Isolation and Molecular Identification of Virulence, Antimicrobial and Heavy Metal Resistance Genes in Livestock-Associated Methicillin-Resistant Staphylococcus aureus
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
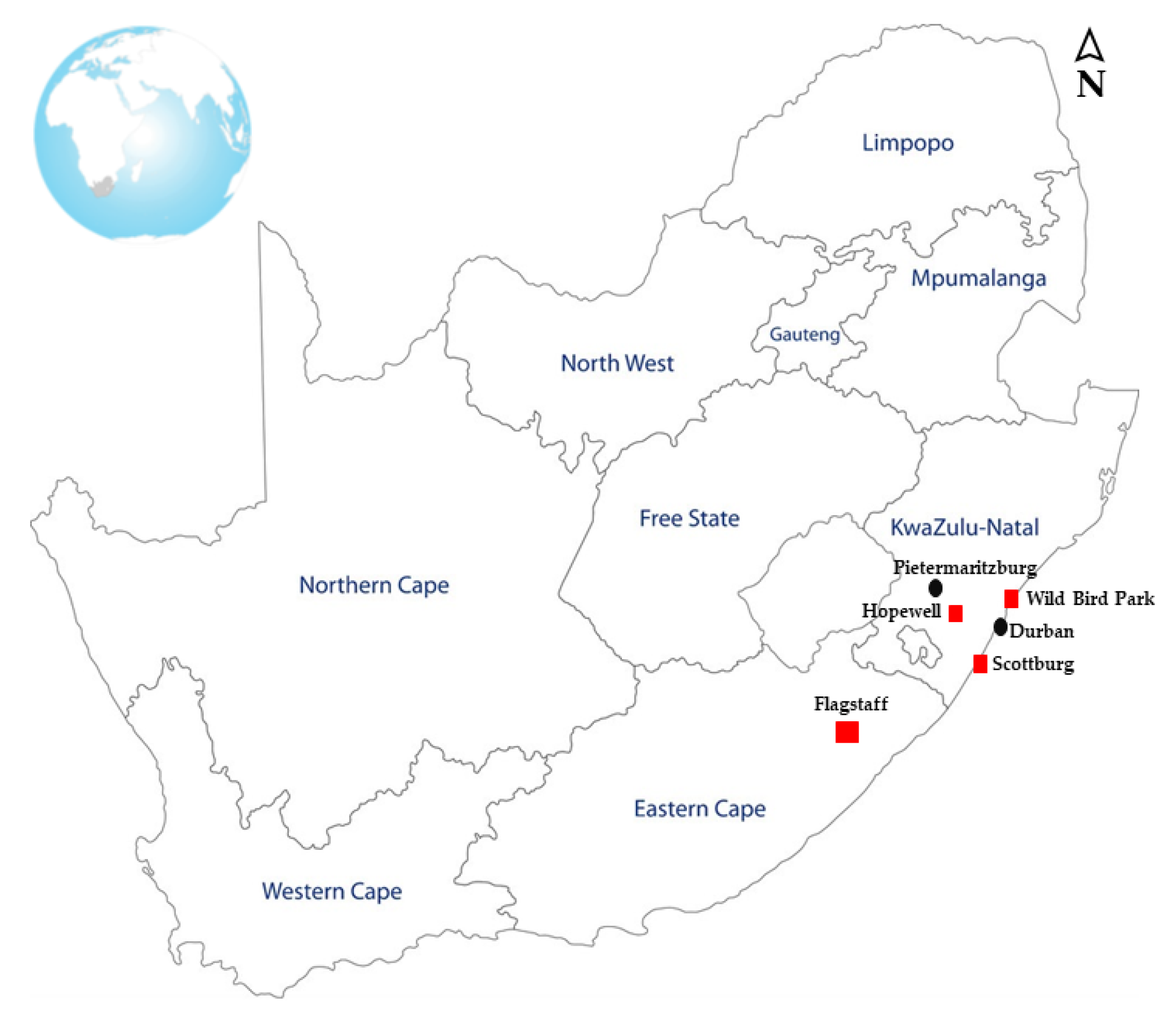
2.2. Sample Collection
2.3. Sample Processing and Bacterial Isolation
2.4. DNA Extraction and Molecular Identification of S. aureus
2.5. Antimicrobial and Heavy Metal Susceptibility Testing
2.6. Molecular Detection of Virulence, Antimicrobial and Heavy Metal Resistance Genes
2.7. Statistical Analysis
3. Results
3.1. Prevalence of S. aureus
3.2. Antimicrobial and Heavy Metal Resistance Susceptibility Testing
3.3. Virulence, Antimicrobial and Heavy Metal Resistance Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wertheim, H.F.L.; Melles, D.C.; Vos, M.C.; Van Leeuwen, W.; Van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The nasal carriage in Staphyloccocus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- van Hal, S.J.; Jensen, S.O.; Vaska, V.L.; Espedido, B.A.; Paterson, D.L.; Gosbell, I.B. Predictors of mortality in Staphylococcus aureus bacteremia. Clin. Microbiol. Rev. 2012, 25, 362–386. [Google Scholar] [CrossRef] [PubMed]
- Ercoli, L.; Gallina, S.; Nia, Y.; Auvray, F.; Primavilla, S.; Guidi, F.; Pierucci, B.; Graziotti, C.; Decastelli, L.; Scuota, S. Investigation of a Staphylococcal Food Poisoning Outbreak from a Chantilly Cream Dessert, in Umbria (Italy). Foodborne Pathog. Dis. 2017, 14, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention Healthcare-Associated Infections-Community Interface ( HAIC ): Emerging Infections Program ( EIP ) Network Report Invasive Staphylococcus aureus, 2016. US CDC 1600 Clifton Rd, Atlanta, Georgia, USA. Available online: https://www.cdc.gov/hai/eip/pdf/2016-%0AMRSA-Report-P.pdf (accessed on 2 April 2019).
