Pyrazinoic Acid Inhibits the Bifunctional Enzyme (Rv2783) in Mycobacterium tuberculosis by Competing with tmRNA
Abstract
1. Introduction
2. Results
2.1. The M. tuberculosis Rv2783cA2104C Mutant is Associated with PZA Resistance
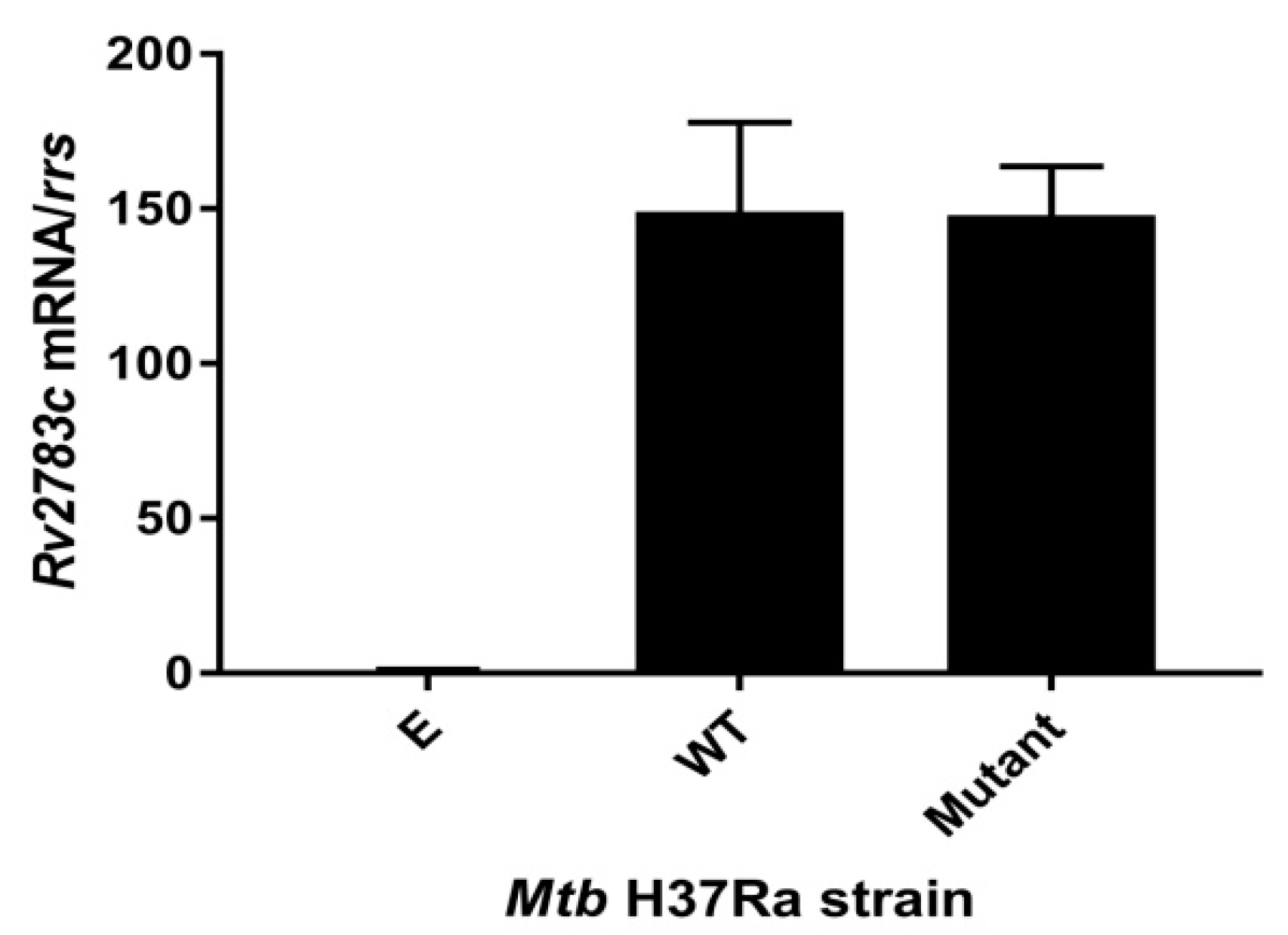
2.2. Expression of Rv2783cA2104C in M. tuberculosis H37Ra Causes PZA Resistance
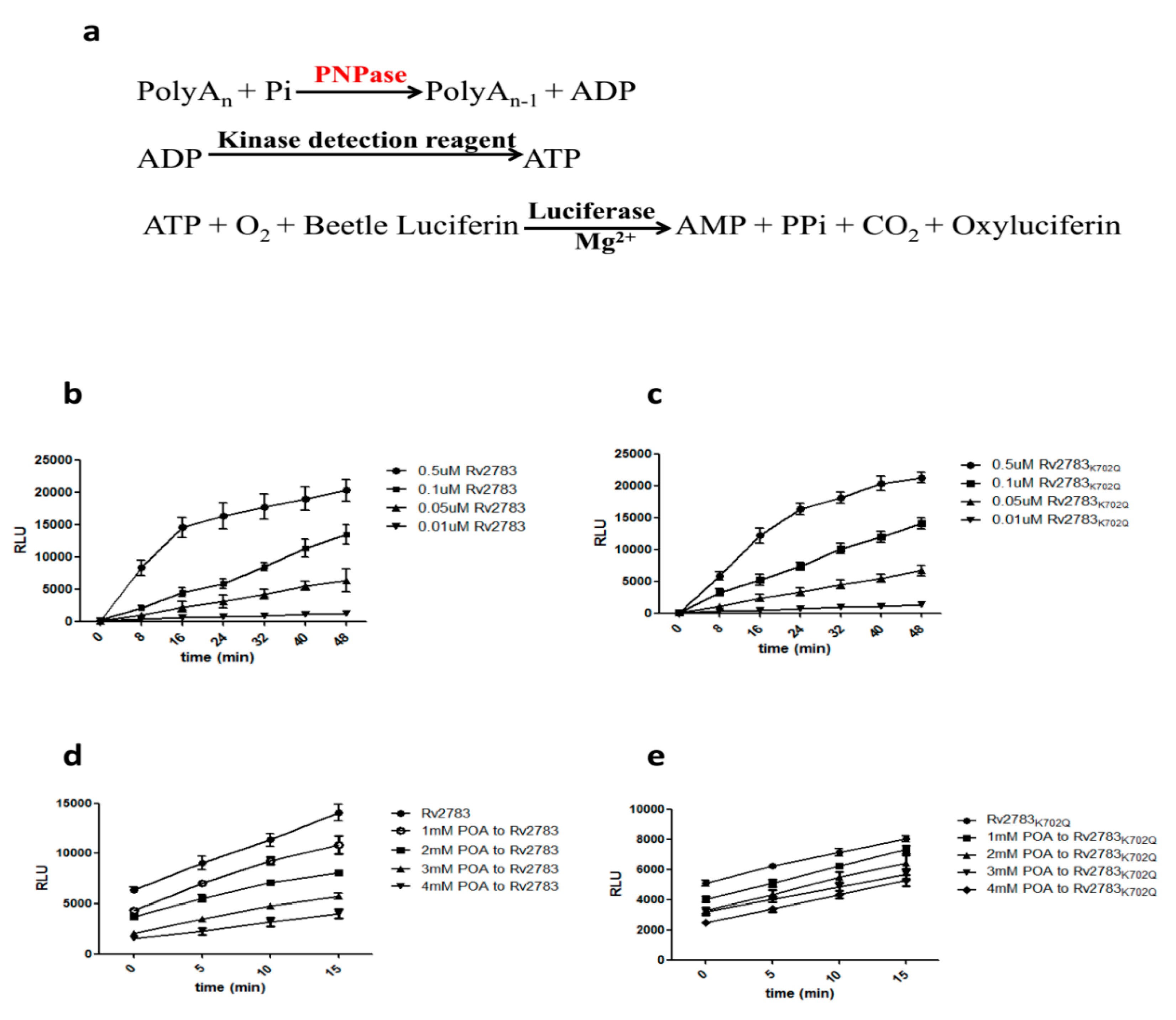
2.3. Rv2783K702Q Mutant Protein Retains PNPase Activity But Loses Ability to Bind POA
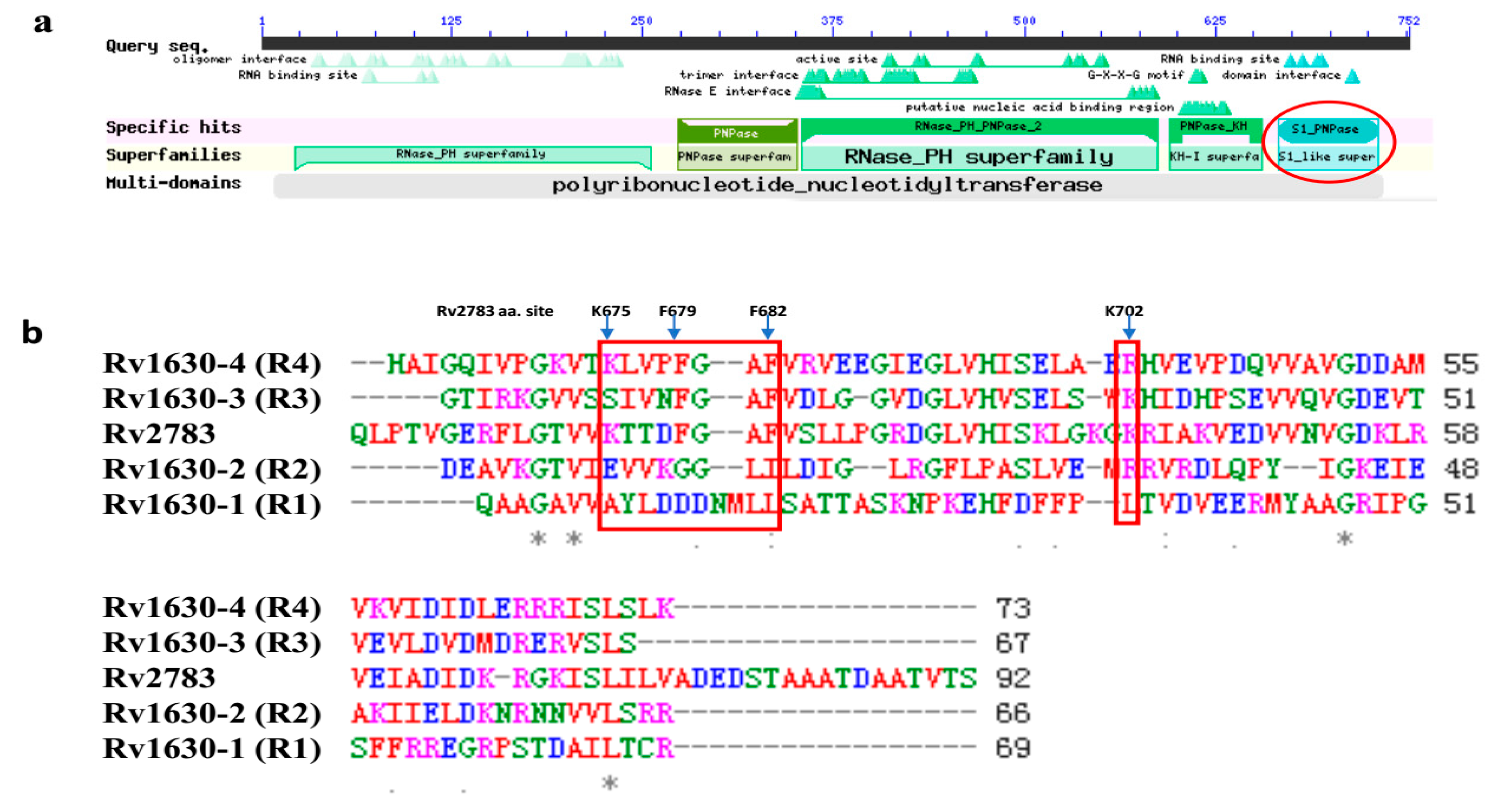
2.4. Domain Alignment of Rv2783 to Predict Possible Essential Sites for POA and tmRNA Binding
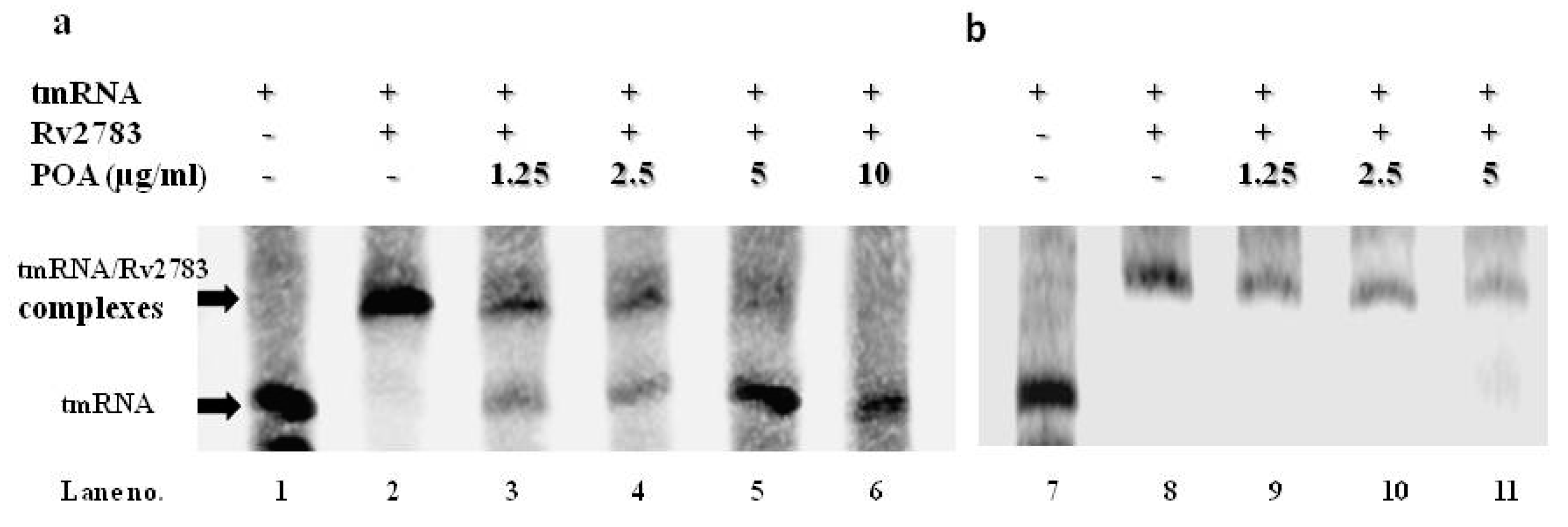
2.5. POA Competes with tmRNA to Bind Wild-Type Rv2783 and Not Rv2783K702Q Mutant Protein
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. PZA Susceptibility Testing
4.3. PZase Activity Determination
4.4. DNA Isolation, PCR, and DNA Sequencing of Rv2783c
4.5. Rv2783c and Rv2783cA2104C Mutant Gene Expression in M. tuberculosis
4.6. qRT-PCR Verification of Expression of Rv2783c Gene
4.7. Drug Susceptibility Testing (DST)
4.8. Cloning, Sequencing, Expression, and Purification of M. tuberculosis Rv2783
4.9. In Vitro Enzyme Inhibition
4.10. Synthesis and Purification of tmRNA
4.11. RNA Electrophoretic Mobility Shift Assay (REMSA)
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mitchison, D.A. The action of antituberculosis drugs in short-course chemotherapy. Tubercle 1985, 66, 219–225. [Google Scholar] [CrossRef]
- Zhang, Y.; Mitchison, D. The curious characteristics of pyrazinamide: A review. Int. J. Tuberc. Lung Dis. 2003, 7, 6–21. [Google Scholar] [PubMed]
