Occurrence of Campylobacter jejuni and Campylobacter coli in Cattle and Sheep in Northern Spain and Changes in Antimicrobial Resistance in Two Studies 10-years Apart
Abstract
1. Introduction
2. Results
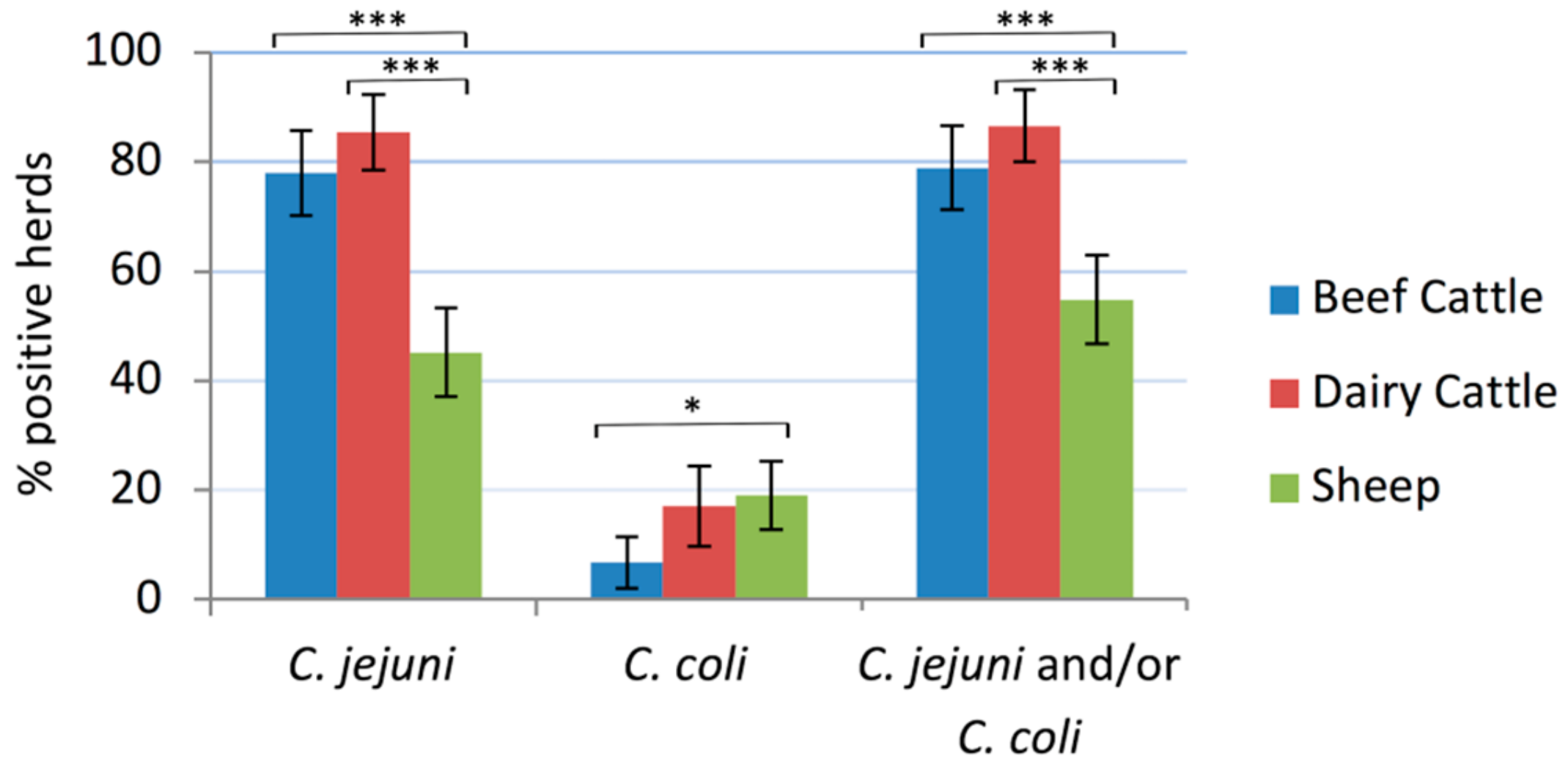
2.1. Campylobacter Herd Prevalence
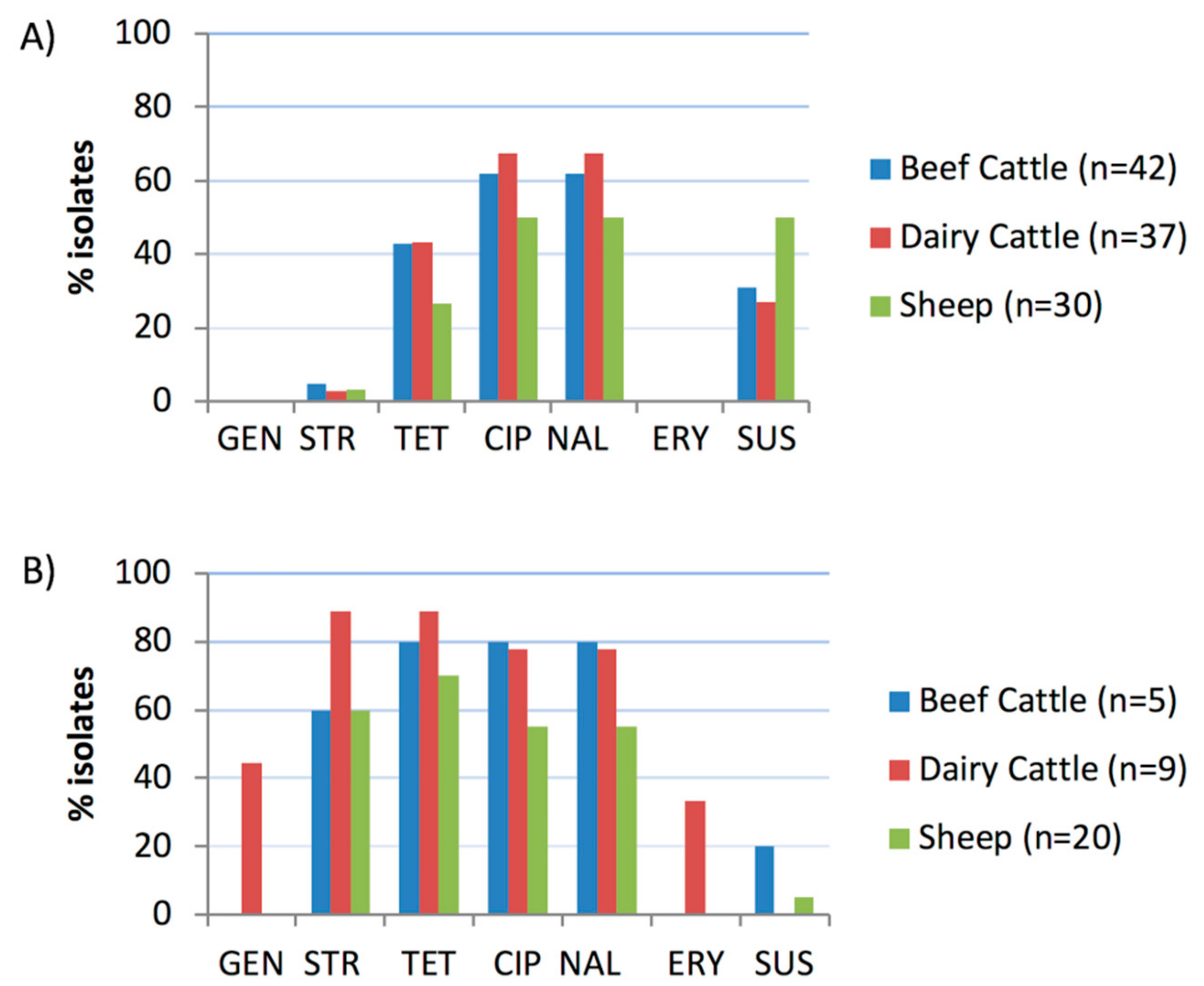
2.2. Antimicrobial Susceptibility Tests
2.3. Real-Time PCR Discrimination of Single Nucleotide Polymorphisms (SNPs) Associated to Quinolone and Macrolide Resistance
2.4. Isolation of Macrolide-Resistant Campylobacter Using a PCR-Based Screening Method Followed by Selective Isolation
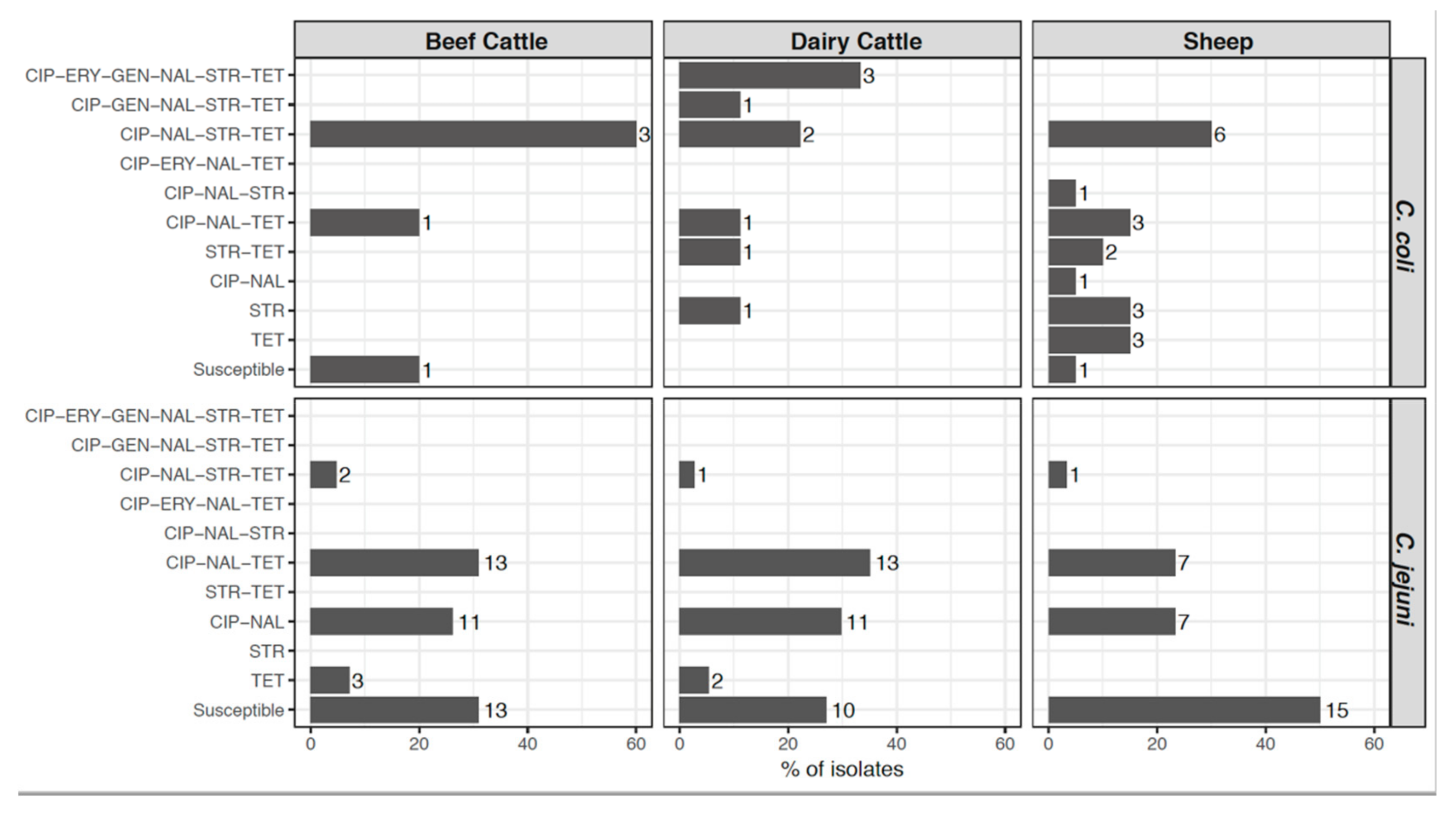
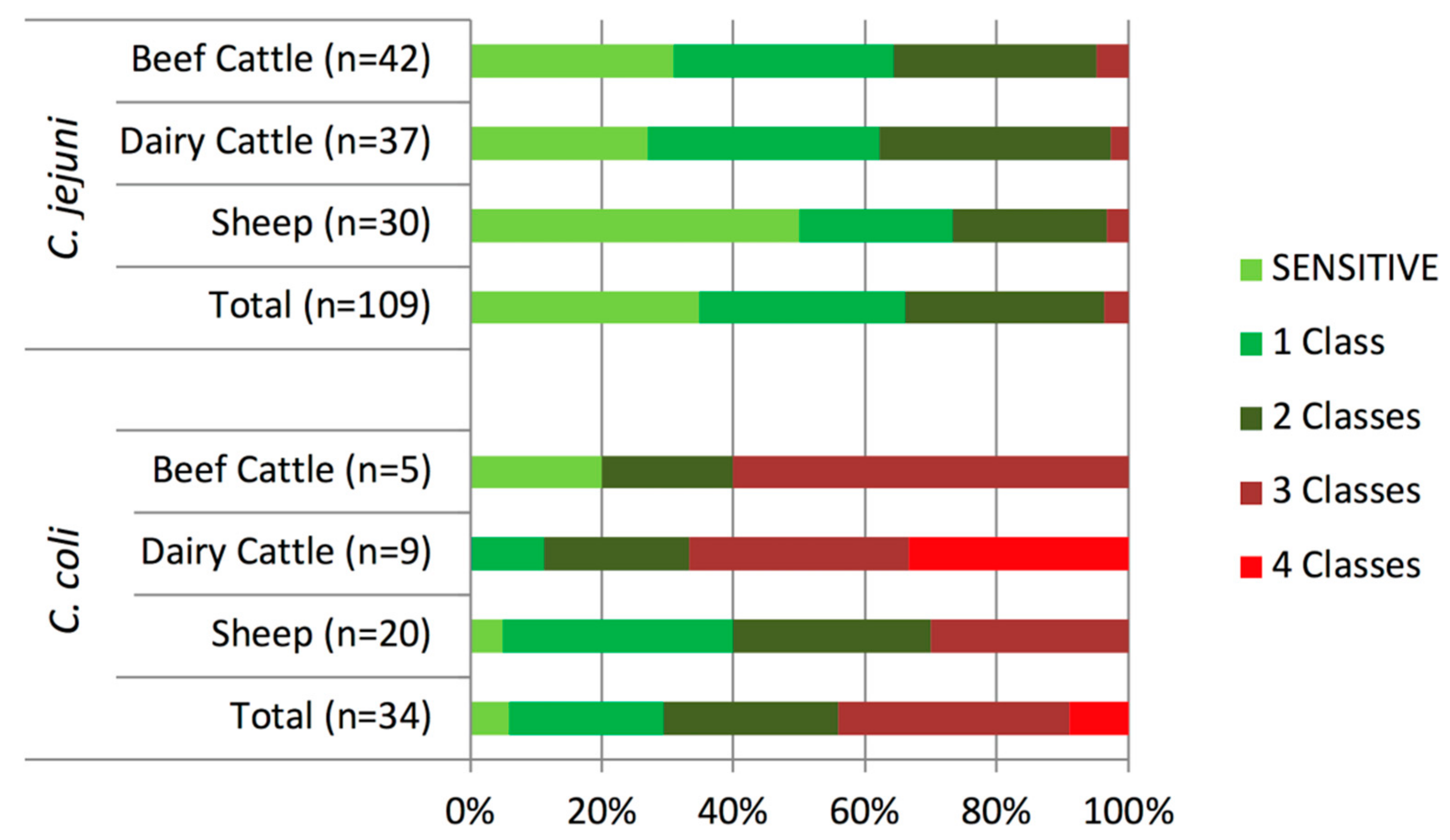
2.5. Changes in Campylobacter Antimicrobial Resistance Profiles in Two Studies Carried Out 10-Years Apart
2.6. Campylobacter coli Strain Characterization by Multilocus Sequence Typing (MLST)
3. Discussion
4. Materials and Methods
4.1. Sampling Design
4.2. Campylobacter Isolation and Identification
4.3. Antimicrobial Resistance: Broth Microdilution Tests and SNP Discrimination by Real-Time PCR
4.4. PCR-Based Screening Method for the Isolation of Macrolide-Resistant Campylobacter
4.5. Assessment of Changes in Campylobacter Antimicrobial Resistance Profiles in Two Studies Carried Out 10-Years Apart
4.6. Campylobacter coli Strain Characterization by Multilocus Sequence Typing (MLST)
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, 5500. [Google Scholar]
- Anonymous. Informe 2017: Salud Pública y Adicciones; Servicio Central de Publicaciones del Gobierno Vasco: Vitoria-Gasteiz, Spain, 2018. [Google Scholar]