- Thapaliya, D.; Kadariya, J.; Capuano, M.; Rush, H.; Yee, C.; Oet, M.; Lohani, S.; Smith, T.C. Prevalence and Molecular Characterization of S. aureus and MRSA on Children’s Playgrounds. Pediatr. Infect. Dis. J. 2018, 38, 1. [Google Scholar]
- Bai, Y.; Zhang, X.; Tian, Y.; Tian, D.; Zhang, B. Incidence of surgical-site infection following open reduction and internal fixation of a distal femur fracture. Medicine (Baltimore). 2019, 98, e14547. [Google Scholar] [CrossRef]
- Smith, T.H.; Fox, L.K.; Middleton, J.R. Outbreak of mastitis caused by one strain of Staphylococcus aureus in a closed dairy herd. J Am Vet Med Assoc 1998, 212, 553–556. [Google Scholar]
- Smith, T.C. Livestock-Associated Staphylococcus aureus: The United States Experience. PLOS Pathog. 2015, 11, e1004564. [Google Scholar] [CrossRef]
- Voss, A.; Loeffen, F.; Bakker, J.; Klaassen, C.; Wulf, M. Methicillin resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 2005, 11, 1965–1966. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus aureus toxins. Curr. Opin. Microbiol. 2014, 17, 32–37. [Google Scholar] [CrossRef]
- Adler, A.; Temper, V.; Block, C.S.; Abramson, N.; Moses, A.E. Panton-Valentine Leukocidin – producing Staphylococcus. Emerg. Infect. Dis. 2006, 12, 1789–1790. [Google Scholar]
- World Health Organization Antibiotic Resistance. Geneava, Switzerland. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 27 February 2019).
- Helperby Therapeutics World Antibiotic Headlines. Helperby Therapeutics ltd, London, UK. Available online: https://www.helperby.com/world-headlines/ (accessed on 25 February 2019).
- Rayner, C.; Munckhof, W.J. Antibiotics currently used in the treatment of infections caused by Staphylococcus aureus. Intern. Med. J. 2005, 35 (Suppl. 2), S3–S16. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef]
- World Health Organization. WHO publishes list of bacteria for which new antibiotics are urgently needed. Geneava, Switzerland. Available online: https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 4 March 2019).
- Bvenura, C.; Afolayan, A. Heavy metal contamination of vegetables cultivated in home gardens in the Eastern Cape. S. Afr. J. Sci. 2012, 108, 1–6. [Google Scholar] [CrossRef]
- Iwegbue, C.M.A.; Nwajei, G.E.; Iyoha, E.H. Heavy metal residues of chicken meat and gizzard and turkey meat consumed in southern Nigeria. Bulg. J. Vetenary Med. 2008, 11, 275–280. [Google Scholar]
- Raikwar, M.; Kumar, P.; Singh, A. Toxic effect of heavy metals in livestock health. Vet. World 2009, 1, 28–30. [Google Scholar] [CrossRef]
- Patra, S.; Das, T.K.; Avila, C.; Cabello, V.; Castillo, F.; Paria, D.S.; Ganguly, S.L.; Jana, B.B. Cadmium tolerance and antibiotic resistance in Escherichia coli isolated from waste stabilization ponds. IJEB 2012, 50, 300–307. [Google Scholar]
- Singh, Y.; Ramteke, P.W.; Tripathy, A.; Shukla, P.K. Isolation and Characterization of Bacillus resistant to multiple heavy metals. Int.J.Curr.Microbiol.App.Sci 2013, 2, 525–530. [Google Scholar]
- Nair, R.; Thapaliya, D.; Su, Y.; Smith, T.C. Resistance to Zinc and Cadmium in Staphylococcus aureus of Human and Animal Origin. Infect. Control Hosp. Epidemiol. 2014, 35, S32–S39. [Google Scholar] [CrossRef]
- Department of Agriculture Forestry and Fisheries. Economic review of the South African Agriculture 2016/2017; Department of Agriculture, Forestry and Fisheries: Pretoria, South Africa, 2017. Available online: https://www.daff.gov.za/daffweb3/Home/Crop-Estimates/Statistical-Information/Economic-Review (accessed on 2 April 2019).
