Putative Periodontal Pathogens, Filifactor alocis and Peptoanaerobacter stomatis, Induce Differential Cytokine and Chemokine Production by Human Neutrophils
Abstract
:1. Introduction
2. Results
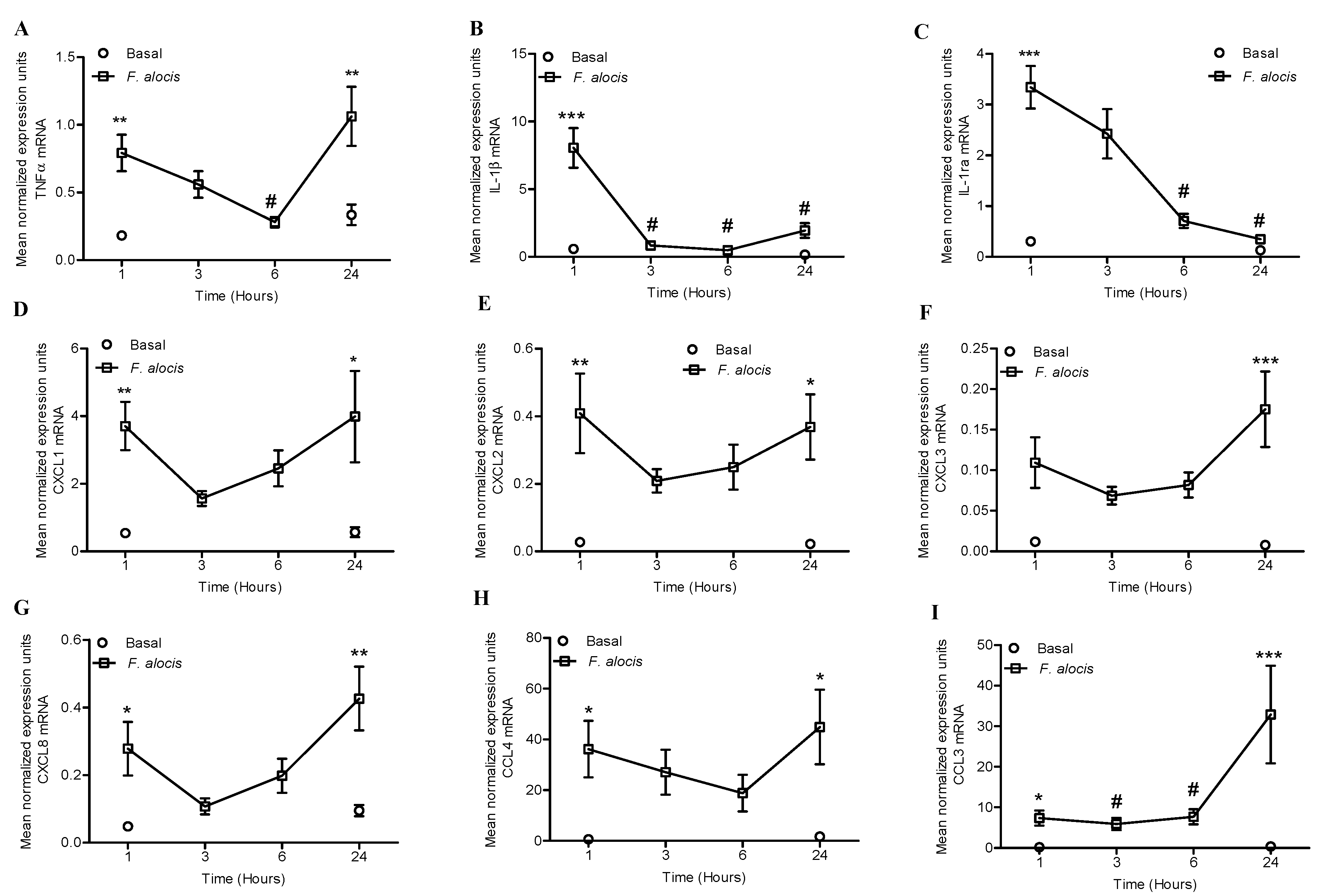
2.1. F. alocis Challenge Induced Both Expression and Release of Neutrophil-Derived Cytokines and Chemokines
2.2. Distinct Pro-Inflammatory and Anti-Inflammatory Cytokine Release by F. alocis Compared to P. gingivalis and P. stomatis
2.3. Both F. alocis and P. stomatis Activated TLR2/6 to a Greater Extent than TLR2/1
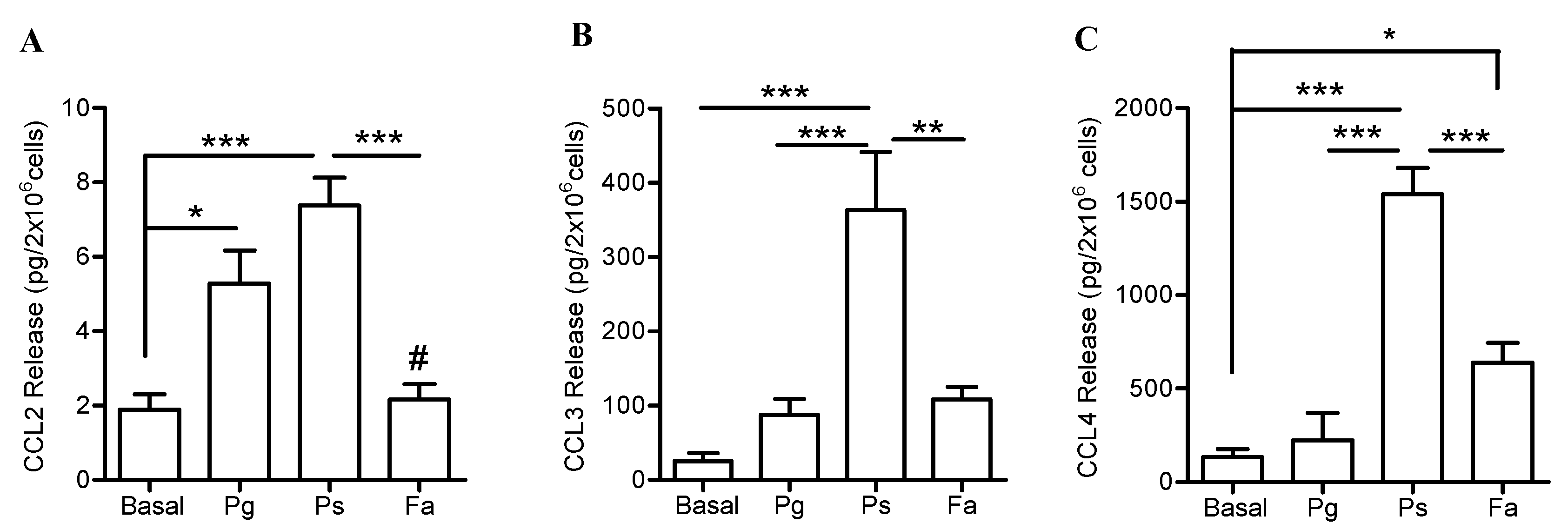
2.4. Higher Release of CXCL1 and CCL Chemokines by Neutrophils when Challenged with P. stomatis Compared to F. alocis or P. gingivalis
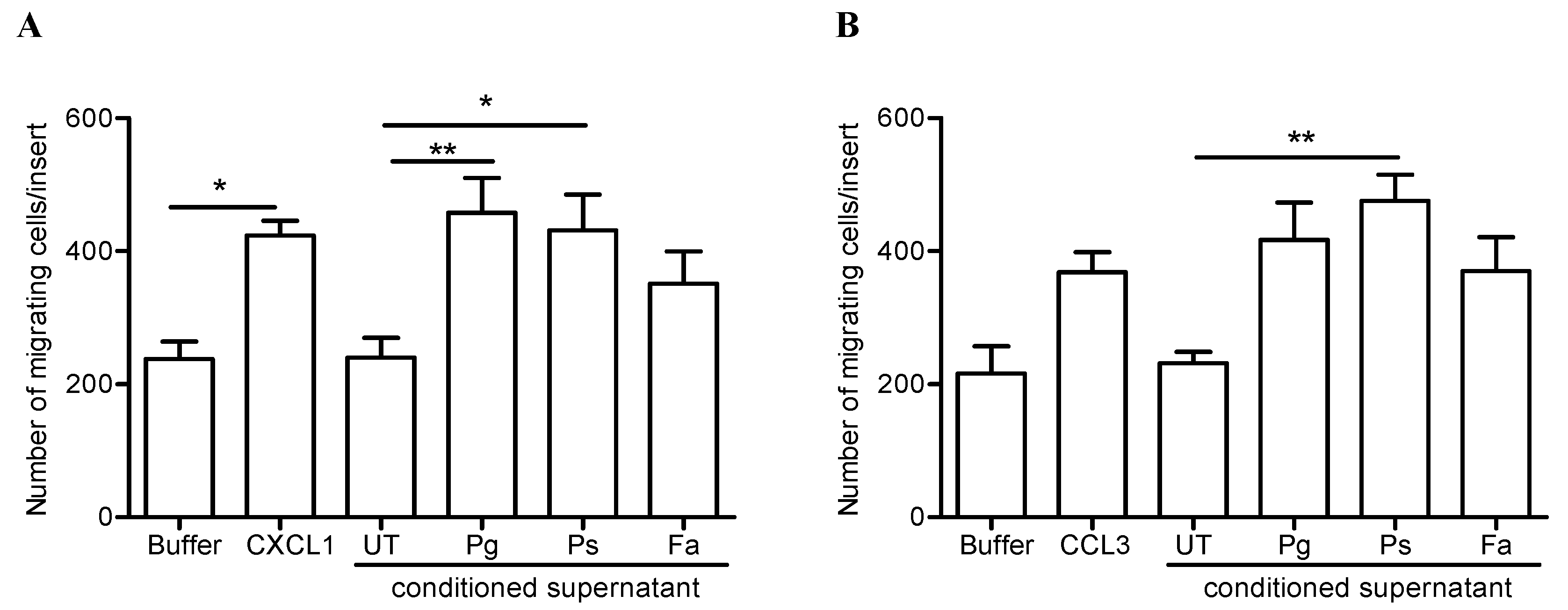
2.5. Release of Neutrophil-Derived Chemokines by P. stomatis Challenge Promotes Leukocyte Migration
3. Discussion
4. Materials and Methods
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Eke, P.I.; Dye, B.A.; Wei, L.; Slade, G.D.; Thornton-Evans, G.O.; Borgnakke, W.S.; Taylor, G.W.; Page, R.C.; Beck, J.D.; Genco, R.J. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J. Periodontol. 2015, 86, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015, 15, 30–44. [Google Scholar] [CrossRef]
- Hajishengallis, G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 2014, 35, 3–11. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Lamont, R.J. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 2012, 27, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef]
- Wade, W.G. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J. Clin. Periodontol. 2011, 38, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Griffen, A.L.; Beall, C.J.; Firestone, N.D.; Gross, E.L.; Difranco, J.M.; Hardman, J.H.; Vriesendorp, B.; Faust, R.A.; Janies, D.A.; Leys, E.J. CORE: A phylogenetically-curated 16S rDNA database of the core oral microbiome. PloS ONE 2011, 6, e19051. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.S.; Griffen, A.L.; Moeschberger, M.L.; Leys, E.J. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 2005, 43, 3944–3955. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.C.; Frick, I.-M. Gram-positive anaerobic cocci—commensals and opportunistic pathogens. FEMS Microbiol. Rev. 2013, 37, 520–553. [Google Scholar] [CrossRef]
- Sizova, M.V.; Chilaka, A.; Earl, A.M.; Doerfert, S.N.; Muller, P.A.; Torralba, M.; McCorrison, J.M.; Durkin, A.S.; Nelson, K.E.; Epstein, S.S. High-quality draft genome sequences of five anaerobic oral bacteria and description of Peptoanaerobacter stomatis gen. nov., sp. nov., a new member of the family Peptostreptococcaceae. Stand. Genomic Sci. 2015, 10, 37. [Google Scholar] [CrossRef]
- Downes, J.; Wade, W.G. Peptostreptococcus stomatis sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 2006, 56, 751–754. [Google Scholar] [CrossRef]
- Dahlen, G.; Leonhardt, A. A new checkerboard panel for testing bacterial markers in periodontal disease. Oral Microbiol. Immunol. 2006, 21, 6–11. [Google Scholar] [CrossRef]
- Moffatt, C.E.; Whitmore, S.E.; Griffen, A.L.; Leys, E.J.; Lamont, R.J. Filifactor alocis interactions with gingival epithelial cells. Mol. Oral Microbiol. 2011, 26, 365–373. [Google Scholar] [CrossRef]
- Maiden, M.F.; Tanner, A.; Macuch, P.J. Rapid characterization of periodontal bacterial isolates by using fluorogenic substrate tests. J. Clin. Microbiol. 1996, 34, 376–384. [Google Scholar]
- Aruni, A.W.; Roy, F.; Fletcher, H.M. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect. Immun. 2011, 79, 3872–3886. [Google Scholar] [CrossRef]
- Schlafer, S.; Riep, B.; Griffen, A.L.; Petrich, A.; Hübner, J.; Berning, M.; Friedmann, A.; Göbel, U.B.; Moter, A. Filifactor alocis—Involvement in periodontal biofilms. BMC Microbiol. 2010, 10, 1–13. [Google Scholar] [CrossRef]
- Wang, Q.; Jotwani, R.; Le, J.; Krauss, J.L.; Potempa, J.; Coventry, S.C.; Uriarte, S.M.; Lamont, R.J. Filifactor alocis infection and inflammatory responses in the mouse subcutaneous chamber model. Infect. Immun. 2014, 82, 1205–1212. [Google Scholar] [CrossRef]
