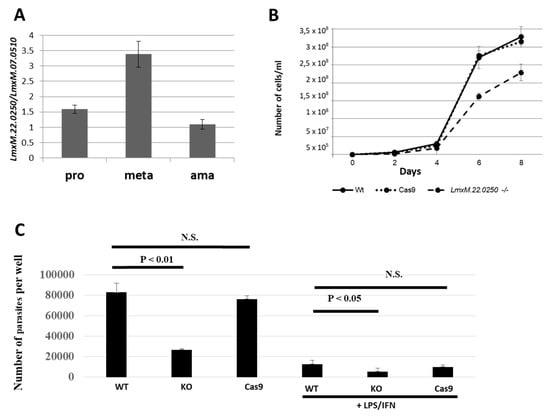
Figure 3.
Expression of LmxM.22.0250 in L. mexicana development, growth kinetics, and macrophage infection in vitro of the wild type (WT), Cas9, and LmDUSP1 KO cells. (A) Quantitative RT-PCR analysis of LmxM.22.0250 expression in the axenically differentiated procyclic promastigotes (pro), metacyclic promastigotes (meta), and amastigotes (ama) of the wild type (Cas9) L. mexicana. (B) Growth curves of the WT, Cas9, and LmDUSP1 KO L. mexicana. (C) The intensity of infection (number of parasites per well) was calculated for WT, LmDUSP1 KO, and Cas9-expressing L. mexicana in non-stimulated or LPS/IFN-γ stimulated primary murine macrophages. Data are summarized from three independent biological replicates. The error bars indicate standard deviation. N.S. indicates a not statistically significant difference.
Next, we studied the effect of LmxM.22.0250 ablation on L. mexicana growth by comparing cell division of the wild type, Cas9, and LmDUSP1 KO strains in vitro. To eliminate the negative effect of continuous in vitro cultivation [30], the procyclic promastigote cultures were started from the cells passaged through animals. Growth kinetics were monitored every 48 hours. Our data demonstrated that the promastigotes of the LmDUSP1 KO strain divide significantly slower compared to their wild type or Cas9 counterparts (Figure 3B). This was reminiscent of the situation previously documented for L. mexicana with ablated LmxM.30.2090, a gene encoding a putative ATP/GTPase ALD1 [31].
In addition, we also checked whether the absence of LmxM.22.0250 can be compensated by overexpression of its paralog, LmxM.32.2840. The expression level of LmxM.32.2840 was assayed by qRT-PCR in Leishmania developmental stages (Figure S2). No statistically significant difference was detected between the wild type (Cas9) and LmDUSP1 KO cells, suggesting that LmxM.32.2840 does not compensate for the loss of LmxM.22.0250 function.
2.5. Experimental Infection of Lutzomyia longipalpis
In total, 215 Lutzomyia longipalpis females were dissected, out of which 68 were infected with the wild type, 76 with Cas9, and 71 with LmDUSP1 KO L. mexicana. On day 2 PBM (post blood meal), infection rates and intensity were similar between the tested lines (p = 0.85 and p = 0.92, respectively; Figure S3A). In all lines, procyclic promastigotes were localized inside the endoperitrophic space, within the bloodmeal surrounded by the peritrophic matrix (Figure S3B). On day 8 PBM, fully developed late infections were documented in all three lines, with similar infection rates ranging from 84% to 91% (p = 0.56) (Figure S3A). Heavy infections (62%) dominated in females harbouring the wild type strain, while moderate infections were predominant in females infected with Cas9 (53%) and LmDUSP1 KO (48%) Leishmania strains. No differences were documented in the localization of infections on day 8. Parasites of all tested strains were always present in the abdominal and/or thoracic midgut. Colonization of the stomodeal valve was observed in a similar percentage in all three strains: 83%, 82%, and 86% for the wild type, Cas9, and LmDUSP1 KO, respectively (Figure S3B).
2.6. Macrophage Infection In Vitro
We also investigated whether LmDUSP1 KO can manifest virulence defects in cultured primary murine macrophages. Non-activated and classically stimulated macrophages were compared side-by-side in infection experiments using wild type, Cas9 control, and LmDUSP1 KO strains of L. mexicana. In both cases, the levels of the LmDUSP1 KO infections were significantly lower compared to those of wild type or Cas9 flagellates (Figure 3C). Thus, we concluded that wild type and LmDUSP1 KO strains differ in their ability to infect or develop in the primary murine macrophages in vitro.