- Ribeiro, J.C.; Tamanini, R.; Soares, B.F.; De Oliveira, A.M.; De Godoi Silva, F.; Da Silva, F.F.; Augusto, N.A.; Beloti, V. Efficiency of boiling and four other methods for genomic DNA extraction of deteriorating spore-forming bacteria from milk. Semin. Agrar. 2016, 37, 3069–3078. [Google Scholar] [CrossRef]
- Stuhlmeier, R.; Stuhlmeier, K.M. Fast, simultaneous, and sensitive detection of Staphylococci. J. Clin. Pathol. 2003, 56, 782–785. [Google Scholar] [CrossRef][Green Version]
- Stegger, M.; Andersen, P.S.; Kearns, A.; Pichon, B.; Holmes, M.A.; Edwards, G.; Laurent, F.; Teale, C.; Skov, R.; Larsen, A.R. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin. Microbiol. Infect. 2012, 18, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Gomes, R.M.; Bomfim, M.R.Q. Potential Spread of Methicillin-Resistant Staphylococcus aureus Recovered from Patients with Bloodstream Infection. Chemother. Open Access 2015, 4. [Google Scholar] [CrossRef]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 2001, 15, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Kock, M.M.; Ehlers, M.M. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: Genetic diversity and inter-species host transmission. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Mkize, N.; Zishiri, O.T.; Mukaratirwa, S. Genetic characterisation of antimicrobial resistance and virulence genes in Staphylococcus aureus isolated from commercial broiler chickens in the Durban metropolitan area, South Africa. J. S. Afr. Vet. Assoc. 2017, 88, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Omoe, K.; Hu, D.L.; Takahashi-Omoe, H.; Nakane, A.; Shinagawa, K. Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiol. Lett. 2005, 246, 191–198. [Google Scholar] [CrossRef]
- Pinto, B.; Chenoll, E.; Aznar, R. Identification and typing of food-borne Staphylococcus aureus by PCR-based techniques. Syst. Appl. Microbiol. 2005, 28, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Cariou, N.; Salandre, O.; Le Guennec, J.; Nemeghaire, S.; Butaye, P. Genotyping and antimicrobial resistance of Staphylococcus aureus isolates from diseased turkeys. Avian Pathol. 2013, 42, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Supplement M100. In Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- Waters, A.E.; Contente-Cuomo, T.; Buchhagen, J.; Liu, C.M.; Watson, L.; Pearce, K.; Foster, J.T.; Bowers, J.; Driebe, E.M.; Engelthaler, D.M.; et al. Multidrug-Resistant Staphylococcus aureus in US Meat and Poultry. Clin. Infect. Dis. 2011, 52, 1227–1230. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Adekanmbi, A.O.; Falodun, O.I. Heavy Metals and Antibiotics Susceptibility Profiles of Staphylococcus aureus Isolated from Several Points Receiving Daily Input from the Bodija Abattoir in Ibadan, Oyo State, Nigeria. Adv. Microbiol. 2015, 5, 871–880. [Google Scholar] [CrossRef]
- Massawe, H.F.; Mdegela, R.H.; Kurwijila, L.R. Antibiotic resistance of Staphylococcus aureus isolates from milk produced by smallholder dairy farmers in Mbeya Region, Tanzania. Int. J. One Heal. 2019, 5, 31–37. [Google Scholar] [CrossRef]
- Rong, D.; Wu, Q.; Xu, M.; Zhang, J.; Yu, S. Prevalence, virulence genes, antimicrobial susceptibility, and genetic diversity of Staphylococcus aureus from retail aquatic products in China. Front. Microbiol. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Oh, D.H.; Song, B.R.; Heo, E.J.; Lim, J.S.; Moon, J.S.; Park, H.J.; Wee, S.H.; Sung, K. Molecular Characterization, Antibiotic Resistance, and Virulence Factors of Methicillin-Resistant Staphylococcus aureus Strains Isolated from Imported and Domestic Meat in Korea. Foodborne Pathog. Dis. 2015, 12, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Klein, E. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463. [Google Scholar] [CrossRef] [PubMed]
- Dweba, C.C.; Zishiri, O.T.; El Zowalaty, M.E. Methicillin-resistant Staphylococcus aureus: Livestock-associated, antimicrobial, and heavy metal resistance. Infect. Drug Resist. 2018, 11, 2497–2509. [Google Scholar] [CrossRef]
- Orrell, C.; Cohen, K.; Conradie, F.; Zeinecker, J.; Ive, P.; Sanne, I.; Wood, R. Efavirenz and rifampicin in the South African context: Is there a need to dose-increase efavirenz with concurrent rifampicin therapy? Antivir. Ther. 2011, 16, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Zehra, A.; Singh, R.; Kaur, S.; Gill, J.P.S. Molecular characterization of antibiotic-resistant Staphylococcus aureus from livestock (bovine and swine). Vet. World 2017, 10, 598–604. [Google Scholar] [CrossRef]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Köck, R.; Schaumburg, F.; Mellmann, A.; Köksal, M.; Jurke, A.; Becker, K.; Friedrich, A.W. Livestock-Associated Methicillin-Resistant Staphylococcus aureus (MRSA) as Causes of Human Infection and Colonization in Germany. PLoS ONE 2013, 8, e55040. [Google Scholar] [CrossRef]
