Abstract
This study aimed to identify the bacterial community in two wastewater treatment plants (WWTPs) and to determine the occurrence and reduction of Arcobacter, along with virulence genes (ciaB and pldA). A total of 48 samples (24 influent and 24 effluent) were collected at two WWTPs in southern Arizona in the United States, monthly from August 2011 to July 2012. Bacterial DNA extract was utilized for 16S rRNA metagenomic sequencing. Quantification of Arcobacter 16S rRNA gene was conducted using a recently developed SYBR Green-based quantitative PCR assay. Among 847 genera identified, 113 (13%) were identified as potentially pathogenic bacteria. Arcobacter 16S rRNA gene was detected in all influent samples and ten (83%) and nine (75%) effluent samples at each plant, respectively. Log reduction ratios of Arcobacter 16S rRNA gene in Plant A and Plant B were 1.7 ± 0.9 (n = 10) and 2.3 ± 1.5 (n = 9), respectively. The ciaB gene was detected by quantitative PCR in eleven (92%) and twelve (100%) of 12 influent samples from Plant A and Plant B, respectively, while the pldA gene was detected in eight (67%) and six (50%) influent samples from Plant A and Plant B, respectively. The prevalence of potentially pathogenic bacteria in WWTP effluent indicated the need for disinfection before discharge into the environment.
1. Introduction
Wastewater treatment plants (WWTPs) collect and treat wastewater. Water reclaimed after treatment can be utilized for various purposes, including agricultural irrigation [1,2], recreational purposes [3], to reduce pollution in rivers [4,5], and as a drinking water source [6], as a part of integrated and sustainable water resource management.
Arcobacter spp. were first detected in 1991 and are gram-negative, non-spore forming curved or helical rod-shaped bacteria of the family Campylobacteraceae [7,8]. Arcobacter has been detected at WWTPs in multiple countries, including Canada [9,10], China [11,12,13,14,15], Germany [16], Italy [17], Saudi Arabia [18], South Africa [19], Spain [6,20,21,22,23,24], Turkey [25], the United Kingdom [26], and the United States [27,28,29]. Various types of methods, such as quantitative PCR (qPCR), most probable number-qPCR, multiplex PCR, 16S rDNA restriction fragment length polymorphism, culture, high-throughput sequencing, and fluorescent in situ hybridization [9,10,16,17,18,19], have been adopted in these studies and have demonstrated high abundances of Arcobacter spp. in WWTPs. However, most studies have not provided quantitative data on the removal efficiency of Arcobacter by WWTPs. There are only a few studies that attempted quantitative detection, but these were limited to only some species of Arcobacter or used culture methods [9,18].
Among the studies done at WWTPs in the United States, McLellan et al. (2010) reported a high proportion of Arcobacter, along with other bacteria identified by pyrosequencing [28]. Millar and Raghavan (2017) determined the bacterial diversity in WWTP samples and found that Arcobacter cryaerophilus contained multiple antibiotic resistant genes and was a major constituent of the sewage microbiome [27]. Sigala and Unc (2013) estimated diversity of antibiotic resistant Arcobacter and Escherichia coli through pyrosequencing [29]. In Ohio (USA), a waterborne outbreak affected about 1450 persons, and it was believed that groundwater contamination was linked to human waste originating from both wastewater and septic tanks [30]. Arcobacter, along with indicator bacteria, pathogenic bacteria, coliphages, and viruses were recovered from associated groundwater wells [30]. Despite various attempts to examine the prevalence, abundance, and persistence of Arcobacter as a dominant genus in WWTPs, quantification of this taxon in treatment plants is important for better understanding the incidence of Arcobacter and its removal.
In our previous study, a SYBR Green-based qPCR assay targeting a wide range of Arcobacter spp. was developed [31]. The designed primers were highly specific to most of the known Arcobacter species, could quantify between 1.0 × 101 and 6.4 × 106 copies per reaction, and detect as few as three copies per reaction. This qPCR assay was successful in quantifying the Arcobacter 16S rRNA gene in groundwater and surface water samples in Nepal [31].
This study aimed to characterize the bacterial community in the influent and effluent of two WWTPs located in southern Arizona every month over the course of a one-year period. We also investigated the occurrence of Arcobacter and associated virulence genes (the invasion gene ciaB and the phospholipase gene pldA) to determine reduction by two different wastewater treatment processes.