2.7. Infection of Mice with LmDUSP1 KO L. mexicana
All BALB/c mice inoculated with the wild type, Cas9, and LmDUSP1 KO strains developed lesions. In the LmDUSP1 KO strain, lesions were somewhat smaller than in the wild type or Cas9 controls starting from week 7 post infection, although the variation between individual animals (14 per strain, 42 in total) did not allow us to assign statistical significance to this observation (Figure 4).
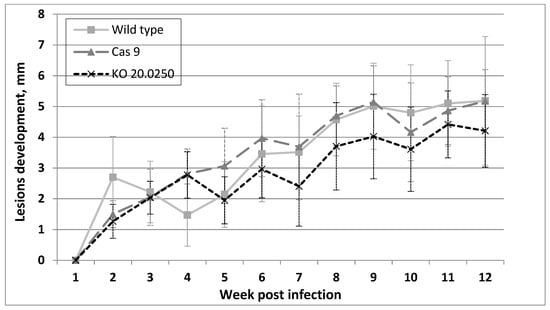
Figure 4.
Development of clinical symptoms in inoculated mice ears. Diameter of the lesions from seven independent experiments per Leishmania strain, measured weekly. Data for one representative experiment (out of two) are presented.
The effect of LmxM.22.02250 ablation remains to be investigated in other animal models (for example, hamster, dog, non-human primate, or wild rodent [32]) available to researchers in the field.
3. Discussion
Protein phosphorylation/dephosphorylation is a vital regulatory mechanism controlling cellular metabolism, growth, trafficking, and other important physiological processes. The sets of cellular kinases and phosphatases perform reversible phosphorylation of proteins on Ser, Thr, and Tyr. Protein Tyr phosphatases are a large superfamily that encompasses the most divergent dual specificity phosphatases [33]. Numerous pathogens successfully use kinases and phosphatases to internalize, replicate, and survive, modifying the host′s phosphorylation profile or signal transduction pathways [34]. Multiple phosphatases and kinases from diverse bacterial pathogens such as Salmonella, Yersinia, Listeria, and Mycobacterium spp. have been previously identified as virulence factors in human infections [22,35,36,37].
Recently, protein tyrosine phosphatases have been implicated in Leishmania pathogenesis [38,39]. In this work, we identified and characterized a novel putative virulence factor of Leishmania infection. This dual specificity protein/lipid phosphatase (LmDUSP1) can dephosphorylate two types of molecules; in proteins, it recognizes preferentially P-Tyr, but also P-Ser. The LmDUSP1-encoding gene (LmxM.22.0250 in L. mexicana) was acquired from bacteria via horizontal gene transfer. Importantly, its orthologues have been implicated in virulence in several bacterial species, e.g. M. tuberculosis [21] and L. monocytogenes [22].
In trypanosomatids, these genes were completely lost from some basal lineages (most notably, Trypanosoma and Blechomonas), while retained in others (monoxenous and dixenous Leishmaniinae, Phytomonas spp., Strigomonadinae, and early-branching Paratrypanosoma) [2]. Moreover, Strigomonadinae and Leishmaniinae harbor multiple anciently-diverged paralogues in the genome, suggesting a significant separation of DUSPs′ functions. Our data, demonstrating that ablation of LmxM.22.0250 does not change the expression of the paralogous LmxM.32.2840 gene, are in agreement with this hypothesis. We also noted that the DUSP1-encoding gene has been pseudogenized in members of the subgenus Leishmania (Viannia), suggesting that these parasites rely on other factors determining their virulence [40,41].
Leishmania mexicana with ablated LmxM.22.0250 demonstrated attenuated virulence in mouse macrophages, suggesting that this gene is important for infectivity in vertebrates. Despite significant upregulation of LmxM.22.0250 expression in metacyclic promastigotes (Figure 3A), its ablation did not affect the ability of mutant cells to differentiate into the virulent stages. In contrast to the "ancestral" bacterial secreted phosphatases, no signal peptide sequence was found in LmDUSP1. This is reminiscent of the situation of another Leishmania protein tyrosine phosphatase implicated in virulence, LPTP1. This protein also does not encode a signal peptide and is not secreted [39]. Another possibility is that LmDUSP1-encoded phosphatase is secreted by means of the non-classical secretion pathways, which might play a major role in yet understudied leishmanial protein secretion [42].
It remains to be investigated which specific biochemical pathways involve LmDUSP1 and how this facilitates parasites survival in the mammalian host. One of the interesting possibilities is that LmDUSP1 may target substrate(s) of the host, thereby affecting its signal transduction pathways.