- Ryder, M.I. Comparison of neutrophil functions in aggressive and chronic periodontitis. Periodontol. 2000 2010, 53, 124–137. [Google Scholar] [CrossRef]
- Scott, D.A.; Krauss, J.L. Neutrophils in periodontal inflammation. Front. Oral Biol. 2012, 15, 56–83. [Google Scholar]
- Amulic, B.; Cazalet, C.; Hayes, G.L.; Metzler, K.D.; Zychlinsky, A. Neutrophil function: from mechanisms to disease. Annu. Rev. Immunol. 2012, 30, 459–489. [Google Scholar] [CrossRef]
- Edmisson, J.S.; Tian, S.; Armstrong, C.L.; Vashishta, A.; Klaes, C.K.; Miralda, I.; Jimenez-Flores, E.; Le, J.; Wang, Q.; Lamont, R.J.; Uriarte, S.M. Filifactor alocis modulates human neutrophil antimicrobial functional responses. Cell. Microbiol. 2018, 20, e12829. [Google Scholar] [CrossRef] [PubMed]
- Jimenez Flores, E.; Tian, S.; Sizova, M.; Epstein, S.S.; Lamont, R.J.; Uriarte, S.M. Peptoanaerobacter stomatis primes human neutrophils and induces granule exocytosis. Infect. Immun. 2017, 85, e01043-16. [Google Scholar] [CrossRef] [PubMed]
- Tamassia, N.; Bianchetto-Aguilera, F.; Arruda-Silva, F.; Gardiman, E.; Gasperini, S.; Calzetti, F.; Cassatella, M.A. Cytokine production by human neutrophils: Revisiting the “dark side of the moon”. Eur. J. Clin. Invest. 2018, 48, e12952. [Google Scholar] [CrossRef]
- Scapini, P.; Lapinet-Vera, J.A.; Gasperini, S.; Calzetti, F.; Bazzoni, F.; Cassatella, M.A. The neutrophil as a cellular source of chemokines. Immunol. Rev. 2000, 177, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Scapini, P.; Carletto, A.; Nardelli, B.; Calzetti, F.; Roschke, V.; Merigo, F.; Tamassia, N.; Pieropan, S.; Biasi, D.; Sbarbati, A.; Sozzani, S.; Bambara, L.; Cassatella, M.A. Proinflammatory mediators elicit secretion of the intracellular B-lymphocyte stimulator pool (BLyS) that is stored in activated neutrophils: implications for inflammatory diseases. Blood 2005, 105, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, C.L.; Miralda, I.; Neff, A.C.; Tian, S.; Vashishta, A.; Perez, L.; Le, J.; Lamont, R.J.; Uriarte, S.M. Filifactor alocis promotes neutrophil degranulation and chemotactic activity. Infect. Immun. 