2. Results
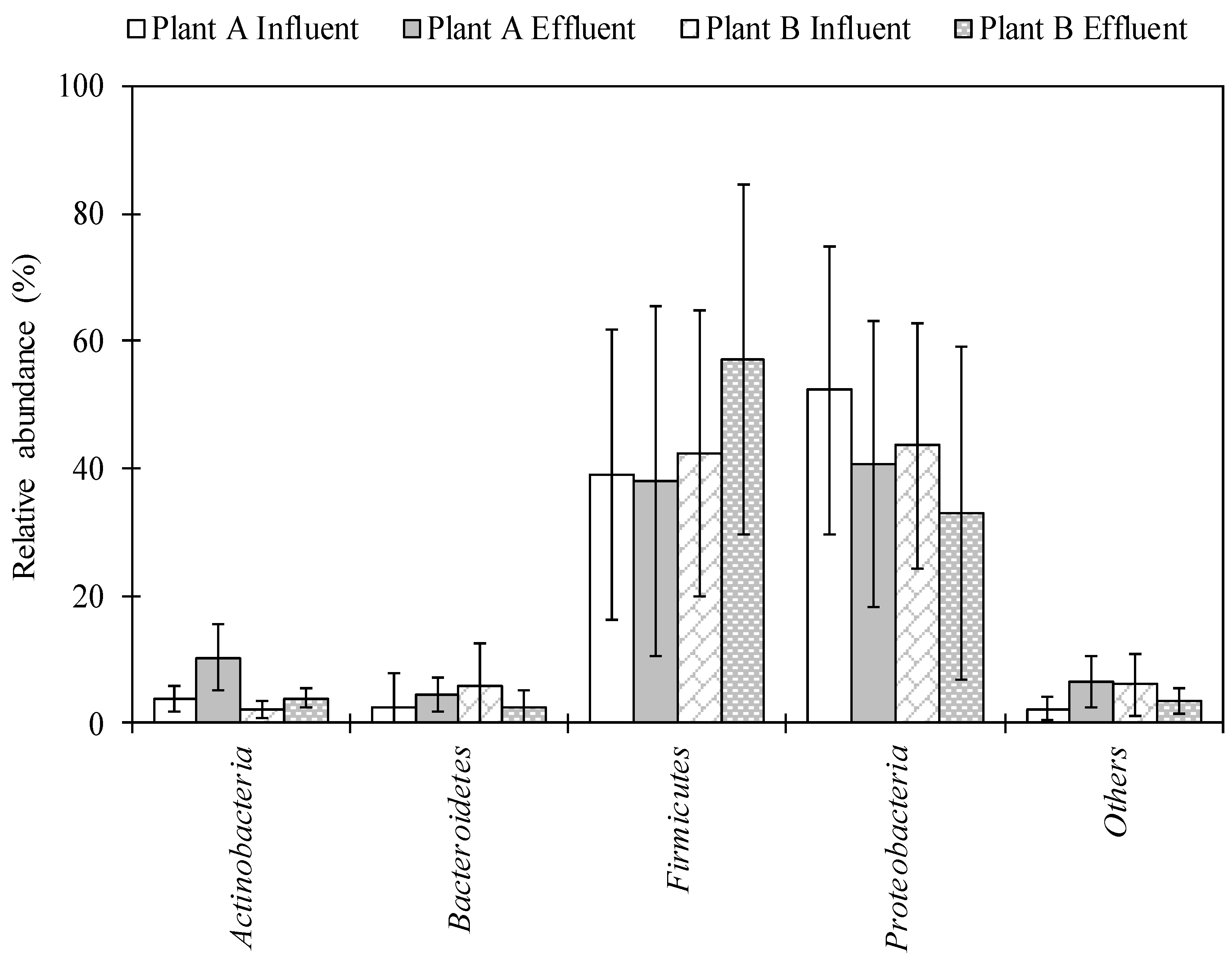
2.1. Characterization of Bacterial Community Using Next-Generation Sequencing (NGS) in Wastewater Samples
The total numbers of sequences detected using NGS analysis ranged from 4533 to 233,125 and from 60,010 to 272,204 in influent and effluent samples collected in Plant A, respectively. A total of 64,078–262,343 and 63,136–279,359 sequence reads were obtained in influent and effluent samples collected in Plant B, respectively. A total of 29 phyla were detected. Proteobacteria and Firmicutes were identified in high abundance, ranging from 3.2–89.4% and 4.0–90.8% in Plant A and 3.9–88.9% and 9.6–91.1% in Plant B, respectively (Figure 1). A total of 155–558 and 336–561 genera were identified in the influent and effluent samples of Plant A, respectively. For Plant B, 335–576 and 334–603 genera were identified in the influent and effluent samples, respectively. Among the 847 genera identified, 55 genera, including Acinetobacter, Arcobacter, Bacillus, and Pseudomonas, were detected in all influent and effluent wastewater samples from both Plant A and Plant B.
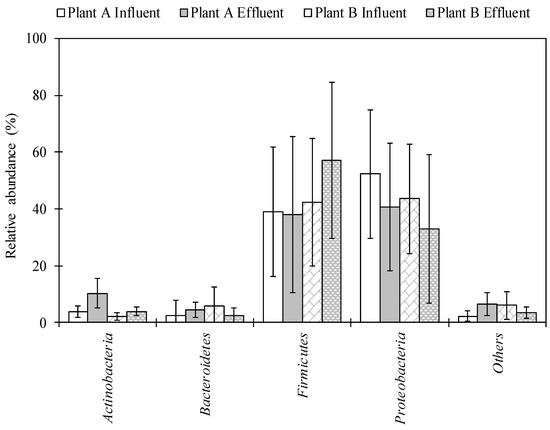
Figure 1.
Bacterial composition in wastewater samples of Plant A and Plant B at the phylum level. Bars and error bar represent an average of twelve samples and standard deviation, respectively.
Among the 113 potentially pathogenic bacteria identified, 42 genera were detected at abundances of >1% in at least one of the tested samples (Table 1). In the influent of Plant A, Bacillus, Pseudomonas, and Erwinia were in high abundance (18.6 ± 15.4%, 9.8 ± 24.3%, and 8.5 ± 12.0%, respectively), whereas in effluent samples Bacillus, Mycobacterium, and Acinetobacter were detected in high abundance (25.8 ± 24.7%, 6.5 ± 4.0%, and 3.2 ± 8.0%, respectively). In Plant B, Bacillus, Pseudomonas, and Streptococcus were detected at high abundances (18.9 ± 18.7%, 8.7 ± 22.8%, and 5.1 ± 5.0%, respectively) in influent samples, whereas in effluent samples Bacillus, Pseudomonas, and Acinetobacter were observed to be in high abundances (33.2 ± 25.3%, 5.1 ± 17.3%, and 4.0 ± 11.8%, respectively). There was no significant difference in abundances of most of the potentially pathogenic bacteria between influent and effluent water samples. In Plant A and Plant B, 29 and 34 out of 42 potentially pathogenic bacteria did not show any significant differences in abundance between influent and effluent water samples, respectively (t-test, p > 0.05).