- Havelaar, A.H.; Ivarsson, S.; Löfdahl, M.; Nauta, M.J. Estimating the true incidence of campylobacteriosis and salmonellosis in the European Union, 2009. Epidemiol. Infect. 2013, 141, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, S.K.; Dallas, J.F.; Strachan, N.J.C.; MacRae, M.; McCarthy, N.D.; Wilson, D.J.; Gormley, F.J.; Falush, D.; Ogden, I.D.; Maiden, M.C.J.; et al. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 2009, 48, 1072–1078. [Google Scholar] [CrossRef]
- Roux, F.; Sproston, E.; Rotariu, O.; MacRae, M.; Sheppard, S.K.; Bessell, P.; Smith-Palmer, A.; Cowden, J.; Maiden, M.C.J.; Forbes, K.J.; et al. Elucidating the aetiology of human Campylobacter coli infections. PLoS ONE 2013, 8, e64504. [Google Scholar] [CrossRef]
- Jacobs-Reitsma, W.; Wagenaar, J. Campylobacter in the food supply. In Campylobacter; Nachamkin, I., Szymanski, C., Blaser, M., Eds.; ASM Press: Washington, DC, USA, 2008; ISBN 978-1-55581-437-3. [Google Scholar]
- Heuvelink, A.E.; van Heerwaarden, C.; Zwartkruis-Nahuis, A.; Tilburg, J.J.H.C.; Bos, M.H.; Heilmann, F.G.C.; Hofhuis, A.; Hoekstra, T.; de Boer, E. Two outbreaks of campylobacteriosis associated with the consumption of raw cows’ milk. Int. J. Food Microbiol. 2009, 134, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.G.; Price, L.; Ahmed, R.; Woodward, D.L.; Melito, P.L.; Rodgers, F.G.; Jamieson, F.; Ciebin, B.; Li, A.; Ellis, A. Characterization of waterborne outbreak–associated Campylobacter jejuni, Walkerton, Ontario. Emerg. Infect. Dis. 2003, 9, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- Luangtongkum, T.; Jeon, B.; Han, J.; Plummer, P.; Logue, C.M.; Zhang, Q. Antibiotic resistance in Campylobacter Emergence, transmission and persistence. Future Microbiol. 