2016, 84, 3423–3433. [Google Scholar] [CrossRef] [PubMed]
- Tecchio, C.; Micheletti, A.; Cassatella, M.A. Neutrophil-derived cytokines: facts beyond expression. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Tecchio, C.; Cassatella, M.A. Neutrophil-derived chemokines on the road to immunity. Semin. Immunol. 2016, 28, 119–128. [Google Scholar] [CrossRef]
- Prince, L.R.; Whyte, M.K.; Sabroe, I.; Parker, L.C. The role of TLRs in neutrophil activation. Curr. Opin. Pharmacol. 2011, 11, 397–403. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Lambris, J.D. Microbial manipulation of receptor crosstalk in innate immunity. Nat. Rev. Immunol. 2011, 11, 187–200. [Google Scholar] [CrossRef]
- Hajishengallis, G. Immune evasion strategies of Porphyromonas gingivalis. J. Oral Biosci. 2011, 53, 233–240. [Google Scholar] [CrossRef]
- Uriarte, S.M.; Edmisson, J.S.; Jimenez-Flores, E. Human neutrophils and oral microbiota: a constant tug-of-war between a harmonious and a discordant coexistence. Immunol. Rev. 2016, 273, 282–298. [Google Scholar] [CrossRef]
- Cassatella, M.A. Neutrophil-derived proteins: selling cytokines by the pound. Adv. Immunol. 1999, 73, 369–509. [Google Scholar]
- Yoshimura, A.; Hara, Y.; Kaneko, T.; Kato, I. Secretion of IL-1β, TNF-α, IL-8 and IL-1ra by human polymorphonuclear leukocytes in response to lipopolysaccharides from periodontopathic bacteria. J. Periodontal Res. 1997, 32, 279–286. [Google Scholar] [CrossRef]
- Ling, M.R.; Chapple, I.L.; Matthews, J.B. Peripheral blood neutrophil cytokine hyper-reactivity in chronic periodontitis. Innate Immun. 2015, 21, 714–725. [Google Scholar] [CrossRef]