Table 1.
Abundance ratios of potential pathogenic bacteria in influent and effluent water samples from Plant A and Plant B.
2.2. Occurrence of Total Bacteria, Arcobacter and Associated Virulence Genes in Wastewater Samples
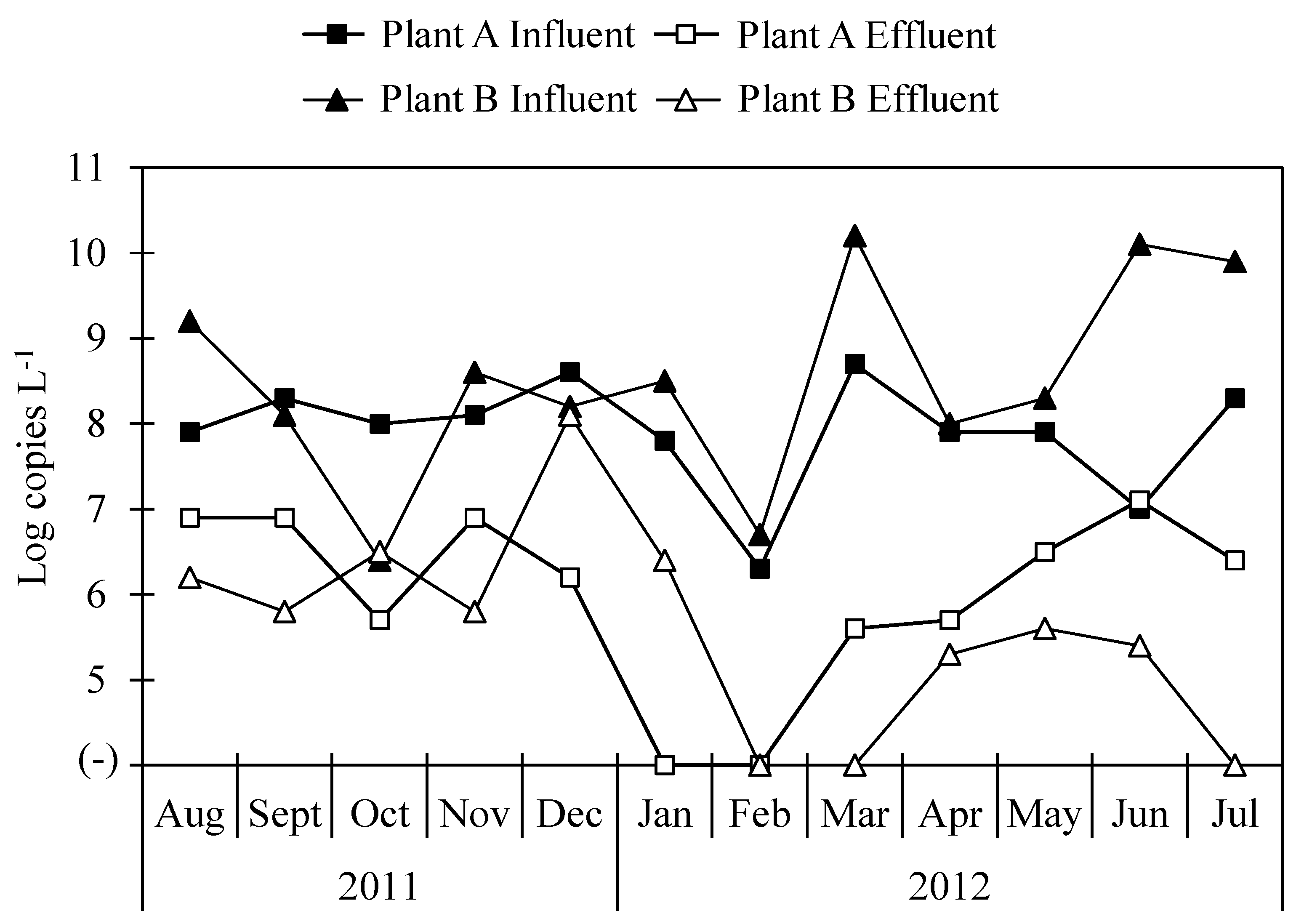
The concentrations of total bacterial 16S rRNA gene in influent samples from both Plant A and Plant B were 9.7 ± 0.3 and 9.7 ± 0.4 log copies L−1, respectively, and in effluent samples, the concentrations of total bacterial 16S rRNA gene were 8.0 ± 0.4 and 8.3 ± 0.2 log copies L−1 in Plant A and Plant B, respectively. The concentrations of Arcobacter 16S rRNA gene in wastewater samples collected from Plant A and Plant B are shown in Figure 2. Arcobacter was detected in all influent samples from both Plant A and Plant B and in ten (83%) and nine (75%) of twelve effluent samples at each plant. Average concentrations of Arcobacter 16S rRNA gene were 7.9 ± 0.7 and 8.5 ± 1.2 log copies L−1 in influent samples of Plant A and Plant B, respectively. Those in effluent samples were 6.4 ± 0.6 and 6.1 ± 0.9 log copies L−1 in Plant A and Plant B, respectively.