- Pirolo, M.; Gioffrè, A.; Visaggio, D.; Gherardi, M.; Pavia, G.; Samele, P.; Ciambrone, L.; Di Natale, R.; Spatari, G.; Casalinuovo, F.; et al. Prevalence, molecular epidemiology, and antimicrobial resistance of methicillin-resistant Staphylococcus aureus from swine in southern Italy. BMC Microbiol. 2019, 19, 51. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Liu, Y.; Han, C.; Chen, Z.; Ye, X. Phenotypic and molecular characteristics of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolated from pigs: Implication for livestock-association markers and vaccine strategies. Infect. Drug Resist. 2018, 11, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Köck, R.; Harlizius, J.; Bressan, N.; Laerberg, R.; Wieler, L.H.; Witte, W.; Deurenberg, R.H.; Voss, A.; Becker, K.; Friedrich, A.W. Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) among pigs on German farms and import of livestock-related MRSA into hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 1375–1382. [Google Scholar] [CrossRef] [PubMed]
- Mulders, M.N.; Haenen, A.P.J.; Geenen, P.L.; Vesseur, P.C.; Poldervaart, E.S.; Bosch, T.; Huijsdens, X.W.; Hengeveld, P.D.; Dam-Deisz, W.D.C.; Graat, E.A.M.; et al. Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in the Netherlands. Epidemiol. Infect. 2010, 138, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Feßler, A.T.; Kadlec, K.; Hassel, M.; Hauschild, T.; Eidam, C.; Ehricht, R.; Monecke, S.; Schwarz, S. Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl. Environ. Microbiol. 2011, 77, 7151–7157. [Google Scholar] [CrossRef] [PubMed]
- Feltrin, F.; Alba, P.; Kraushaar, B.; Ianzano, A.; Argudín, A.; Matteo, D. Staphylococcus aureus Clonal Complex 97 Lineage Spreading in Dairy Cattle and Pigs in Italy. Appl. Environ. Microbiol. 2016, 82, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Locatelli, C.; Cremonesi, P.; Caprioli, A.; Carfora, V.; Ianzano, A.; Barberio, A.; Morandi, S.; Casula, A.; Castiglioni, B.; Bronzo, V.; et al. Occurrence of methicillin-resistant Staphylococcus aureus in dairy cattle herds, related swine farms, and humans in contact with herds. J. Dairy Sci. 2016, 100, 608–619. [Google Scholar] [CrossRef]
- Spohr, M.; Rau, J.; Friedrich, A.; Klittich, G.; Fetsch, A.; Guerra, B.; Hammerl, J.A.; Tenhagen, B.-A. Methicillin-Resistant Staphylococcus aureus (MRSA) in Three Dairy Herds in Southwest Germany. Zoonoses Public Health 2011, 58, 252–261. [Google Scholar] [CrossRef]
- Kaspar, U.; von Lützau, K.; Schlattmann, A.; Rösler, U.; Köck, R.; Becker, K. Zoonotic multidrug-resistant microorganisms among non-hospitalized horses from Germany. One Heal. 2019, 7, 100091. [Google Scholar] [CrossRef]
- Giacinti, G.; Carfora, V.; Caprioli, A.; Sagrafoli, D.; Marri, N.; Giangolini, G.; Amoruso, R.; Iurescia, M.; Stravino, F.; Dottarelli, S.; et al. Prevalence and characterization of methicillin-resistant Staphylococcus aureus carrying mecA or mecC and methicillin-susceptible Staphylococcus aureus in dairy sheep farms in central Italy. J. Dairy Sci. 2017, 100, 7857–7863. [Google Scholar] [CrossRef]
- Carfora, V.; Giacinti, G.; Sagrafoli, D.; Marri, N.; Giangolini, G.; Alba, P.; Feltrin, F.; Sorbara, L.; Amoruso, R.; Caprioli, A.; et al. Methicillin-resistant and methicillin-susceptible Staphylococcus aureus in dairy sheep and in-contact humans: An intra-farm study. J. Dairy Sci. 2016, 99, 4251–4258. [Google Scholar] [CrossRef] [PubMed]
- Gharsa, H.; Ben Slama, K.; Lozano, C.; Gómez-Sanz, E.; Klibi, N.; Ben Sallem, R.; Gómez, P.; Zarazaga, M.; Boudabous, A.; Torres, C. Prevalence, antibiotic resistance, virulence traits and genetic lineages of Staphylococcus aureus in healthy sheep in Tunisia. Vet. Microbiol. 2012, 156, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Yu, C.; Lee, Y.; Su, Y. Genetically divergent methicillin-resistant Staphylococcus aureus and sec-dependent mastitis of dairy goats in Taiwan. BMC Vet. Res. 2012, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Cortimiglia, C.; Bianchini, V.; Franco, A.; Caprioli, A.; Battisti, A.; Colombo, L.; Stradiotto, K.; Vezzoli, F.; Luini, M. Short communication: Prevalence of Staphylococcus aureus and methicillin-resistant S. aureus in bulk tank milk from dairy goat farms in Northern Italy. J. Dairy Sci. 2015, 98, 2307–2311. [Google Scholar] [CrossRef] [PubMed]