- Wright, H.J.; Chapple, I.L.C.; Matthews, J.B.; Cooper, P.R. Fusobacterium nucleatum regulation of neutrophil transcription. J. Periodontal Res. 2011, 46, 1–12. [Google Scholar] [CrossRef]
- McDonald, P.P.; Gasperini, S.; Calzetti, F.; Cassatella, M.A. Modulation by interferon-γ of the production and gene expression of IL-1 receptor antagonist in human neutrophils. Cell. Immunol. 1998, 184, 45–50. [Google Scholar] [CrossRef]
- Maekawa, T.; Krauss, J.L.; Abe, T.; Jotwani, R.; Triantafilou, M.; Triantafilou, K.; Hashim, A.; Hoch, S.; Curtis, M.A.; Nussbaum, G.; et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe 2014, 15, 768–778. [Google Scholar] [CrossRef]
- Uriarte, S.M.; Rane, M.J.; Luerman, G.C.; Barati, M.T.; Ward, R.A.; Nauseef, W.M.; McLeish, K.R. Granule exocytosis contributes to priming and activation of the human neutrophil respiratory burst. J. Immunol. 2011, 187, 391–400. [Google Scholar] [CrossRef]
- SenGupta, S.; Hittle, L.E.; Ernst, R.K.; Uriarte, S.M.; Mitchell, T.C. A Pseudomonas aeruginosa hepta-acylated lipid A variant associated with cystic fibrosis selectively activates human neutrophils. J. Leukoc. Biol. 2016, 100, 1047–1059. [Google Scholar] [CrossRef]
- Uriarte, S.M.; Molestina, R.E.; Miller, R.D.; Bernabo, J.; Farinati, A.; Eiguchi, K.; Ramirez, J.A.; Summersgill, J.T. Effects of fluoroquinolones on the migration of human phagocytes through Chlamydia pneumoniae-infected and tumor necrosis factor alpha-stimulated endothelial cells. Antimicrob. Agents Chemother. 2004, 48, 2538–2543. [Google Scholar] [CrossRef]
- Arruda-Silva, F.; Bianchetto-Aguilera, F.; Gasperini, S.; Polletti, S.; Cosentino, E.; Tamassia, N.; Cassatella, M.A. Human neutrophils produce CCL23 in response to various TLR-agonists and TNFα. Front. Cell. Infect. Microbiol. 2017, 7, 176. [Google Scholar] [CrossRef]



| Cytokines | Basal | Non-op-F. alocis | Op-F. alocis |
|---|---|---|---|
| IL-1β (pg/0.5 × 106 cells) | 1.30 ± 0.736 | 5.125 ± 0.417 ** | 5.464 ± 0.720 ** |
| TNFα (pg/0.5 × 106 cells) | 6.448 ± 1.302 | 46.759 ± 6.382 ** | 35.514 ± 7.401 * |