2009, 4, 189–200. [Google Scholar] [CrossRef]
- Wieczorek, K.; Osek, J. Antimicrobial resistance mechanisms among Campylobacter. Biomed. Res. Int. 2013, 2013, 340605. [Google Scholar] [CrossRef]
- Mughini Gras, L.; Smid, J.H.; Wagenaar, J.A.; de Boer, A.G.; Havelaar, A.H.; Friesema, I.H.M.; French, N.P.; Busani, L.; van Pelt, W. Risk factors for campylobacteriosis of chicken, ruminant, and environmental origin: A combined case-control and source attribution analysis. PLoS ONE 2012, 7, e42599. [Google Scholar] [CrossRef]
- Mullner, P.; Spencer, S.E.F.; Wilson, D.J.; Jones, G.; Noble, A.D.; Midwinter, A.C.; Collins-Emerson, J.M.; Carter, P.; Hathaway, S.; French, N.P. Assigning the source of human campylobacteriosis in New Zealand: A comparative genetic and epidemiological approach. Infect. Genet. Evol. 2009, 9, 1311–1319. [Google Scholar] [CrossRef]
- Oporto, B.; Esteban, J.I.; Aduriz, G.; Juste, R.A.; Hurtado, A. Prevalence and strain diversity of thermophilic campylobacters in cattle, sheep and swine farms. J. Appl. Microbiol. 2007, 103, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Oporto, B.; Hurtado, A. Emerging thermotolerant Campylobacter species in healthy ruminants and swine. Foodborne Pathog. Dis. 2011, 8, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Oporto, B.; Juste, R.A.; Hurtado, A. Phenotypic and genotypic antimicrobial resistance profiles of Campylobacter jejuni isolated from cattle, sheep, and free-range poultry faeces. Int. J. Microbiol. 2009, 2009, 456573. [Google Scholar] [CrossRef] [PubMed]