4. Materials and Methods
4.1. In Silico Analyses
For the identification of the LmxM.22.0250 homologs, BLASTP and TBLASTN searches were performed using this protein as a query for a local database containing all trypanosomatid annotated proteins and genome assemblies available in the TriTrypDB (release 35) [14] and NCBI Genomes, as well as the genome of the eubodonid Bodo saltans (Table S1). The closest homologs outside Kinetoplastea were identified using a BLASTP search against NCBI nr database. Duplicates and sequences from organisms without clear taxonomic identification were removed from the best 500 collected hits. The remaining 139 protein sequences were combined with the trypanosomatid hits and aligned using the L-INS-i algorithm in MAFFT v. 7.4 [43]. Poorly aligned regions were removed using trimAl v. 1.4 [44] with the default settings, resulting in the final alignment containing 189 positions. The maximum likelihood phylogenetic tree was inferred in IQ-TREE v. 1.68 [45] with the LG + I + G4 model, as selected by the built-in ModelFinder module [46], and 1,000 standard bootstrap replicates. Bayesian inference was conducted using MrBayes v. 3.2.6 [47] with analysis run for 1 million generations, sampling every 100th of them. The model of the site heterogeneity (I + G) was set based on the ModelFinder analysis, while the optimal model of the amino acid substitutions (Wag) was selected in MrBayes using the mixed amino acid model prior. Domain prediction was carried out using a search against the Pfam database [48].
4.2. Axenic Cultivation and Growth Kinetics
Leishmania mexicana (isolate MNYC/BZ/62/M379) culture was maintained in M199 medium (Sigma-Aldrich, St. Louis, USA) supplemented with 2 μg/mL Biopterin (Sigma-Aldrich), 2 μg/mL Hemin (Jena Bioscience GmbH, Jena, Germany), 25mM HEPES, 50 units/mL of penicillin, 50 μg/mL of streptomycin, and 10% fetal bovine serum (FBS) (all from Life Technologies, Carlsbad, USA) at 23 °C. Analysis of growth kinetics was done as described previously [9]. All cell lines were passaged through mice before further analyses. For this, one mouse per each strain was intradermaly infected by injection into the base of tail. At week 9 post infection, all three Leishmania mexicana strains (wild type, LmDUSP1 KO, and Cas 9) were re-isolated into the culture. Expression of LmxM.22.0250 and LmxM.32.2840 was analyzed by RT-qPCR using primer pairs LmxM.22.0250_qPCR_f2 and LmxM.22.0250_qPCR_r2, and LmxM.32.2840_qPCR_f and LmxM.32.2840_qPCR_r, respectively (Table S2) and normalized as in [29].
4.3. Genetic Manipulations of Leishmania mexicana: Conventional Approach
In order to ablate LmxM.22.0250 in L. mexicana, two alleles were sequentially replaced with selectable markers for Nourseothricin (Sat) and Hygromycin (Hyg). Targeting constructs were generated by fusion PCR [49]. In the first round of PCR, 5′ and 3′ arms of homology were amplified from the L. mexicana genomic DNA using primers A/B and D/F, respectively (Table S2). The ORFs of the Sat- and Hyg-resistance genes were amplified from the plasmids pF4T7polNLS1.4sat and pF4TR1.4hyg [50], using primer pairs SAT_5′f/SAT_3′r and Hyg_5′f/Hyg_3′r, respectively [9]. In the fusion PCR reaction, 5′ and 3′ arms of homology were combined with either the Sat- or Hyg-resistance gene and amplified with nested primers G and H (Table S2). L. mexicana promastigotes were transfected with 5 μg of the targeting constructs as described previously using a BTX ECM 630 electroporator (Harvard Apparatus Inc, Holliston, USA) [51]. The first allele knockout cell line was isolated in complete M199 medium containing 100 μg/mL of Sat (Jena Bioscience). The second allele knockout L. mexicana clones were selected on solid M199 medium supplemented as above with additional 100 μg/mL of Sat and 100 μg/mL of Hyg. Correct integration was confirmed by Southern blot [52]. In brief, total genomic DNA was isolated using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), digested with EcoRV overnight, separated on 0.75% agarose gel, and transferred to a ZetaProbe blotting membrane (Bio-Rad, Hercules, USA). The following primers (Table S2) were used to amplify probes: SBp_SAT_f and SBp_SAT_r (for Sat), SBp_Hyg_f and SBp_Hyg_r (for Hyg), South5_LmxM.22.0250_f and South5_LmxM.22.0250_r (for 5′ UTR). The probes were labelled with radioactive 32P using the DecaLabel DNA Labeling kit (Thermo Fisher Scientific, Waltham, USA).