- Heifets, L.; Lindholm-Levy, P. Pyrazinamide sterilizing activity in vitro against semidormant Mycobacterium tuberculosis bacterial populations. Am. Rev. Respir. Dis 1992, 145, 1223–1225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shi, W.; Zhang, W.; Mitchison, D. Mechanisms of Pyrazinamide Action and Resistance. Microbiol. Spectr. 2013, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Scorpio, A.; Zhang, Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 1996, 2, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Zhang, X.; Jiang, X.; Yuan, H.; Lee, J.S.; Barry, C.E.; Wang, H.; Zhang, W.; Zhang, Y. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 2011, 333, 1630–1632. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Y.; Bi, J.; Cai, Q.; Liao, X.; Li, W.; Guo, C.; Zhang, Q.; Lin, T.; Zhao, Y.; et al. Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide. Mol. Microbiol. 2015, 95, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Simons, S.O.; Mulder, A.; van Ingen, J.; Boeree, M.J.; van Soolingen, D. Role of rpsA gene sequencing in diagnosis of pyrazinamide resistance. J. Clin. Microbiol. 2013, 51, 382. [Google Scholar] [CrossRef] [PubMed]
- Werngren, J.; Alm, E.; Mansjö, M. Non-pncA gene-mutated but pyrazinamide-resistant Mycobacterium tuberculosis: why is that? J. Clin. Microbiol. 2017, 55, 1920–1927. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.C.; Ma, J.H.; Guthrie, J.L.; Blair, J.; Chedore, P.; Jamieson, F.B. Gene sequencing for routine verification of pyrazinamide resistance in Mycobacterium tuberculosis: A role for pncA but not rpsA. J. Clin. Microbiol. 2012, 50, 3726–3728. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Busby, S.M.; Valafar, F. Systematic review of mutations in pyrazinamidase associated with pyrazinamide resistance in Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 2015, 59, 5267–5277. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Zhao, L.L.; Li, F.; Fan, Y.M.; Chen, Y.Y.; Wu, B.B.; Liu, Z.W.; Pan, A.Z.; Zhu, M. Phenotypic and genotypic characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates in Zhejiang, China. Antimicrob. Agents Chemother. 2015, 59, 1690–1695. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Yu, X.; Jiang, G.; Wang, X.; Ma, Y.; Li, Y.; Huang, H. Pyrazinamide resistance among multidrug-resistant tuberculosis clinical isolates in a national referral center of China and its correlations with pncA, rpsA, and panD gene mutations. Diagn. Microbiol. Infect. Dis. 2016, 84, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Busby, S.M.; Rodwell, T.C.; Fink, L.; Catanzaro, D.; Jackson, R.L.; Pettigrove, M.; Catanzaro, A.; Valafar, F. A Multinational Analysis of Mutations and Heterogeneity in PZase, RpsA, and PanD Associated with Pyrazinamide Resistance in M/XDR Mycobacterium tuberculosis. Sci. Rep. 2017, 7, 3790. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.T.; Malik, S.I.; Bhatti, A.I.; Ali, S.; Khan, A.S.; Zeb, M.T.; Nadeem, T.; Fazal, S. Pyrazinamide-resistant mycobacterium tuberculosis isolates from Khyber Pakhtunkhwa and rpsA mutations. J. Biol. Regul. Homeost. Agents 2018, 32, 705–709. [Google Scholar] [PubMed]
- Zhang, S.; Chen, J.; Shi, W.; Liu, W.; Zhang, W.; Zhang, Y. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg. Microb. Infect. 2013, 2, e34. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Chen, J.; Feng, J.; Cui, P.; Zhang, S.; Weng, X.; Zhang, W.; Zhang, Y. Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosis. Emerg. Microb. Infect. 2014, 3, e58. [Google Scholar] [CrossRef] [PubMed]