- Englen, M.D.; Hill, A.E.; Dargatz, D.A.; Ladely, S.R.; Fedorka-Cray, P.J. Prevalence and antimicrobial resistance of Campylobacter in US dairy cattle. J. Appl. Microbiol. 2007, 102, 1570–1577. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.; Menzies, P.; Reid-Smith, R.J.; Avery, B.P.; Mcewen, S.A.; Moon, C.S.; Berke, O. Antimicrobial resistance in Campylobacter spp. isolated from Ontario sheep flocks and associations between antimicrobial use and antimicrobial resistance. Zoonoses Public Health 2012, 59, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Rotariu, O.; Dallas, J.F.; Ogden, I.D.; MacRae, M.; Sheppard, S.K.; Maiden, M.C.J.; Gormley, F.J.; Forbes, K.J.; Strachan, N.J.C. Spatiotemporal homogeneity of Campylobacter subtypes from cattle and sheep across northeastern and southwestern Scotland. Appl. Environ. Microbiol. 2009, 75, 6275–6281. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sproston, E.L.; Ogden, I.D.; MacRae, M.; Dallas, J.F.; Sheppard, S.K.; Cody, A.J.; Colles, F.M.; Wilson, M.J.; Forbes, K.J.; Strachan, N.J.C. Temporal variation and host association in the Campylobacter population in a longitudinal ruminant farm study. Appl. Environ. Microbiol. 2011, 77, 6579–6586. [Google Scholar] [CrossRef] [PubMed]