- Alzohairy, M.A. Colonization and antibiotic susceptibility pattern of methicillin resistance Staphylococcus aureus ( MRSA ) among farm animals in Saudi Arabia. J. Bacteriol. Res. 2011, 3, 63–68. [Google Scholar]
- Rinsky, J.L.; Nadimpalli, M.; Wing, S.; Hall, D.; Baron, D.; Price, L.B.; Larsen, J.; Stegger, M.; Stewart, J.; Heaney, C.D. Livestock-Associated Methicillin and Multidrug Resistant Staphylococcus aureus Is Present among Industrial, Not Antibiotic-Free Livestock Operation Workers in North Carolina. Plos ONE 2013, 8, e67641. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.F.; Pisanic, N.; Rhodes, S.M.; Brown, A.; Keller, H.; Nadimpalli, M.; Christ, A.; Ludwig, S.; Ordak, C.; Spicer, K.; et al. Occurrence of Staphylococcus aureus in swine and swine workplace environments on industrial and antibiotic-free hog operations in North Carolina, USA: A One Health pilot study. Environ. Res. 2018, 163, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Wu, Z.; Li, L.; Li, F.; Wang, Y.; Zhao, X. Coexistence of heavy metal and antibiotic resistance within a novel composite staphylococcal cassette chromosome in a Staphylococcus haemolyticus isolate from bovine mastitis milk. Antimicrob. Agents Chemother. 2015, 59, 5788–5792. [Google Scholar] [CrossRef]
- Wales, A.; Davies, R. Co-Selection of Resistance to Antibiotics, Biocides and Heavy Metals, and Its Relevance to Foodborne Pathogens. Antibiotics 2015, 4, 567–604. [Google Scholar] [CrossRef]
- Aonghusa, C.N.; Gray, N.F. Laundry detergents as a source of heavy metals in Irish domestic wastewater. J. Environ. Sci. Heal. Part A Toxic/Hazardous Subst. Environ. Eng. 2002, 37, 1–6. [Google Scholar] [CrossRef]
- Sani, A.; Shehu, A. Determination of some heavy metals concentration in selected detergents used in Kano Metropolis, Nigeria. Environ. Toxicol. Stud. ournal 2018, 2, 1–4. [Google Scholar]
- Thompson, J.M.; Gündoǧdu, A.; Stratton, H.M.; Katouli, M. Antibiotic resistant Staphylococcus aureus in hospital wastewaters and sewage treatment plants with special reference to methicillin-resistant Staphylococcus aureus (MRSA). J. Appl. Microbiol. 2013, 114, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.; Riboli, D.F.; Pereira, C.V.; De Souza da Cunha, M. de L.R. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated from a Brazilian university hospital. Brazilian J. Infect. Dis. 2014, 18, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Elhassan, M.M.; Ozbak, H.A.; Hemeg, H.A.; Elmekki, M.A.; Ahmed, L.M. Absence of the mecA gene in methicillin resistant Staphylococcus aureus isolated from different clinical specimens in Shendi City, Sudan. Biomed Res. Int. 2015, 2015. [Google Scholar] [CrossRef]
- Antonios, Z.; Theofilos, P.; Ioannis, M.; Georgios, S.; Georgios, V.; Evridiki, B.; Loukia, E.; Kyriaki, M.; Athanasios, A.; Vasiliki, L. Prevalence, genetic diversity, and antimicrobial susceptibility profiles of Staphylococcus aureus isolated from bulk tank milk from Greek traditional ovine farms. Small Rumin. Res. 2015, 125, 120–126. [Google Scholar] [CrossRef]
- Garcia-Alvarez, L.; Holden, M.; Lindsay, H.; Webb, C.; Brown, D.; Curran, M. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet 2011, 11, 595–603. [Google Scholar] [CrossRef]
- Klibi, A.; Maaroufi, A.; Torres, C.; Jouini, A. Detection and characterization of methicillin-resistant and susceptible coagulase-negative Staphylococci in milk from cows with clinical mastitis in Tunisia. Int. J. Antimicrob. Agents 2018, 52, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Lozano, C.; Gharsa, H.; Ben Slama, K.; Zarazaga, M.; Torres, C. Staphylococcus aureus in Animals and Food: Methicillin Resistance, Prevalence and Population Structure. A Review in the African Continent. Microorganisms 2016, 4, 12. [Google Scholar] [CrossRef]
- Perovic, O.; Iyaloo, S.; Kularatne, R.; Lowman, W.; Bosman, N.; Wadula, J.; Seetharam, S.; Duse, A.; Mbelle, N.; Bamford, C.; et al. Prevalence and trends of Staphylococcus aureus bacteraemia in hospitalized patients in South Africa, 2010 to 2012: Laboratory-based surveillance mapping of antimicrobial resistance and molecular epidemiology. PLoS ONE 2015, 10, e0145429. [Google Scholar] [CrossRef]
- Abdulgader, S.M.; Shittu, A.O.; Nicol, M.P.; Kaba, M. Molecular epidemiology of Methicillin-resistant Staphylococcus aureus in Africa: A systematic review. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Singh-Moodley, A.; Marais, E.; Perovic, O. Discrepancies in the identification of methicillin-resistant Staphylococcus aureus and the absence of mecC in surveillance isolates in South Africa. South. African J. Infect. Dis. 2015, 30, 122–124. [Google Scholar]