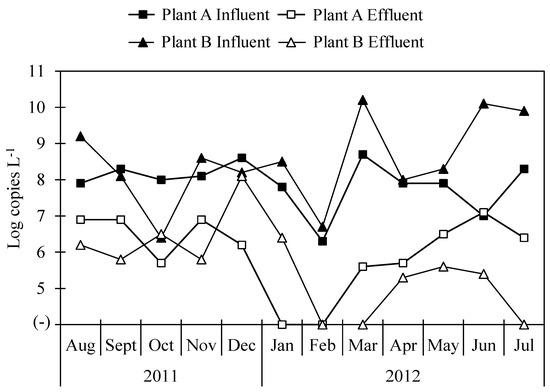
Figure 2.
Concentration of Arcobacter 16S rRNA gene in influent and effluent samples.
In our previous study, concentrations of total bacterial 16S rRNA gene obtained using qPCR and abundances of Arcobacter obtained using NGS analysis were utilized to compare results obtained from the two methodologies [31]. Here, concentrations of Arcobacter 16S rRNA gene were calculated similarly for both Plant A and Plant B. The correlation coefficient of Arcobacter 16S rRNA gene concentrations calculated by the two different methods were −0.95 and −0.99 for Plant A and Plant B, respectively.
The ciaB gene was detected in eleven (92%) and twelve (100%) of twelve influent samples collected from both Plant A and Plant B, respectively, while the pldA gene was detected in eight (67%) and six (50%) influent samples from Plant A and Plant B, respectively. Average concentrations of ciaB and pldA genes in influent samples were 7.3 ± 0.7 and 5.9 ± 0.2 log copies L−1 in Plant A, respectively, whereas those in influent samples were 7.8 ± 1.2 and 6.8 ± 0.8 log copies L−1 in Plant B, respectively (Table 2).

Table 2.
Concentrations of ciaB and pldA genes of Arcobacter in influent and effluent samples.
2.3. Reduction Ratios of Arcobacter and Virulence Genes During Wastewater Treatment
The log reduction ratios of 16S rRNA genes of total bacteria and Arcobacter in Plant A, where a conventional activated sludge process is utilized, were 1.6 ± 0.4 (n = 12) and 1.7 ± 0.9 (n = 10), respectively. Those of total bacteria and Arcobacter at Plant B, which utilizes a biological trickling filter process, were 1.5 ± 0.4 (n = 12) and 2.3 ± 1.5 (n = 9), respectively. For Arcobacter, the highest reduction ratios in Plant A and Plant B were obtained in March (3.1 log) and June (4.7 log), respectively. Even though the methodologies for the treatment of wastewater were different between locations, there was no significant difference in log reductions of total bacteria or Arcobacter between Plant A and Plant B. The log reduction ratio of the ciaB gene was 1.7 ± 1.0 (n = 8) and 2.1 ± 1.8 (n = 7) in Plant A and Plant B, respectively (Table 3). The log reduction ratio of the pldA gene was 1.1 ± 0.1 (n = 2) in Plant A.

Table 3.
Reduction of total bacteria and Arcobacter at WWTPs.
3. Discussion
The overall diversity and abundance of bacterial genera were identified in influent and effluent of wastewater samples using NGS. The phylum Proteobacteria was the most abundant phylum, followed by Firmicutes, Bacteroidetes, and Actinobacteria in both WWTPs (Figure 1). These data are in agreement with results of previous studies that tested untreated sewage [27,28] and WWTP samples [29] in the United States. This is also in agreement with the study by Zhang et al. (2012), in which Proteobacteria were the most abundant phylum at 14 WWTPs in samples collected from Asia (China, Hong Kong, and Singapore) and North America (Canada and the United States) [32]. Average abundance of Bacillus in both plant locations was found to be highest in the effluent (among potentially pathogenic bacteria) (Table 1). Lee et al. (2008) found that concentrations of Bacillus cereus were highest in final effluents (disinfected by chlorination and UV radiation) in two WWTPs in Canada [33]. The genus Pseudomonas was also abundant and has commonly been detected in wastewater samples collected in the United States [29]. Pseudomonas is ubiquitous in the environment, and pathogenic species of Pseudomonas can cause infections in hospital patients and/or those with weakened immune systems, such as pneumonia and blood infections [34]. The abundance of Mycobacterium significantly increased from influent to effluent samples in Plant A. This taxon has also been found in effluent and activated sludge of a WWTP in Hong Kong via metagenomic analysis [35,36].
The occurrence of potentially pathogenic bacteria in influent and effluent samples emphasizes their ability to persist and be discharged in the environment. WWTPs have been considered a potential hub for evolution and dissemination of antibiotic resistance and virulence genes [37]. Focusing on construction and maintenance of treatment plants, treatment methodologies, and disinfection processes before releasing treated wastewater into the environment will help reduce the spread of potentially pathogenic bacteria into the water environment.