- Oporto, B.; Juste, R.A.; López-Portolés, J.A.; Hurtado, A. Genetic diversity among Campylobacter jejuni isolates from healthy livestock and their links to human isolates in Spain. Zoonoses Public Health 2011, 58, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, S.K.; Didelot, X.; Jolley, K.A.; Darling, A.E.; Pascoe, B.; Meric, G.; Kelly, D.J.; Cody, A.; Colles, F.M.; Strachan, N.J.C.; et al. Progressive genome-wide introgression in agricultural Campylobacter coli. Mol. Ecol. 2013, 22, 1051–1064. [Google Scholar] [CrossRef] [PubMed]
- Kahlmeter, G.; Brown, D.F.J.; Goldstein, F.W.; MacGowan, A.P.; Mouton, J.W.; Österlund, A.; Rodloff, A.; Steinbakk, M.; Urbaskova, P.; Vatopoulos, A. European harmonization of MIC breakpoints for antimicrobial susceptibility testing of bacteria. J. Antimicrob. Chemother. 2003, 52, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Englen, M.D.; Fedorka-Cray, P.J.; Ladely, S.R.; Dargatz, D.A. Antimicrobial resistance patterns of Campylobacter from feedlot cattle. J. Appl. Microbiol. 2005, 99, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Châtre, P.; Haenni, M.; Meunier, D.; Botrel, M.A.; Calavas, D.; Madec, J.Y. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from cattle between 2002 and 2006 in France. J. Food Prot. 2010, 73, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Bywater, R.; Deluyker, H.; Deroover, E.; de Jong, A.; Marion, H.; McConville, M.; Rowan, T.; Shryock, T.; Shuster, D.; Thomas, V.; et al. A European survey of antimicrobial susceptibility among zoonotic and commensal bacteria isolated from food-producing animals. J. Antimicrob. Chemother. 2004, 54, 744–754. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019, 17, 5598. [Google Scholar]