- Marais, E.; Aithma, N.; Perovic, O.; Oosthuysen, W.F.; Musenge, E.; Duse, A.G. Antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolates from South Africa. South African Med. J. 2009, 99, 170–173. [Google Scholar]
- Khairalla, A.S.; Wasfi, R.; Ashour, H.M. Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care personnel, patients, and environment. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Dekker, D.; Wolters, M.; Mertens, E.; Boahen, K.G.; Krumkamp, R.; Eibach, D.; Schwarz, N.G.; Adu-Sarkodie, Y.; Rohde, H.; Christner, M.; et al. Antibiotic resistance and clonal diversity of invasive Staphylococcus aureus in the rural Ashanti Region, Ghana. BMC Infect. Dis. 2016, 16, 720. [Google Scholar] [CrossRef] [PubMed]
- Shittu, A.O.; Lin, J. Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal province, South Africa. BMC Infect. Dis. 2006, 6, 125. [Google Scholar] [CrossRef]
- Pekana, A.; Green, E. Antimicrobial Resistance Profiles of Staphylococcus aureus Isolated from Meat Carcasses and Bovine Milk in Abattoirs and Dairy Farms of the Eastern Cape, South Africa. Int. J. Environ. Res. Public Health 2018, 15, 2223. [Google Scholar] [CrossRef]
- Haenni, M.; Châtre, P.; Dupieux, C.; Métayer, V.; Maillard, K.; Bes, M.; Madec, J.Y.; Laurent, F. mecC-positive MRSA in horses. J. Antimicrob. Chemother. 2015, 70, 3401–3402. [Google Scholar] [CrossRef]
- Walther, B.; Wieler, L.H.; Vincze, S.; Antão, E.; Brandenburg, A.; Stamm, I.; Kopp, P.A.; Kohn, B. MRSA Variant Animals. Emerg. Infect. Dis. 2012, 18, 2017–2020. [Google Scholar] [CrossRef]
- Paterson, G.K.; Harrison, E.M.; Holmes, M.A. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014, 22, 42–47. [Google Scholar] [CrossRef]
- Bortolami, A.; Verin, R.; Chantrey, J.; Corrò, M.; Ashpole, I.; Lopez, J.; Timofte, D. Characterization of Livestock-Associated Methicillin-Resistant Staphylococcus aureus CC398 and mecC -positive CC130 from Zoo Animals in the United Kingdom. Microb. Drug Resist. 2017, 23, 908–914. [Google Scholar] [CrossRef]
- Garcia-Garrote, F.; Cercenado, E.; Marin, M.; Bal, M.; Trincado, P.; Corredoira, J.; Ballesteros, C.; Pita, J.; Alonso, P.; Vindel, A. Methicillin-resistant Staphylococcus aureus carrying the mecC gene: emergence in Spain and report of a fatal case of bacteraenia. J. Antimicrob. Chemother. 2014, 69, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Llarrull, L.I.; Fisher, J.F.; Mobashery, S. Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new β-lactams that meet the challenge. Antimicrob. Agents Chemother. 2009, 53, 4051–4063. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Dai, M.; Wang, Y.; Huang, L.; Yuan, Z. Key genetic elements and regulation systems in methicillin-resistant Staphylococcus aureus. Future Microbiol. 2012, 7, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fan, X.; Xiong, Y.; Zhong, Z.; Tang, H.; Feng, P.; Zhou, T. A Study of the Regulating Gene of femA from Methicillin-resistant Staphylococcus aureus Clinical Isolates. J. Int. Med. Res. 2013, 36, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Huys, G.; D’Haene, K.; Eldere, J.; Von Holy, A.; Swings, J. Molecular diversity and characterization of tetracycline-resistant Staphylococcus aureus isolates fro a poultry processing plant. Appl. Environ. Microbiol. 2005, 71, 574–579. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Argudin, M.; Lauzat, B.; Kraushaar, B.; Alba, P.; Agerso, Y.; Cavaco, L.; Butaye, P.; Porrero, M.; Battisti, A.; Tenhagen, B.; et al. Heavy metal and disinfectant resistance genes among livestock-associated methicillin resistant Staphylococcus aureus isolates. Vet. Microbiol. 2016, 191, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sanz, E.; Kadlec, K.; Feßler, A.T.; Zarazaga, M.; Torres, C.; Schwarz, S. Novel erm(T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrob. Agents Chemother. 2013, 57, 3275–3282. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.; Liu, W.; Huang, W.; Fu, Q.; Li, M. Molecular characteristic and virulence gene profiles of community-associated methicillin-resistant Staphylococcus aureus isolates from pediatric patients in Shanghai, China. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Hoseini Alfatemi, S.M.; Motamedifar, M.; Hadi, N.; Ebrahim Saraie, H.S. Analysis of virulence genes among methicillin resistant Staphylococcus aureus (MRSA) strains. Jundishapur J. Microbiol. 2014, 7, 1–10. [Google Scholar] [CrossRef]