- Gopal, P.; Nartey, W.; Ragunathan, P.; Sarathy, J.; Kaya, F.; Yee, M.; Setzer, C.; Manimekalai, M.S.S.; Dartois, V.; Gruber, G.; et al. Pyrazinoic Acid Inhibits Mycobacterial Coenzyme A Biosynthesis by Binding to Aspartate Decarboxylase PanD. ACS Infect. Dis. 2017, 3, 807–819. [Google Scholar] [CrossRef] [PubMed]
- Njire, M.; Wang, N.; Wang, B.; Tan, Y.; Cai, X.; Liu, Y.; Mugweru, J.; Guo, J.; Hameed, H.M.A.; Tan, S.; et al. Pyrazinoic Acid Inhibits a Bifunctional Enzyme in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2017, 61, e00070-17. [Google Scholar] [CrossRef] [PubMed]
- Engman, J.; Negrea, A.; Sigurlasdottir, S.; Georg, M.; Eriksson, J.; Eriksson, O.S.; Kuwae, A.; Sjolinder, H.; Jonsson, A.B. Neisseria meningitidis Polynucleotide Phosphorylase Affects Aggregation, Adhesion, and Virulence. Infect. Immun. 2016, 84, 1501–1513. [Google Scholar] [CrossRef] [PubMed]
- Mikulík, K.; Palecková, P.; Felsberg, J.; Bobek, J.; Zídková, J.; Halada, P. SsrA genes of streptomycetes and association of proteins to the tmRNA during development and cellular differentiation. Proteomics 2008, 8, 1429–1441. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E., 3rd; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.E.; Gawronski, J.D.; Dejesus, M.A.; Ioerger, T.R.; Akerley, B.J.; Sassetti, C.M. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011, 7, e1002251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Ioerger, T.R.; Huttenhower, C.; Long, J.E.; Sassetti, C.M.; Sacchettini, J.C.; Rubin, E.J. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 2012, 8, e1002946. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Shen, Y.; Fan, X.; Diao, N.; Wang, F.; Wang, S.; Weng, X.; Zhang, W. Underestimation of the resistance of Mycobacterium tuberculosis to second-line drugs by the new GenoType MTBDRsl test. J. Mol. Diagn. JMD 2013, 15, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Aono, A.; Hirano, K.; Hamasaki, S.; Abe, C. Evaluation of BACTEC MGIT 960 PZA medium for susceptibility testing of Mycobacterium tuberculosis to pyrazinamide (PZA): Compared with the results of pyrazinamidase assay and Kyokuto PZA test. Diagn. Microbiol. Infect. Dis. 2002, 44, 347–352. [Google Scholar] [CrossRef]
- Wayne, L.G. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am. Rev. Respir. Dis. 1974, 109, 147–151. [Google Scholar] [PubMed]
- Garbe, T.R.; Barathi, J.; Barnini, S.; Zhang, Y.; Abou-Zeid, C.; Tang, D.; Mukherjee, R.; Young, D.B. Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology 1994, 140, 133–138. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wei, J.; Dahl, J.L.; Moulder, J.W.; Roberts, E.A.; O’Gaora, P.; Young, D.B.; Friedman, R.L. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J. Bacteriol. 2000, 182, 377–384. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wang, X.; Cui, P.; Jin, J.; Chen, J.; Zhang, W.; Zhang, Y. ubiA (Rv3806c) encoding DPPR synthase involved in cell wall synthesis is associated with ethambutol resistance in Mycobacterium tuberculosis. Tuberculosis 2015, 95, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Garbe, T.; Young, D. Transformation with katG restores isoniazid-sensitivity in Mycobacterium tuberculosis isolates resistant to a range of drug concentrations. Mol. Microbiol. 1993, 8, 521–524. [Google Scholar] [CrossRef] [PubMed]
- Colangeli, R.; Helb, D.; Vilchèze, C.; Hazbón, M.H.; Lee, C.G.; Safi, H.; Sayers, B.; Sardone, I.; Jones, M.B.; Fleischmann, R.D.; et al. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 2007, 3, e87. [Google Scholar] [CrossRef] [PubMed]