- European Medicines Agency; European Surveillance of Veterinary Antimicrobial Consumption. Sales of Veterinary Antimicrobial Agents in 30 European Countries in 2016 (EMA/275982/2018); European Medicines Agency: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Cha, W.; Mosci, R.E.; Wengert, S.L.; Vargas, C.V.; Rust, S.R.; Bartlett, P.C.; Grooms, D.L.; Manning, S.D. Comparing the genetic diversity and antimicrobial resistance profiles of Campylobacter jejuni recovered from cattle and humans. Front. Microbiol. 2017, 8, 818. [Google Scholar] [CrossRef] [PubMed]
- Sproston, E.L.; Wimalarathna, H.M.L.; Sheppard, S.K. Trends in fluoroquinolone resistance in Campylobacter. Microb. Genomics 2018, 4, e00198. [Google Scholar] [CrossRef] [PubMed]
- Jonas, R.; Kittl, S.; Overesch, G.; Kuhnert, P. Genotypes and antibiotic resistance of bovine Campylobacter and their contribution to human campylobacteriosis. Epidemiol. Infect. 2015, 143, 2373–2380. [Google Scholar] [CrossRef]
- Klein-Jöbstl, D.; Sofka, D.; Iwersen, M.; Drillich, M.; Hilbert, F. Multilocus sequence typing and antimicrobial resistance of Campylobacter jejuni isolated from dairy calves in Austria. Front. Microbiol. 2016, 7, 72. [Google Scholar] [CrossRef]
- Danish Integrated Antimicrobial Resistance Monitoring and Research Programme (DANMAP). Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark 2016; National Food Institute, Technical University of Denmark and Statens Serum Institut: Copenhagen, Denmark, 2017; Available online: www.danmap.org (accessed on 1 July 2019).