- Nimmo, G.; Bergh, H.; Nakos, J.; Whiley, D.; Marquess, J.; Huygens, F. Replacement of healthcare-associated MRSA by community-associated MRSA in Queensland: Confirmation by genotyping. J. Infect. 2013, 67, 439–447. [Google Scholar] [CrossRef]
- Velasco, V.; Buyukcangaz, E.; Sherwood, J.S.; Stepan, R.M.; Koslofsky, R.J.; Logue, C.M. Characterization of Staphylococcus aureus from humans and a comparison with isolates of animal origin, in North Dakota, United States. PLoS ONE 2015, 10, e0140497. [Google Scholar] [CrossRef] [PubMed]
- Alba, P.; Feltrin, F.; Cordaro, G.; Porrero, M.C.; Kraushaar, B.; Argudín, M.A.; Nykäsenoja, S.; Monaco, M.; Stegger, M.; Aarestrup, F.M.; et al. Livestock-associated methicillin resistant and methicillin susceptible Staphylococcus aureus sequence type (CC)1 in European farmed animals: High genetic relatedness of isolates from Italian cattle herds and humans. PLoS ONE 2015, 10, e0137143. [Google Scholar] [CrossRef] [PubMed]
- Kulangara, V.; Nair, N.; Sivasailam, A.; Sasidharan, S.; Kollannur, J.D.; Syam, R. Genotypic and phenotypic β-lactam resistance and presence of PVL gene in Staphylococci from dry bovine udder. PLoS ONE 2017, 12, e0187277. [Google Scholar] [CrossRef] [PubMed]
- Richardson, E.J.; Bacigalupe, R.; Harrison, E.M.; Weinert, L.A.; Lycett, S.; Vrieling, M.; Robb, K.; Hoskisson, P.A.; Holden, M.T.G.; Feil, E.J.; et al. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2018, 2, 1468–1478. [Google Scholar] [CrossRef]



| Target | Gene | Primer Sequence | Tm (°C) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|---|
| Thermonuclease | nuc | F-GCGATTGATGGTGATACGGTT R-AGCCAAGCCTTGACGAACTAAAGC | 68 | 270 | [25] |
| Methicillin | mecC | F- GAAAAAAAGGCTTAGAACGCCTC R- GAAGATCTTTTCCGTTTTCAGC | 54 | 138 | [26] |
| Aminoglycosides | aac(6′)-aph(2″) | F-TAATCCAAGAGCAATAAGGGC R-GCCACACTATCATAACCACTA | 61 | 227 | [27] |
| Tetracycline | tetK | F-TCGATAGGAACAGCAGTA R-CAGCAGATCCTACTCCTT | 57 | 169 | [28] |
| Vancomycin | vanB | F- GTGACAAACCGGAGCGAGGA R- CCGCCATCCTCCTGCAAAAAA | 46 | 433 | [29] |
| Leukocidin | LukS/F-PV | F-ATCATTAGGTAAAATGTCTGGACATGATCCA R-GCATCAAGTGTATTGGATAGCAAAAGC | 56 | 443 | [30] |
| Others | spa | F-CAAGCACCAAAAGAGGAA R-CACCAGGTTTAACGACAT | 57 | 180 | |
| coa | F-CGAGACCAAGATTCAACAAG R-AAAGAAAACCACTCACATCA | 61 | 730 | ||
| Enterotoxins | sea | F- CCTTTGGAAACGGTTAAAACG R-TCTGAACCTTCCCATCAAAAAC | 56 | 127 | [31] |
| see | F-TAGATAAAGTTAAAAAACAAGC R-TAACTTACCGTGGACCCTTC | 46 | 170 | [32] | |
| Copper | copB | F-TAGTGGCCATGCACATCATC R-CCACCAGACAAGAACGGTTT | 60 | 201 | [33] |
| Animal Host | Flagstaff | Scottburg | Hopewell | Total | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Oral | Fecal | Feed | Soil | Water | Oral | Fecal | Feed | Soil | Water | Oral | Fecal | Feed | Soil | Water | Other * | ||
| Chicken | 0 | 0 | 0 | 0 | 0 | 31/40 | 29/40 | - | 3/5 | 3/5 | 0 | 0 | 0 | 0 | 0 | 0 | 66/90 |
| Ducks | 0 | 4/10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4/10 |
| Cow | 0 | 4/5 | 0 | 5/5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9/10 |
| Goats | 0/1 | 7/9 | 0 | 5/6 | 2/6 | 13/15 | 10/14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 37/51 |
| Sheep | 10/12 | 7/10 | 0 | 6/6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 23/28 |
| Horses | 0 | 2/5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2/5 |
| Pigs | 13/17 | 9/17 | 4/9 | 9/9 | 5/6 | 0 | 0 | 0 | 0 | 0 | 2/10 | 7/35 | 4/18 | 0 | 0 | 6/46 | 59/167 |
| Total | 23/30 | 33/56 | 4/9 | 25/26 | 7/12 | 44/55 | 39/54 | 0 | 3/5 | 3/5 | 2/10 | 7/35 | 4/18 | 0 | 0 | 6/46 | 200/361 |
| Wild Bird Species | Durban | |||
|---|---|---|---|---|
| Oral | S. aureus | Fecal | S. aureus | |
| Scarlet ibis | 8 | 4 | 8 | 4 |
| African Spoonbill | 3 | 2 | 3 | 0 |
| Fulvis Whistling duck | 2 | 0 | 2 | 0 |
| Carolina duck | 2 | 1 | 2 | 2 |
| Bahama pintail | 2 | 1 | 2 | 0 |
| Fireback pheasant | 1 | 0 | 1 | 1 |
| Whiteface whistling duck | 1 | 1 | 1 | 0 |
| Mandrin duck | 1 | 1 | 1 | 0 |
| Yellow bale duck | 1 | 0 | 1 | 0 |
| Total | 21 | 10 | 21 | 7 |
| Antibiotic Class | Antibiotic | Resistance Phenotypes | ||
|---|---|---|---|---|
| Resistant (R) | Intermediate (I) | Susceptible (S) | ||
| β-lactam | Penicillin G (10 IU) | 98.1 | 0 | 1.9 |
| Cefoxitin (30 µg) | 94.5 | 0 | 5.5 | |
| Aminoglycoside | Gentamicin (10 µg) | 19 | 21 | 60 |
| Quinolone | Ciprofloxacin (5 µg) | 14.3 | 16.6 | 69.1 |
| Macrolides | Erythromycin (15 µg) | 76.9 | 5.1 | 18.0 |
| Tetracycline | Tetracycline (30 µg) | 79.6 | 1.4 | 19 |
| Phenicols | Chloramphenicol (30 µg) | 30.7 | 15.3 | 54 |
| Sulfonamides | Trimethoprim-sulfamethoxazole (25 µg) | 60.9 | 4.7 | 34.4 |