Prevalence of Arcobacter has been found to be higher in wastewater compared to other aquatic environments, such as lakes, river, recreational beaches, groundwater, seawater, and drinking water [38]. This is also supported by metagenomic analysis of wastewater samples from various locations where Arcobacter has been detected as one of the most abundant genera [19,27,28]. Arcobacter is ubiquitous in the environment, and this taxon has been associated with both humans and animals and can cause gastroenteritis, septicemia, mastitis, reproductive disorders, and abortion in livestock [22,39]. Arcobacter was detected in all influent samples tested in the current study, and detection in the effluent of both plant locations indicates a high tolerance capability of Arcobacter, which can ultimately lead to persistence and spread of the pathogenic bacteria.
Wastewater samples tested in this study have been previously tested for viruses and protozoa. Most of the viruses and protozoa tested did not show any significant differences in log reductions between Plant A and Plant B [40,41]. Similarly, there was no significant difference in the reduction of Arcobacter in either Plant A or Plant B. The efficiencies of either plant, operating the activated sludge process or the biological trickling filter process, were not effective in removing pathogens; therefore, they must be improved. When less effectively treated water is used for agricultural or recreational uses, there is a high risk of contamination with Arcobacter, as one of the routes of transmission of this taxon is water. Seasonal variations, especially during extreme rainfall, can affect the transportation of Arcobacter from WWTPs to groundwater [38]. The concentration of Arcobacter was lowest in February but increased in March in Plant A and Plant B. In this study, sample collection for a year may not capture the seasonal variation in Arcobacter in WWTPs, and a longer survey period could help gain a better understanding of Arcobacter dynamics.
In this study, the ciaB gene was detected more frequently than the pldA gene in wastewater samples of both plant locations (Table 2). This is likely due to a higher detection frequency for the ciaB gene in all Arcobacter strains compared to the pldA gene [42]. These virulence genes have been detected in Arcobacter strains obtained from a variety of species, including humans, chickens, pigs, cattle, sheep, horses, dogs, clams, mussels, and in milk [42,43,44]. These genes have also been identified from isolates of Arcobacter recovered from fecal samples originating from humans and from animals [45]. It is evident from these reports that these virulence genes are ubiquitous in the environment. The detection of ciaB and pldA genes in effluent samples may influence nearby environmental microbial communities, and the dispersal and fate of these genes in water environments suggests negative impacts associated with contaminated wastewater effluent on the gene content of water bodies [46]. The presence of potentially pathogenic organisms and their virulence genes in water demonstrates the importance of monitoring effluent water samples before release into the water bodies and the need for disinfection of the effluent before discharge.
In summary, the characterization of the bacterial community in two WWTPs via NGS detected 113 (13%) of 847 genera as potential pathogenic bacteria. In all influent samples of both plant locations, the Arcobacter 16S rRNA gene was quantified and detected in ten and nine of twelve effluent samples at Plant A and Plant B, respectively. There was no significant difference in reduction ratios of the Arcobacter 16S rRNA gene between the two plants. Virulence genes, ciaB and pldA, were also detected in both influent and effluent samples from Plant A and Plant B. The presence of potential pathogenic bacteria and quantification of Arcobacter and its virulence genes in effluent samples of WWTPs demonstrate the need for disinfection before discharge into the environment.
4. Materials and Methods
4.1. Collection of WWTP Samples
As described previously [40], during a 12-month period between August 2011 and July 2012, monthly sampling of influent and effluent wastewater was conducted at two WWTPs in southern Arizona. Wastewater samples from Plants A (conventional activated sludge process) and B (biological trickling filter process) were collected as grab samples. Each sampling was conducted at ~10:00.
4.2. Bacterial DNA Extraction
Water samples (100 mL of influent and 1000 mL of effluent) were used to measure viral concentrations using a mixed cellulose ester membrane (pore size: 0.45 µm, diameter: 90 mm; Merck Millipore, Cat. No. HAWP-090-00, Billerica, MA, USA) as described previously [40]. The membrane filter was used for extraction of bacterial DNA after virus elution. In brief, the membrane filter was cut in half, and one piece was mixed with 10 mL of surfactant-based elution buffer in a 50-mL tube. The tube was vortexed vigorously for ~5 min, and the eluate was transferred to a new tube. This step was repeated by adding 5 mL of the elution buffer to the original tube, resulting in ~15 mL of eluate. The tube was centrifuged at 2000× g for 10 min at 4 °C, and the supernatant was removed. A volume of 200 µL phosphate buffered saline was added to the tube containing the pellet, mixed, and transferred into a new 2-mL microtube. This step was repeated until the final volume of the bacterial concentrate reached 1 mL. Bacterial DNA (200 μL) was extracted from 200 μL of the concentrate using a QIAamp DNA mini kit (QIAGEN, Hilden, Germany).
4.3. NGS for Characterization of Bacterial Communities
The bacterial DNA extracts of wastewater samples were used for metagenomic sequencing via a MiSeq gene sequencer (Illumina, San Diego, CA, USA) as described previously [47]. Operational taxonomic units obtained were analyzed based on the bacterial domain, phylum, family, and genus. A genus was considered as a potentially pathogenic bacterium if any one species of the genus was categorized as biosafety level 2 or 3 by the American Biological Safety Association (https://my.absa.org/tiki-index.php?page=Riskgroups) as described previously [48]. The raw sequences obtained were registered in the NCBI Sequence Read Archive under the accession number PRJNA525124.