- Food, E.; Authority, S. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018, 16, 5182. [Google Scholar]
- De Briyne, N.; Atkinson, J.; Borriello, S.P.; Pokludová, L. Antibiotics used most commonly to treat animals in Europe. Vet. Rec. 2014, 175, 325–332. [Google Scholar] [CrossRef]
- Zeitouni, S.; Kempf, I. Fitness cost of fluoroquinolone resistance in Campylobacter coli and Campylobacter jejuni. Microb. Drug Resist. 2011, 17, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, M.; Deng, F.; Shen, Z.; Wu, C.; Zhang, J.; Zhang, Q.; Shen, J. Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob. Agents Chemother. 2014, 58, 5405–5412. [Google Scholar] [CrossRef] [PubMed]
- Hurtado, A.; Ocejo, M.; Oporto, B. Salmonella spp. and Listeria monocytogenes shedding in domestic ruminants and characterization of potentially pathogenic strains. Vet. Microbiol. 2017, 210, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Zirnstein, G.; Helsel, L.; Li, Y.; Swaminathan, B.; Besser, J. Characterization of gyrA mutations associated with fluoroquinolone resistance in Campylobacter coli by DNA sequence analysis and MAMA PCR. FEMS Microbiol. Lett. 2000, 190, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dingle, K.E.; Colles, F.M.; Wareing, D.R.A.; Ure, R.; Fox, A.J.; Bolton, F.E.; Bootsma, H.J.; Willems, R.J.L.; Urwin, R.; Maiden, A.M.C.J. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 2001, 39, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.G.; On, S.L.W.; Wang, G.; Fontanoz, S.; Lastovica, A.J.; Mandrell, R.E. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 2005, 43, 2315–2329. [Google Scholar] [CrossRef] [PubMed]
| Antimicrobial Class | Antimicrobial Agent | Campylobacter Species | % Resistance | No. of Isolates at the Indicated MIC (mg/L) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TOTAL | Beef Cattle | Dairy Cattle | Sheep | 0.064 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | |||
| Aminoglycoside | Gentamicin | C. jejuni | 0.0 | 0.0 | 0.0 | 0.0 | 3 | 12 | 87 | 7 | |||||||||
| C. coli | 11.8 | 0.0 | 44.4 | 0.0 | 15 | 11 | 4 | 4 | |||||||||||
| Aminoglycoside | Streptomycin | C. jejuni | 3.7 | 4.8 | 2.7 | 3.3 | 1 | 17 | 84 | 3 | 4 | ||||||||
| C. coli | 67.7 | 60.0 | 88.9 | 60.0 | 8 | 3 | 5 | 1 | 17 | ||||||||||
| Macrolide | Erythromycin | C. jejuni | 0.0 | 0.0 | 0.0 | 0.0 | 109 | ||||||||||||
| C. coli | 8.8 | 0.0 | 33.3 | 0.0 | 31 | 3 | |||||||||||||
| Quinolone | Nalidixic acid | C. jejuni | 60.6 | 61.9 | 67.6 | 50.0 | 1 | 31 | 11 | 6 | 60 | ||||||||
| C. coli | 64.7 | 80.0 | 77.8 | 55.0 | 3 | 9 | 3 | 19 | |||||||||||
| (Fluoro)Quinolone | Ciprofloxacin | C. jejuni | 60.6 | 61.9 | 67.6 | 50.0 | 43 | 1 | 41 | 24 | |||||||||
| C. coli | 64.7 | 80.0 | 77.8 | 55.0 | 11 | 1 | 6 | 9 | 7 | ||||||||||
| Tetracycline | Tetracycline | C. jejuni | 38.5 | 42.9 | 43.2 | 26.7 | 67 | 2 | 2 | 6 | 23 | 9 | |||||||