- Marco, M.L.; Kleerebezem, M. Assessment of real-time RT-PCR for quantification of Lactobacillus plantarum gene expression during stationary phase and nutrient starvation. J. Appl. Microbiol. 2008, 104, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Abate, G.; Mshana, R.N.; Miorner, H. Evaluation of a colorimetric assay based on 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) for rapid detection of rifampicin resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 1998, 2, 1011–1016. [Google Scholar] [PubMed]
- Vincent, H.A.; Ziehr, B.; Moorman, N.J. Mechanism of Protein Kinase R Inhibition by Human Cytomegalovirus pTRS1. J. Virol. 2017, 91, e01574-16. [Google Scholar] [CrossRef] [PubMed]
- Rio, D.C. Electrophoretic mobility shift assays for RNA-protein complexes. Cold Spring Harb. Protoc. 2014, 2014, 435–440. [Google Scholar] [CrossRef] [PubMed]
| Genotype in Rv2783c | PZA-Resistant 2 | PZA-Susceptible | |||||
|---|---|---|---|---|---|---|---|
| NT 1 mutations | AA 1 mutations | n | # | % | n | # | % |
| A2104C | K702Q | 56 | 2 | 4 | 42 | 0 | 0 |
| Strain | PZA (pH 5.5) | INH | Kan |
|---|---|---|---|
| H37Ra | 100 | 0.06 | 2 |
| H37Ra/pOLYG | 100 | 0.06 | 2 |
| H37Ra/pOLYG+Rv2783c | 200 | 0.06 | 2 |
| H37Ra/pOLYG+Rv2783cA2104C | 600 | 0.06 | 2 |
| Inhibitor | Rv2783 | Rv2783K702Q |
|---|---|---|
| 1mM POA | 0.85 | 1.03 |
| 2mM POA | 0.56 | 1.00 |
| 3mM POA | 0.48 | 0.85 |
| 4mM POA | 0.32 | 0.94 |
| Primer | Sequence |
|---|---|
| Primers for PCR and DNA sequencing of Rv2783c | |
| A (F1) | 5′-GCGTCACAGTCGGAAACCTAG-3′ |
| B (R2) | 5′-GTGCTCGGCTACACCAGGAC-3′ |
| Primers for construction of Rv2783c expression in Mtb H37Ra strain | |
| C (F) | 5′-GCTCTAGAATGTCTGCCGCTGAAATTGAC-3′(XbaI3) |
| D (R) | 5′-CCAAGCTTTCAGCTGGTGACCGTCGCGGCATCG-3′(HindIII) |