4.4. Genetic Manipulations in Leishmania mexicana: CRISPR-Cas9
In order to ablate LmxM.22.0250, we used a strategy described earlier [9] with gRNA gtgatgaagcaggtatcgat|cgg. The U6 promoter, gRNA with tracrRNA, and U6 terminator were amplified using the following primers, respectively (Table S2): A_U6_prom_LmxM_f and B_LmxM.22.0250-sgRNA281_r, C_U6_term_LmxM_f and D_U6_term_LmxM_r, E_LmxM.22.0250-sgRNA281_f and F_sgRNA_mCh396_r. These fragments were fused with primers G_sgRNA_NotI_f and H_sgRNA_NcoI_r, and cloned into pLEXSY-SAT2 (Jena Bioscience). The donor sequence encoding the puromycin (Puro) resistance gene was amplified from pLS6-PFR2 [10] with 30 bp of LmxM.22.0250 sequences flanking double-stranded break site using primers I_donor_LmxM.22.0250-281_f and J_donor_LmxM.22.0250-281_r. The construct containing gRNA and the donor construct were transfected into the CRISR-Cas9-expressing L. mexicana [9]. Clones were selected on solid complete M199 medium supplemented with 100 μg/mL of Hyg, 100 μg/mL of Sat, and 20 μg/mL of Puro. Ablation of LmxM.22.0250 was verified by PCR with primers LmxM.22.0250_check_1f and LmxM.22.0250_check_1r (wild type size is 792 bp; after donor insertion the size is 2,045 bp), and by Southern blot as described earlier with probes 5′ UTR-, 3′ UTR-, and Puro. The following primers were used for probes amplification: Puromycin probe: South_Puro_f and South_Puro_r; 5’UTR probe: South5_LmxM.22.0250_f and South5_LmxM.22.0250_r, 3’UTR probe: South6_LmxM.22.0250_f and South6_LmxM.22.0250_r (Table S2).
4.5. Infection of Macrophages
Parasites were harvested in the stationary phase, washed three times in the 0.9% saline solution (5 min, 6,000 rpm), re-suspended in the complete macrophage´s medium (see below), and their concentration was determined by hemocytometer.
Bone marrow was obtained by flushing the tibiae and femurs of euthanized BALB/c mice. The differentiation from bone marrow precursor cells to bone marrow-derived macrophages proceeded for 7 days at 37 °C with 5% CO2 in the presence of the L929 fibroblast cell culture supernatant (20%), which served as a source of the macrophage colony-stimulating factor. After differentiation, the macrophages were cultured as above in the complete RPMI-1640 medium supplemented with 10% FBS, 50 units/mL penicillin, 50 μg/mL streptomycin, 2 mM L-glutamine, and 0.05 mM 2-mercapto-ethanol (all from Sigma-Aldrich).
The bone marrow-derived macrophages were plated into CellStar 24-wells (Greiner Bio-One GmbH, Kremsmünster, Austria) at a concentration of 4 × 105 cells/ml. The stationary-phase Leishmania cells were added at a parasite-to-macrophage ratio of 6:1. After two hours, the cells were washed and incubated either in the complete RPMI 1640 or in the media combined with 50 U/mL IFN-γ (Bio-Rad) and 0.5 µg/mL LPS (Sigma-Aldrich) (classically stimulated macrophages). At 72 hours post infection, macrophages were lysed in 0.016% SDS (Sigma-Aldrich) and washed with 0.9% saline solution, and amastigotes were spun down by centrifugation, re-suspended in the complete RPMI medium, and counted with a hemocytometer. All experiments were performed in two independent biological replicates, and samples were analyzed in triplicate.
4.6. Experimental Infection of Lutzomyia longipalpis
For all the experiments, a well-established sand fly colony of Lutzomyia longipalpis (originated from Brazil) was used. This colony was maintained under standard conditions as described previously [53,54]. At 24 hours prior to the infection feeding, groups of approximately 150 sand fly females (5–7 days old) were separated and deprived of sucrose food. Promastigotes from log-phase cultures were resuspended in heat-inactivated sheep blood at concentration of 106 cells/mL and offered to sand flies through the chicken skin. Engorged females were separated, provided with sucrose, and maintained under standard conditions until the end of the experiment.
Dissections were performed before defecation (early stage of infection) on day 2 post blood meal (PBM) and after defecation (late stage of infection) on day 8 PBM. Abundance and localization of the flagellates in the sand fly gut were examined by light microscopy. Parasite loads were graded as light, moderate/medium, and heavy (≤100, 100–1000, and ≥1000 parasites per gut, respectively) as described previously [55]. Experimental feeding of L. longipalpis was repeated three times for each L. mexicana line.