4.4. qPCR of Total Bacteria and Arcobacter
For total bacterial 16S rRNA gene, qPCR was performed using 515F and U806R primers [49,50] with the thermal conditions, qPCR mixture components, and qPCR reaction conditions as previously described [48]. Arcobacter 16S rRNA gene was quantified via qPCR using 2 µL of template DNA, 12.5 µL of a MightyAmp for Real Time (SYBR Plus) (Takara Bio, Kusatsu, Japan), 0.1 µL each of 1 µM of Arco-F and Arco-R-rev primers [31], and 10.3 µL of ultrapure water. For the virulence genes ciaB and pldA, 2 µL of template DNA, 12.5 µL of a SYBR Premix Ex Taq II (Tli RNase Plus) (Takara Bio), 0.1 µL each of 1 µM of ciaB-F and ciaB-R primers (for the ciaB gene) [42] or pldA-F and pldA-R primers (for the pldA gene) [42], and 10.3 µL of ultrapure water were used. qPCR was performed with a Thermal Cycler Dice Real Time System Single TP850 (Takara Bio) under the following thermal conditions: for Arcobacter 16S rRNA gene, 98 °C for 2 min, followed by 35 cycles at 98 °C for 10 s, 55 °C for 30 s, and 68 °C for 40 s; for the ciaB gene, 95 °C for 30 s, followed by 35 cycles at 94 °C for 15 s, 55 °C for 30 s, and 72 °C for 20 s; and for the pldA gene, 95 °C for 30s, followed by 35 cycles at 94 °C for 15 s, 56 °C for 45 s, and 72 °C for 20 s. A melting curve analysis was performed to confirm the generation of specific qPCR products.
4.5. Statistical Analysis
Student’s t-tests were performed with Microsoft Excel 2018 (Microsoft Corporation, Redmond, WA, USA) to determine the difference in abundance of bacteria in influent and effluent wastewater samples. The test was also used to determine whether log reductions of total bacteria and Arcobacter at Plants A and B were statistically different. Differences were considered statistically significant if the resulting p value was <0.05. The log reduction of Arcobacter was calculated from the samples, which were qPCR-positive for both influent and effluent samples.
Author Contributions
conceptualization, M.K., Y.T., C.P.G., and E.H.; methodology, R.G.S., Y.T., and E.H.; software, R.G.S.; validation, Y.T. and E.H.; formal analysis, R.G.S.; investigation, R.G.S., S.P.S., M.K.; data curation, Y.T. and E.H.; writing—original draft preparation, R.G.S.; writing—review and editing, S.P.S., M.K., Y.T., C.P.G., and E.H.; visualization, R.G.S.; supervision, E.H.; project administration, M.K., C.P.G., and E.H.; funding acquisition, E.H.
Funding
This research was supported by a research grant from University of Yamanashi, Japan.
Acknowledgements
The authors thank Bikash Malla and Sarmila Tandukar (University of Yamanashi, Japan) for their invaluable support during laboratory analysis.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Norton-Brandão, D.; Scherrenberg, S.M.; van Lier, J.B. Reclamation of used urban waters for irrigation purposes–a review of treatment technologies. J. Environ. Manag. 2013, 122, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Lazarova, V.; Levine, B.; Sack, J.; Cirelli, G.; Jeffrey, P.; Muntau, H.; Salgot, M.; Brissaud, F. Role of water reuse for enhancing integrated water management in Europe and Mediterranean countries. Water Sci. Technol. 2001, 43, 25–33. [Google Scholar] [CrossRef]
- Asano, T.; Cotruvo, J.A. Groundwater recharge with reclaimed municipal wastewater: Health and regulatory considerations. Water Res. 2004, 38, 1941–1951. [Google Scholar] [CrossRef] [PubMed]
- Angelakis, A.N.; Durham, B. Water recycling and reuse in EUREAU countries: Trends and challenges. Desalination 2008, 218, 3–12. [Google Scholar] [CrossRef]
- Bixio, D.; Thoeye, C.; Wintgens, T.; Hochstrat, R.; Melin, T.; Chikurel, H.; Aharoni, A.; Durham, B. Wastewater reclamation and reuse in the European Union and Israel: Status quo and future prospects. Int. Rev. Environ. Strateg. 2006, 6, 251–268. [Google Scholar]
- Rodriguez-Manzano, J.; Alonso, J.L.; Ferrús, M.A.; Moreno, Y.; Amorós, I.; Calgua, B.; Hundesa, A.; Guerrero-Latorre, L.; Carratala, A.; Rusiñol, M.; et al. Standard and new faecal indicators and pathogens in sewage treatment plants, microbiological parameters for improving the control of reclaimed water. Water Sci. Technol. 2012, 66, 2517–2523. [Google Scholar] [CrossRef]
- Vandamme, P.; Falsen, E.; Rossau, R.; Hoste, B.; Segers, P.; Tytgat, R.; De Ley, J. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: Emendation of generic descriptions and proposal of Arcobacter gen. nov. Int. J. Syst. Bacteriol. 1991, 41, 88–103. [Google Scholar] [CrossRef]
- Vandamme, P.; Vancanneyt, M.; Pot, B.; Mels, L.; Hoste, B.; Dewettinck, D.; Vlaes, L.; Van Den Borre, C.; Higgins, R.; Hommez, J.; et al. Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov., an aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 1992, 42, 344–356. [Google Scholar] [CrossRef][Green Version]