| Primers for site-directed mutagenesis | |
| E (K702Q-F) | 5′-GACTAGCAGCGGGTGGAGATCG-3′ |
| F (K702Q-R) | 5′-GCAGCTTGTCACCGACATTGACAAC-3′ |
| Primers for qRT-PCR | |
| G (RT-F) | 5′-CTACAACTTCCCGCCGTTCT-3′ |
| H (RT-R) | 5′-ATACGGGAATTCCTCGACGC-3′ |
| I (rrs-F) | 5′-GCGATACGGGCAGACTAGAG-3′ |
| J (rrs-R) | 5′-AAGGAAGGAAACCCACACCT-3′ |
| Primers for construction of Rv2783c expression E. coli strains | |
| K (F) | 5′-GGCTAGC ATGTCTGCCGCTGAAATTGAC-3′(NheI) |
| L (R) | 5′-CCAAGCTTTCAGCTGGTGACCGTCGCGGCATCG-3′(HindIII) |
| Primers for PCR amplication of tmRNA | |
| M (L) | 5′-TAATACGACTCACTATAGGATCTGACCGGGAAGTTAATGGC-3′ |
| N (R) | 5′-GATCAGATCCGGACGATCGGCATCG-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Cui, P.; Shi, W.; Li, Q.; Zhang, W.; Li, M.; Zhang, Y. Pyrazinoic Acid Inhibits the Bifunctional Enzyme (Rv2783) in Mycobacterium tuberculosis by Competing with tmRNA. Pathogens 2019, 8, 230. https://doi.org/10.3390/pathogens8040230
He L, Cui P, Shi W, Li Q, Zhang W, Li M, Zhang Y. Pyrazinoic Acid Inhibits the Bifunctional Enzyme (Rv2783) in Mycobacterium tuberculosis by Competing with tmRNA. Pathogens. 2019; 8(4):230. https://doi.org/10.3390/pathogens8040230
Chicago/Turabian StyleHe, Lei, Peng Cui, Wanliang Shi, Qiong Li, Wenhong Zhang, Min Li, and Ying Zhang. 2019. "Pyrazinoic Acid Inhibits the Bifunctional Enzyme (Rv2783) in Mycobacterium tuberculosis by Competing with tmRNA" Pathogens 8, no. 4: 230. https://doi.org/10.3390/pathogens8040230
APA StyleHe, L., Cui, P., Shi, W., Li, Q., Zhang, W., Li, M., & Zhang, Y. (2019). Pyrazinoic Acid Inhibits the Bifunctional Enzyme (Rv2783) in Mycobacterium tuberculosis by Competing with tmRNA. Pathogens, 8(4), 230. https://doi.org/10.3390/pathogens8040230