4.7. Mice Infection
In order to investigate the ability of parasites to infect the mammalian host, BALB/c mice were experimentally infected with the freshly isolated wild type, LmDUSP1 knock-out (KO), and Cas 9 L. mexicana cells. Suspensions of 5 × 105 parasites in 5 µl of saline solution were injected intradermally into the ear pinnae of mice anesthetized by ketamine/xylazin (62.5 mg/kg and 25 mg/kg, respectively). The experiments were repeated twice, and seven mice for each Leishmania strain were used every time. Development of clinical symptoms and size of nodular lesions were measured weekly. Mice were sacrificed at the 14th week post infection in the first experiment and 12th week post infection in the second experiment. Their infected ears were dissected, and parasites were re-isolated into the culture.
Ethics statement: Animals were maintained and handled in the animal facility of Charles University in Prague in accordance with institutional guidelines and Czech legislation (Act No. 246/1992 and 359/2012 coll. on protection of animals against cruelty in present statutes at large), which complies with all relevant EU guidelines. All the experiments were approved by the Committee on the Ethics of Laboratory Experiments of the Charles University and were performed under permission No. MSMT-31114/2015-13 of the Czech Ministry of the Environment. All efforts were made to minimize the number and suffering of experimental animals during the study.
4.8. Statistical Analysis
Statistical analysis was carried out using R software (http://cran.r-project.org). Infection rates and intensity of infection were analysed using Fisher´s exact test and proportion test, respectively. A p-value of <0.05 was considered statistically significant.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/8/4/241/s1, Figure S1: Conventional ablation of LmxM.22.0250. A: Schematic representation of the WT and recombined alleles after replacement with Sat- and Hyg-resistant genes; annealing positions of the probes and expected fragment sizes are shown. B: Southern blot analysis of the EcoRV digested L. mexicana genomic DNA of the wild type (WT) and LmxM.22.0250 ablated strains (clonal cultures 1 and 2) with Sat, Hyg, and LmxM.22.0250 5’ UTR probes. Indicated sizes are in kb. Figure S2: Expression of LmxM.32.2840 in L. mexicana developmental stages. Figure S3: Experimental infection of Lutzomyia longipalpis. A: Infection rates and intensity in L. longipalpis harbouring different strains of L. mexicana; B: Localization of parasites inside L. longipalpis gut after infection with different strains of L. mexicana. Table S1: Kinetoplastid genome sequences analyzed in this study. Table S2: List of primers used in this study for amplification of the LmxM.22.0250 gene specific targeting sequences, selectable markers, fusion PCR products, Southern blot probes, and PCR analysis of correct integration.
Author Contributions
Conceptualization, V.Y. and J.V.; methodology, A.I., N.K., J.V., and P.V.; investigation, N.K., T.L., A.I., K.M., A.B., S.V., J.B., A.C., T.S., A.Y.K., and J.V.; data curation, A.B., A.Y.K., P.V., and V.Y.; writing—original draft preparation, V.Y.; writing—review and editing, V.Y., J.V., A.B., P.V., J.L., N.K., J.B., and A.Y.K.; supervision, V.Y., J.V., P.V., and J.L.; funding acquisition, V.Y., J.V., P.V., and J.L.
Funding
This research was funded by the Grant Agency of Czech Republic (grant 17-10656S to V.Y. and J.V.); Russian Science Foundation (grant 19-15-00054 to V.Y., bioinformatics analysis of the novel putative virulence factors); European Research Council (CZ LL1601 to J.L.); European Regional Funds (project "Centre for Research of Pathogenicity and Virulence of Parasites" CZ.02.1.01/0.0/0.0/16_019/0000759 to V.Y., J.L., P.V., J.V., N.K., A.Y.K., T.L., T.S., and J.S.); Grant Agency of Charles University (UNCE 20472 to T.L.), and a grant from the University of Ostrava (to A.C. and V.Y.).
Acknowledgments
We thank members of our laboratories for stimulating discussions.