- Banting, G.S.; Braithwaite, S.; Scott, C.; Kim, J.; Jeon, B.; Ashbolt, N.; Ruecker, N.; Tymensen, L.; Charest, J.; Pintar, K.; et al. Evaluation of various Campylobacter -specific qPCR assays for detection and enumeration of Campylobacteraceae in irrigation water and wastewater using a miniaturized MPN-qPCR assay. Appl. Environ. Microbiol. 2016, 82, 4743–4756. [Google Scholar] [CrossRef]
- Webb, A.L.; Taboada, E.N.; Selinger, L.B.; Boras, V.F.; Inglis, G.D. Efficacy of wastewater treatment on Arcobacter butzleri density and strain diversity. Water Res. 2016, 105, 291–296. [Google Scholar] [CrossRef]
- Gao, P.; Xu, W.; Sontag, P.; Li, X.; Xue, G. Correlating microbial community compositions with environmental factors in activated sludge from four full-scale municipal wastewater treatment plants in Shanghai, China. Appl. Microbiol. Biotechnol. 2016, 100, 4663–4673. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Zhang, X.; Miao, Y.; Zhao, Y.; Ye, L.; Li, B.; Zhang, T. Fate of antibiotic resistance genes and their associations with bacterial community in livestock breeding wastewater and its receiving river water. Water Res. 2017, 124, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Ju, F.; Li, B.; Ma, L.; Wang, Y.; Huang, D.; Zhang, T. Antibiotic resistance genes and human bacterial pathogens: Co- occurrence, removal, and enrichment in municipal sewage sludge digesters. Water Res. 2016, 91, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Lu, P. Characterization of bacterial communities in sediments receiving various wastewater effluents with high-throughput sequencing analysis. Microb. Ecol. 2014, 67, 612–623. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, X.-X.; Wang, Z.; Huang, K.; Wang, Y.; Liang, W.; Tan, Y.; Liu, B.; Tang, J. Bacterial pathogens and community composition in advanced sewage treatment systems revealed by metagenomics analysis based on high-throughput sequencing. PLoS ONE 2015, 10, e0125549. [Google Scholar] [CrossRef]
- Snaidr, J.; Amann, R.; Huber, I.; Ludwig, W. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 1997, 63, 2884–2896. [Google Scholar]
- Stampi, S.; De Luca, G.; Varoli, O.; Zanetti, F. Occurrence, removal and seasonal variation of thermophilic Campylobacters and Arcobacter in sewage sludge. Zentralblatt für Hygiene und Umweltmedizin 1999, 202, 19–27. [Google Scholar] [CrossRef]
- Al-jassim, N.; Ansari, M.I.; Harb, M.; Hong, P. Removal of bacterial contaminants and antibiotic resistance genes by conventional wastewater treatment processes in Saudi Arabia: Is the treated wastewater safe to reuse for agricultural irrigation? Water Res. 2015, 73, 277–290. [Google Scholar] [CrossRef]
- Diergaardt, S.M.; Venter, S.N.; Spreeth, A.; Theron, J.; Brözel, V.S. The occurrence of campylobacters in water sources in South Africa. Water Res. 2004, 38, 2589–2595. [Google Scholar] [CrossRef]
- Collado, L.; Inza, I.; Guarro, J.; Figueras, M.J. Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution. Environ. Microbiol. 2008, 10, 1635–1640. [Google Scholar] [CrossRef]
- Collado, L.; Kasimir, G.; Perez, U.; Bosch, A.; Pinto, R.; Saucedo, G.; Huguet, J.M.; Jose, M. Occurrence and diversity of Arcobacter spp. along the Llobregat River catchment, at sewage effluents and in a drinking water treatment plant. Water Res. 2010, 44, 3696–3702. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Botella, S.; Montes, R.M.; Moreno, Y.; Ferrús, M.A. Direct detection and identification of Arcobacter species by multiplex PCR in chicken and wastewater samples from Spain. J. Food Prot. 2007, 70, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Levican, A.; Collado, L.; Figueras, M.J. The use of two culturing methods in parallel reveals a high prevalence and diversity of Arcobacter spp. in a wastewater treatment plant. BioMed. Res. Int. 2016, 2016, 8132058. [Google Scholar] [CrossRef] [PubMed]
- Moreno, Y.; Botella, S.; Alonso, J.L.; Ferrus, M.A.; Hernandez, M.; Hernandez, J. Specific detection of Arcobacter and Campylobacter strains in water and sewage by PCR and Fluorescent In Situ Hybridization. Appl. Environ. Microbiol. 2003, 69, 1181–1186. [Google Scholar] [CrossRef]
- Akinglouchu, F. Isolation of Arcobacter Species from Different Water Sources and Characterization of Isolated Species by Molecular Techniques. Master’s Thesis, Graduate School of Engineering and Sciences of Izmir Institute of Technology, Izmir, Turkey, 2011. [Google Scholar]
- Merga, J.Y.; Royden, A.; Pandey, A.K.; Williams, N.J. Arcobacter spp. isolated from untreated domestic effluent. Lett. Appl. Microbiol. 2014, 59, 122–126. [Google Scholar] [CrossRef]
- Millar, J.A.; Raghavan, R. Accumulation and expression of multiple antibiotic resistance genes in Arcobacter cryaerophilus that thrives in sewage. PeerJ 2017, 5, e3269. [Google Scholar] [CrossRef]
- McLellan, S.L.; Huse, S.M.; Mueller-Spitz, S.R.; Andreishcheva, E.N.; Sogin, M.L. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ. Microbiol. 2010, 12, 378–392. [Google Scholar] [CrossRef]
- Sigala, J.; Unc, A. Pyrosequencing estimates of the diversity of antibiotic resistant bacteria in a wastewater system. Water Sci. Technol. 2013, 67, 1534–1543. [Google Scholar] [CrossRef]