| C. coli | 76.5 | 80.0 | 88.9 | 70.0 | 8 | 3 | 2 | 21 | |||||||||||
| AMR a | Sheep | Cattle | Beef Cattle | Dairy Cattle | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | |
| CIP/NAL | 1.22 (0.39–3.80) | 0.729 | 2.01 (0.52–7.82) | 0.312 | 2.46 (0.25–24.02) | 0.438 | 1.68 (0.30–9.34) | 0.553 |
| ERY b | NA | NA | NA | NA | ||||
| TET | 6.42 (1.83–22.46) | 0.004 | 7.94 (1.67–37.86) | 0.005 | 5.33 (0.55–51.88) | 0.149 | 10.50 (1.19–92.72) | 0.034 |
| GEN b | NA | NA | NA | NA | ||||
| STR | 43.5 (4.89–386.74) | 0.001 | 92.89 (16.62–519.09) | <0.001 | 30.00 (3.06–294.56) | 0.004 | 288.00 (16.23–5108.64) | <0.001 |
| MDR | 12.43 (1.36–113.41) | 0.025 | 46.60 (9.30–223.47) | <0.001 | 30.00 (3.06–294.56) | 0.004 | 72.00 (6.39–811.79) | 0.001 |
| Target | Name | Sequences (5’ → 3’) | C (nM) | Reference |
|---|---|---|---|---|
| C. jejuni gyrA C257T | gyrCj-Fw | GGGTGCTGTTATAGGTCGTTATCA | 900 | [15] |
| gyrCj-Rv | TTGAGCCATTCTAACCAAAGCAT | 900 | ||
| Probe-gyrCj-S | HEX–CAT[+G]GAGAT[+A][+C][+A]GC[+A]GTTT–BHQ1 | 150 | ||
| Probe-gyrCj-R | FAM–CATGGAGATATAGCAGTTT–MGB | 150 | ||
| C. coli gyrA C257T | gyrCc-Fw | GAAGTGCATATAAAAAATCTGCTCGTA | 400 | This study |
| gyrCc-Rv | TGCCATTCTTACTAAGGCATCGT | 400 | ||
| Probe-gyrCc-S | FAM–AACAGCAGTATCGCC–MGB | 150 | ||
| Probe-gyrCc-R | VIC–AACAGCAATATCG–MGB | 150 | ||
| 23S rRNA A2075G | 23S-Fw | CAGTGAAATTGTAGTGGAGGTGAAA | 900 | [15] |
| 23S-Rv | TTCTTATCCAAATAGCAGTGTCAAGCT | 900 | ||
| Probe-23S-S | HEX–CGGGGTC[+T][+T][+T]CCGTCTTG–BHQ1 | 100 | ||
| Probe-23S-R | FAM–CGGGGTC[+T][+C][+T]CCGTCTTG–BHQ1 | 200 | ||
| C. coli gyrA sequencing | GZgyrACcoli3F | TATGAGCGTTATTATCGGTC | 400 | [38] |
| GZgyrACcoli4R | GTCCATCTACAAGCTCGTTA | 400 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ocejo, M.; Oporto, B.; Hurtado, A. Occurrence of Campylobacter jejuni and Campylobacter coli in Cattle and Sheep in Northern Spain and Changes in Antimicrobial Resistance in Two Studies 10-years Apart. Pathogens 2019, 8, 98. https://doi.org/10.3390/pathogens8030098
Ocejo M, Oporto B, Hurtado A. Occurrence of Campylobacter jejuni and Campylobacter coli in Cattle and Sheep in Northern Spain and Changes in Antimicrobial Resistance in Two Studies 10-years Apart. Pathogens. 2019; 8(3):98. https://doi.org/10.3390/pathogens8030098
Chicago/Turabian StyleOcejo, Medelin, Beatriz Oporto, and Ana Hurtado. 2019. "Occurrence of Campylobacter jejuni and Campylobacter coli in Cattle and Sheep in Northern Spain and Changes in Antimicrobial Resistance in Two Studies 10-years Apart" Pathogens 8, no. 3: 98. https://doi.org/10.3390/pathogens8030098
APA StyleOcejo, M., Oporto, B., & Hurtado, A. (2019). Occurrence of Campylobacter jejuni and Campylobacter coli in Cattle and Sheep in Northern Spain and Changes in Antimicrobial Resistance in Two Studies 10-years Apart. Pathogens, 8(3), 98. https://doi.org/10.3390/pathogens8030098