Conflicts of Interest
V.Y. is an Academic Editor of "Pathogens". Other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Bruschi, F.; Gradoni, L. The leishmaniases: Old Neglected Tropical Diseases; Springer: Cham, Switzerland, 2018; p. 245. [Google Scholar]
- Maslov, D.A.; Opperdoes, F.R.; Kostygov, A.Y.; Hashimi, H.; Lukeš, J.; Yurchenko, V. Recent advances in trypanosomatid research: Genome organization, expression, metabolism, taxonomy and evolution. Parasitology 2019, 146, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Lukeš, J.; Butenko, A.; Hashimi, H.; Maslov, D.A.; Votýpka, J.; Yurchenko, V. Trypanosomatids are much more than just trypanosomes: Clues from the expanded family tree. Trends Parasitol. 2018, 34, 466–480. [Google Scholar] [CrossRef] [PubMed]
- Dostálová, A.; Volf, P. Leishmania development in sand flies: Parasite-vector interactions overview. Parasites Vectors 2012, 5, 276. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.A.; Rogers, M.E. New insights into the developmental biology and transmission mechanisms of Leishmania. Curr. Mol. Med. 2004, 4, 601–609. [Google Scholar] [CrossRef]
- Cantacessi, C.; Dantas-Torres, F.; Nolan, M.J.; Otranto, D. The past, present, and future of Leishmania genomics and transcriptomics. Trends Parasitol. 2015, 31, 100–108. [Google Scholar] [CrossRef]
- Beneke, T.; Madden, R.; Makin, L.; Valli, J.; Sunter, J.; Gluenz, E. A CRISPR Cas9 high-throughput genome editing toolkit for kinetoplastids. R. Soc. Open Sci. 2017, 4, 170095. [Google Scholar] [CrossRef]
- Dean, S.; Sunter, J.; Wheeler, R.J.; Hodkinson, I.; Gluenz, E.; Gull, K. A toolkit enabling efficient, scalable and reproducible gene tagging in trypanosomatids. Open Biol. 2015, 5, 140197. [Google Scholar] [CrossRef]
- Ishemgulova, A.; Hlavacova, J.; Majerova, K.; Butenko, A.; Lukes, J.; Votypka, J.; Volf, P.; Yurchenko, V. CRISPR/Cas9 in Leishmania mexicana: A case study of LmxBTN1. PLoS ONE 2018, 13, e0192723. [Google Scholar] [CrossRef]
- Sollelis, L.; Ghorbal, M.; MacPherson, C.R.; Martins, R.M.; Kuk, N.; Crobu, L.; Bastien, P.; Scherf, A.; Lopez-Rubio, J.J.; Sterkers, Y. First efficient CRISPR-Cas9-mediated genome editing in Leishmania parasites. Cell Microbiol. 2015, 17, 1405–1412. [Google Scholar] [CrossRef]
- Zhang, W.W.; Matlashewski, G. CRISPR-Cas9-mediated genome editing in Leishmania donovani. Microbiology 2015, 6, e00861. [Google Scholar] [CrossRef]
- Flegontov, P.; Butenko, A.; Firsov, S.; Kraeva, N.; Eliáš, M.; Field, M.C.; Filatov, D.; Flegontova, O.; Gerasimov, E.S.; Hlaváčová, J.; et al. Genome of Leptomonas pyrrhocoris: A high-quality reference for monoxenous trypanosomatids and new insights into evolution of Leishmania. Sci. Rep. 2016, 6, 23704. [Google Scholar] [CrossRef] [PubMed]
- Inbar, E.; Hughitt, V.K.; Dillon, L.A.; Ghosh, K.; El-Sayed, N.M.; Sacks, D.L. The transcriptome of Leishmania major developmental stages in their natural sand fly vector. Microbiology 2017, 8, e00029-17. [Google Scholar] [CrossRef] [PubMed]
- Aslett, M.; Aurrecoechea, C.; Berriman, M.; Brestelli, J.; Brunk, B.P.; Carrington, M.; Depledge, D.P.; Fischer, S.; Gajri, B.; Gao, X.; et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010, 38, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Opperdoes, F.R.; Butenko, A.; Flegontov, P.; Yurchenko, V.; Lukeš, J. Comparative metabolism of free-living Bodo saltans and parasitic trypanosomatids. J. Eukaryot. Microbiol. 2016, 63, 657–678. [Google Scholar] [CrossRef]
- Soulat, D.; Bogdan, C. Function of macrophage and parasite phosphatases in leishmaniasis. Front. Immunol. 2017, 8, 1838. [Google Scholar] [CrossRef]
- Denu, J.M.; Dixon, J.E. A catalytic mechanism for the dual-specific phosphatases. Proc. Natl. Acad. Sci. USA 1995, 92, 5910–5914. [Google Scholar] [CrossRef]