- Fong, T.T.; Mansfield, L.S.; Wilson, D.L.; Schwab, D.J.; Molloy, S.L.; Rose, J.B. Massive microbiological groundwater contamination associated with a waterborne outbreak in Lake Erie, South Bass Island, Ohio. Environ. Health Perspect. 2007, 115, 856–864. [Google Scholar] [CrossRef]
- Ghaju Shrestha, R.; Tanaka, Y.; Malla, B.; Tandukar, S.; Bhandari, D.; Inoue, D.; Sei, K.; Sherchand, J.B.; Haramoto, E. Development of a quantitative PCR assay for Arcobacter spp. and its application to environmental water samples. Microbes Environ. 2018, 33, 309–316. [Google Scholar] [CrossRef]
- Zhang, T.; Shao, M.F.; Ye, L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012, 6, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Lauder, H.; Cruwys, H.; Falletta, P.; Beaudette, L.A. Development and application of an oligonucleotide microarray and real-time quantitative PCR for detection of wastewater bacterial pathogens. Sci. Total Environ. 2008, 398, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Iglewski, B.H. Pseudomonas. In Medical Microbiology; Baron, S., Ed.; The University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; Chapter 27. [Google Scholar]
- Cai, L.; Ju, F.; Zhang, T. Tracking human sewage microbiome in a municipal wastewater treatment plant. Appl. Microbiol. Biotechnol. 2014, 98, 3317–3326. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Zhang, T. Detecting human bacterial pathogens in wastewater treatment plants by a high-throughput shotgun sequencing technique. Environ. Sci. Technol. 2013, 47, 5433–5441. [Google Scholar] [CrossRef] [PubMed]
- Varela, A.R.; Manaia, C.M. Human health implications of clinically relevant bacteria in wastewater habitats. Environ. Sci. Pollut. Res. 2013, 20, 3550–3569. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.T.D.; Lee, J. Global distribution and prevalence of Arcobacter in food and water. Zoonoses Public Health 2015, 62, 579–589. [Google Scholar] [CrossRef]
- Collado, L.; Figueras, M.J. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 2011, 24, 174–192. [Google Scholar] [CrossRef]
- Kitajima, M.; Haramoto, E.; Iker, B.C.; Gerba, C.P. Occurrence of Cryptosporidium, Giardia, and Cyclospora in influent and effluent water at wastewater treatment plants in Arizona. Sci. Total Environ. 2014, 484, 129–136. [Google Scholar] [CrossRef]
- Kitajima, M.; Iker, B.C.; Pepper, I.L.; Gerba, C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes—Identification of potential viral indicators. Sci. Total Environ. 2014, 488, 290–296. [Google Scholar] [CrossRef]
- Douidah, L.; De Zutter, L.; Baré, J.; De Vos, P.; Vandamme, P.; Vandenberg, O.; Van Den Abeele, A.M.; Houf, K. Occurrence of putative virulence genes in Arcobacter species isolated from humans and animals. J. Clin. Microbiol. 2012, 50, 735–741. [Google Scholar] [CrossRef]
- Tabatabaei, M.; Shirzad Aski, H.; Shayegh, H.; Khoshbakht, R. Occurrence of six virulence-associated genes in Arcobacter species isolated from various sources in Shiraz, Southern Iran. Microb. Pathog. 2014, 66, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Girbau, C.; Guerra, C.; Martínez-Malaxetxebarria, I.; Alonso, R.; Fernández-Astorga, A. Prevalence of ten putative virulence genes in the emerging foodborne pathogen Arcobacter isolated from food products. Food Microbiol. 2015, 52, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Whiteduck-Léveillée, J.; Cloutier, M.; Topp, E.; Lapen, D.R.; Talbot, G.; Villemur, R.; Khan, I.U.H. Development and evaluation of multiplex PCR assays for rapid detection of virulence-associated genes in Arcobacter species. J. Microbiol. Methods 2016, 121, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.T.T.; Petrovich, M.L.; Chaudhary, A.; Wright, D.; Murphy, B.; Wells, G.; Poretsky, R. Metagenomic analysis reveals the impact of wastewater treatment plants on the dispersal of microorganisms. Appl. Environ. Microbiol. 2018, 84, e02168-17. [Google Scholar]
- Xue, J.; Schmitz, B.W.; Caton, K.; Zhang, B.; Zabaleta, J.; Garai, J.; Taylor, C.M.; Romanchishina, T.; Gerba, C.P.; Pepper, I.L.; et al. Assessing the spatial and temporal variability of bacterial communities in two Bardenpho wastewater treatment systems via Illumina MiSeq sequencing. Sci. Total Environ. 2019, 657, 1543–1552. [Google Scholar] [CrossRef]
- Ghaju Shrestha, R.; Tanaka, Y.; Malla, B.; Bhandari, D.; Tandukar, S.; Inoue, D.; Sei, K.; Sherchand, J.B.; Haramoto, E. Next-generation sequencing identification of pathogenic bacterial genes and their relationship with fecal indicator bacteria in different water sources in the Kathmandu Valley, Nepal. Sci. Total Environ. 2017, 601, 278–284. [Google Scholar] [CrossRef]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef]
- Takai, K.; Horikoshi, K. Rapid detection and quantification of members of the archael community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 2000, 66, 5066–5072. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).