| Other | Rifampicin (5 µg) | 76.9 | 3.8 | 19.4 |
| Heavy Metal | No. of Samples with Growth at Each Concentration | ||||
|---|---|---|---|---|---|
| 50 µg/mL | 100 µg/mL | 500 µg/mL | 1000 µg/mL | 1500 µg/mL | |
| Cadmium (Cd) | 200 (92.2%) | 198 (91.2%) | 193 (88.9%) | 193 (88.9%) | 193 (88.9%) |
| Copper (Cu) | 217 (100%) | 217 (100%) | 182 (84%) | 182 (84%) | 182 (84%) |
| Lead (Pb) | 217 (100%) | 217 (100%) | 187 (86.2%) | 187 (86.2%) | 187 (86.2%) |
| Zinc (Zn) | 196 (90%) | 196 (90%) | 194 (89.4%) | 193 (88.9%) | 192 (88.4%) |
| Host | Genetic Determinant | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| see | coa | sea | luks/F-PV | spa | mecC | copB | aac | vanB | tetK | |
| Avian | 1 | 0 | 3 | 1 | 3 | 31 | 5 | 17 | 3 | 37 |
| Goat | 0 | 0 | 0 | 0 | 7 | 7 | 0 | 4 | 0 | 21 |
| Sheep | 0 | 2 | 0 | 0 | 7 | 6 | 0 | 0 | 1 | 5 |
| Cattle | 0 | 0 | 0 | 0 | 1 | 6 | 0 | 2 | 0 | 1 |
| Horse | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| Pig | 1 | 0 | 11 | 1 | 2 | 7 | 8 | 1 | 7 | 12 |
| Total | 2 | 2 | 14 | 2 | 20 | 59 | 13 | 24 | 11 | 76 |
| Variables | Genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| aac | coa | sea | Luks/PV | spa | mecC | see | vanB | copB | tetK | |
| Host Species | 0.001 * | 0.057 ** | 0.07 ** | 1.000 ** | 0.001 * | 0.000 * | 0.005 * | 0.290 ** | 0.083 ** | 0.000 * |
| Sample material | 0.472 ** | 0.554 ** | 0.05 * | 0.189 ** | 0.200 ** | 0.361 ** | 0.042 * | 1.000 ** | 0.045 * | 0.004 * |
| Location | 0.000 * | 0.651 ** | 0.035 * | 1.000 ** | 0.627 ** | 0.075 ** | 0.005 * | 0.45 ** | 0.048 * | 0.00 * |
| aac | coa | sea | Luks/pv | spa | mecC | see | vanB | copB | tetK | |
|---|---|---|---|---|---|---|---|---|---|---|
| aac | 1 | −0.034 (0.618) | −0.093 (0.174) | −0.034 (0.618) | 0.040 (0.558) | 0.082 (0.231) | −0.089 (0.191) | −0.034 (0.618) | −0.081 (0.232) | 0.076 (0.263) |
| coa | −0.034 (0.618) | 1 | −0.025 (0.711) | −0.009 (0.892) | 0.303 ** (0.000) | 0.158 * (0.020) | −0.024 (0.721) | −0.009 (0.892) | −0.022 (0.744) | −0.072 (0.294) |
| sea | −0.093 (0.174) | −0.025 (0.711) | 1 | 0.171 * (0.012) | −0.019 (0.783) | 0.008 (0.905) | 0.329 ** (0.000) | −0.025 (0.711) | 0.110 (0.105) | −0.038 (0.578) |
| Luks/pv | −0.034 (0.618) | −0.009 (0.892) | 0.171 * (0.012) | 1 | 0.136 * (0.045) | −0.059 (0.388) | −0.024 (0.721) | −0.009 (0.892) | −0.022 (0.744) | −0.072 (0.294) |
| spa | 0.040 (0.558) | 0.303 ** (0.000) | −0.019 (0.783) | 0.136 * (0.045) | 1 | 0.020 (0.768) | −0.080 (0.238) | −0.031 (0.653) | −0.001 (0.988) | 0.030 (0.660) |
| mecC | 0.082 (0.231) | 0.158 * (0.020) | 0.008 (0.905) | −0.059 (0.388) | 0.020 (0.768) | 1 | 0.064 (0.348) | −0.059 (0.388) | −0.047 (0.493) | −0.064 (0.352) |
| see | −0.089 (0.191) | −0.024 (0.721) | 0.329 ** (0.000) | −0.024 (0.721) | −0.080 (0.238) | 0.064 (0.348) | 1 | 0.382 ** (0.000) | 0.207 ** (0.002) | −0.025 (0.716) |
| vanB | −0.034 (0.618) | −0.009 (0.892) | −0.025 (0.711) | −0.009 (0.892) | −0.031 (0.653) | −0.059 (0.388) | 0.382 ** (0.000) | 1 | 0.198 ** (0.003) | 0.029 (0.668) |
| copB | −0.081 (0.232) | −0.022 (0.744) | 0.110 (0.105) | −0.022 (0.744) | −0.001 (0.988) | −0.047 (0.493) | 0.207 ** (0.002) | 0.198 ** (0.003) | 1 | −0.127 (0.061) |
| tetK | 0.076 (0.263) | −0.072 (0.294) | −0.038 (0.578) | −0.072 (0.294) | 0.030 (0.660) | −0.064 (0.352) | −0.025 (0.716) | 0.029 (0.668) | −0.127 (0.061) | 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dweba, C.C.; Zishiri, O.T.; El Zowalaty, M.E. Isolation and Molecular Identification of Virulence, Antimicrobial and Heavy Metal Resistance Genes in Livestock-Associated Methicillin-Resistant Staphylococcus aureus. Pathogens 2019, 8, 79. https://doi.org/10.3390/pathogens8020079
Dweba CC, Zishiri OT, El Zowalaty ME. Isolation and Molecular Identification of Virulence, Antimicrobial and Heavy Metal Resistance Genes in Livestock-Associated Methicillin-Resistant Staphylococcus aureus. Pathogens. 2019; 8(2):79. https://doi.org/10.3390/pathogens8020079
Chicago/Turabian StyleDweba, Chumisa C., Oliver T. Zishiri, and Mohamed E. El Zowalaty. 2019. "Isolation and Molecular Identification of Virulence, Antimicrobial and Heavy Metal Resistance Genes in Livestock-Associated Methicillin-Resistant Staphylococcus aureus" Pathogens 8, no. 2: 79. https://doi.org/10.3390/pathogens8020079
APA StyleDweba, C. C., Zishiri, O. T., & El Zowalaty, M. E. (2019). Isolation and Molecular Identification of Virulence, Antimicrobial and Heavy Metal Resistance Genes in Livestock-Associated Methicillin-Resistant Staphylococcus aureus. Pathogens, 8(2), 79. https://doi.org/10.3390/pathogens8020079