- Beresford, N.J.; Saville, C.; Bennett, H.J.; Roberts, I.S.; Tabernero, L. A new family of phosphoinositide phosphatases in microorganisms: Identification and biochemical analysis. BMC Genom. 2010, 11, 457. [Google Scholar] [CrossRef]
- Tautz, L.; Critton, D.A.; Grotegut, S. Protein tyrosine phosphatases: Structure, function, and implication in human disease. Methods Mol. Biol. 2013, 1053, 179–221. [Google Scholar]
- Christensen, S.M.; Dillon, L.A.; Carvalho, L.P.; Passos, S.; Novais, F.O.; Hughitt, V.K.; Beiting, D.P.; Carvalho, E.M.; Scott, P.; El-Sayed, N.M.; et al. Meta-transcriptome profiling of the human Leishmania braziliensis cutaneous lesion. PLOS Negl. Trop. Dis. 2016, 10, e0004992. [Google Scholar]
- Beresford, N.; Patel, S.; Armstrong, J.; Szoor, B.; Fordham-Skelton, A.P.; Tabernero, L. MptpB, a virulence factor from Mycobacterium tuberculosis, exhibits triple-specificity phosphatase activity. Biochem. J. 2007, 406, 13–18. [Google Scholar] [CrossRef]
- Kastner, R.; Dussurget, O.; Archambaud, C.; Kernbauer, E.; Soulat, D.; Cossart, P.; Decker, T. LipA, a tyrosine and lipid phosphatase involved in the virulence of Listeria monocytogenes. Infect. Immun. 2011, 79, 2489–2498. [Google Scholar] [CrossRef] [PubMed]
- Koul, A.; Choidas, A.; Treder, M.; Tyagi, A.K.; Drlica, K.; Singh, Y.; Ullrich, A. Cloning and characterization of secretory tyrosine phosphatases of Mycobacterium tuberculosis. J. Bacteriol. 2000, 182, 5425–5432. [Google Scholar] [CrossRef] [PubMed]
- Beresford, N.J.; Mulhearn, D.; Szczepankiewicz, B.; Liu, G.; Johnson, M.E.; Fordham-Skelton, A.; Abad-Zapatero, C.; Cavet, J.S.; Tabernero, L. Inhibition of MptpB phosphatase from Mycobacterium tuberculosis impairs mycobacterial survival in macrophages. J. Antimicrob. Chemother. 2009, 63, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Rao, V.; Shakila, H.; Gupta, R.; Khera, A.; Dhar, N.; Singh, A.; Koul, A.; Singh, Y.; Naseema, M.; et al. Disruption of mptpB impairs the ability of Mycobacterium tuberculosis to survive in guinea pigs. Mol. Microbiol. 2003, 50, 751–762. [Google Scholar] [CrossRef]
- Rawls, K.A.; Grundner, C.; Ellman, J.A. Design and synthesis of nonpeptidic, small molecule inhibitors for the Mycobacterium tuberculosis protein tyrosine phosphatase PtpB. Organ. Biomol. Chem. 2010, 8, 4066–4070. [Google Scholar] [CrossRef]
- Espinosa, O.A.; Serrano, M.G.; Camargo, E.P.; Teixeira, M.M.; Shaw, J.J. An appraisal of the taxonomy and nomenclature of trypanosomatids presently classified as Leishmania and Endotrypanum. Parasitology 2018, 145, 430–442. [Google Scholar] [CrossRef]
- Kostygov, A.Y.; Yurchenko, V. Revised classification of the subfamily Leishmaniinae (Trypanosomatidae). Folia Parasitol. 2017, 64, 020. [Google Scholar] [CrossRef]
- Ishemgulova, A.; Kraeva, N.; Faktorová, D.; Podešvová, L.; Lukeš, J.; Yurchenko, V. T7 polymerase-driven transcription is downregulated in metacyclic promastigotes and amastigotes of Leishmania mexicana. Folia Parasitol. 2016, 63. [Google Scholar] [CrossRef]
- Sádlová, J.; Svobodová, M.; Volf, P. Leishmania major: Effect of repeated passages through sandfly vectors or murine hosts. Ann. Trop. Med. Parasitol. 1999, 93, 599–611. [Google Scholar] [CrossRef]
- Ishemgulova, A.; Kraeva, N.; Hlavacova, J.; Zimmer, S.L.; Butenko, A.; Podesvova, L.; Lestinova, T.; Lukes, J.; Kostygov, A.; Votypka, J.; et al. A putative ATP/GTP binding protein affects Leishmania mexicana growth in insect vectors and vertebrate hosts. PLOS Negl. Trop. Dis. 2017, 11, e0005782. [Google Scholar] [CrossRef]
- Loría-Cervera, E.N.; Andrade-Narváez, F.J. Animal models for the study of leishmaniasis immunology. Rev. Inst. Med. Trop. Sao Paulo 2014, 56, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Q.; Sun, J.P.; Zhang, Z.Y. An overview of the protein tyrosine phosphatase superfamily. Curr. Top. Med. Chem. 2003, 3, 739–748. [Google Scholar] [CrossRef]
- DeVinney, R.; Steele-Mortimer, O.; Finlay, B.B. Phosphatases and kinases delivered to the host cell by bacterial pathogens. Trends Microbiol. 2000, 8, 29–33. [Google Scholar] [CrossRef]
- Norris, F.A.; Wilson, M.P.; Wallis, T.S.; Galyov, E.E.; Majerus, P.W. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc. Natl. Acad. Sci. USA 1998, 95, 14057–14059. [Google Scholar] [CrossRef] [PubMed]
- Galyov, E.E.; Hakansson, S.; Forsberg, A.; Wolf-Watz, H. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature 1993, 361, 730–732. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Singh, A.; Tyagi, A.K. Deciphering the genes involved in pathogenesis of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2005, 85, 325–335. [Google Scholar] [CrossRef]
- Leitherer, S.; Clos, J.; Liebler-Tenorio, E.M.; Schleicher, U.; Bogdan, C.; Soulat, D. Characterization of the protein tyrosine phosphatase LmPRL-1 secreted by Leishmania major via the exosome pathway. Infect. Immun. 2017, 85, e00084-17. [Google Scholar] [CrossRef]
- Nascimento, M.; Zhang, W.W.; Ghosh, A.; Houston, D.R.; Berghuis, A.M.; Olivier, M.; Matlashewski, G. Identification and characterization of a protein-tyrosine phosphatase in Leishmania: Involvement in virulence. J. Biol. Chem. 2006, 281, 36257–36268. [Google Scholar] [CrossRef]
- Coughlan, S.; Taylor, A.S.; Feane, E.; Sanders, M.; Schonian, G.; Cotton, J.A.; Downing, T. Leishmania naiffi and Leishmania guyanensis reference genomes highlight genome structure and gene evolution in the Viannia subgenus. R. Soc. Open Sci. 2018, 5, 172212. [Google Scholar] [CrossRef]
- Llanes, A.; Restrepo, C.M.; del Vecchio, G.; Anguizola, F.J.; Lleonart, R. The genome of Leishmania panamensis: Insights into genomics of the L. (Viannia) subgenus. Sci. Rep. 2015, 5, 8550. [Google Scholar] [CrossRef]
- Silverman, J.M.; Chan, S.K.; Robinson, D.P.; Dwyer, D.M.; Nandan, D.; Foster, L.J.; Reiner, N.E. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 2008, 9, R35. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martinez, J.M.; Gabaldon, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Merritt, C.; Stuart, K. Identification of essential and non-essential protein kinases by a fusion PCR method for efficient production of transgenic Trypanosoma brucei. Mol. Biochem. Parasitol. 2013, 190, 44–49. [Google Scholar] [CrossRef]
- Kushnir, S.; Gase, K.; Breitling, R.; Alexandrov, K. Development of an inducible protein expression system based on the protozoan host Leishmania tarentolae. Protein Expr. Purif. 2005, 42, 37–46. [Google Scholar] [CrossRef]
- Kraeva, N.; Ishemgulova, A.; Lukeš, J.; Yurchenko, V. Tetracycline-inducible gene expression system in Leishmania mexicana. Mol. Biochem. Parasitol. 2014, 198, 11–13. [Google Scholar] [CrossRef]
- Southern, E.M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 1975, 98, 503–517. [Google Scholar] [CrossRef]
- Volf, P.; Volfová, V. Establishment and maintenance of sand fly colonies. J. Vector Ecol. 2011, 36, S1–S9. [Google Scholar] [CrossRef] [PubMed]
- Lawyer, P.; Killick-Kendrick, M.; Rowland, T.; Rowton, E.; Volf, P. Laboratory colonization and mass rearing of phlebotomine sand flies (Diptera, Psychodidae). Parasite 2017, 24, 42. [Google Scholar] [CrossRef] [PubMed]
- Myšková, J.; Votýpka, J.; Volf, P. Leishmania in sand flies: Comparison of quantitative polymerase chain reaction with other techniques to determine the intensity of infection. J. Med. Entomol. 2008, 45, 133–138. [Google Scholar] [CrossRef] [PubMed][Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).