| IL-1ra (pg/2 × 106 cells) | 38.948 ± 7.785 | 194.527 ± 24.179 *** | 138.451 ± 7.678 ** |
| CXCL Chemokines | |||
| CXCL1 (ng/2 × 106 cells) | 0.004 ± 0.002 | 0.478 ± 0.078 *** | 0.445 ± 0.073 *** |
| CXCL8 (ng/0.1 × 106 cells) | 0.596 ± 0.214 | 4.492 ± 1.166 * | 3.818 ± 0.849 * |
| CCL Chemokines | |||
| CCL3 (pg/2 × 106 cells) | 25.087 ± 10.922 | 108.511 ± 16.580 | 232.526 ± 80.041 * |
| CCL4 (pg/2 × 106 cells) | 133.315 ± 43.227 | 639.611 ± 104.467 | 709.178 ± 203.417 * |
| Genes | Sequence | |
|---|---|---|
| TNF-α | Forward | 5’CAGCCTCTTCTCCTTCCTGAT3’ |
| Reverse | 5’GCCAGAGGGCTGATTAGAGA3’ | |
| IL-1ra | Forward | 5’AACTAGTTGCTGGATACTTGCA3’ |
| Reverse | 5’CCAGACTTGACACAGGACAG3’ | |
| IL-1b | Forward | 5’TACCTGTCCTGCGTGTTGAA3’ |
| Reverse | 5’TCTTTGGGTAATTTTTGGGATCT3’ | |
| CXCL1 | Forward | 5’AACCGAAGTCATAGCCACAC3’ |
| Reverse | 5’CCTCCCTTCTGGTCAGTTG3’ | |
| CXCL2 | Forward | 5’AACCGAAGTCATAGCCACAC3’ |
| Reverse | 5’CTTCTGGTCAGTTGGATTTGC3’ | |
| CXCL3 | Forward | 5’AAGTGTGAATGTAAGGTCCCC3’ |
| Reverse | 5’GTGCTCCCCTTGTTCAGTATC3’ | |
| CXCL8 | Forward | 5’GAGCACTCCATAAGGCACAAA3’ |
| Reverse | 5’ATGGTTCCTTCCGGTGGT3’ | |
| CCL3 | Forward | 5’CGGCAGATTCCACAGAATTTC3’ |
| Reverse | 5’AGGTCGCTGACATATTTCTGG3’ | |
| CCL4 | Forward | 5’TCCTCGCAACTTTGTGGTAG3’ |
| Reverse | 5’TTCAGTTCCAGGTCATACACG3’ | |
| GAPDH | Forward | 5’CTTTGGTATCGTGGAAGGACTC3’ |
| Reverse | 5’GTAGAGGCAGGGATGATGTTC3’ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vashishta, A.; Jimenez-Flores, E.; Klaes, C.K.; Tian, S.; Miralda, I.; Lamont, R.J.; Uriarte, S.M. Putative Periodontal Pathogens, Filifactor alocis and Peptoanaerobacter stomatis, Induce Differential Cytokine and Chemokine Production by Human Neutrophils. Pathogens 2019, 8, 59. https://doi.org/10.3390/pathogens8020059
Vashishta A, Jimenez-Flores E, Klaes CK, Tian S, Miralda I, Lamont RJ, Uriarte SM. Putative Periodontal Pathogens, Filifactor alocis and Peptoanaerobacter stomatis, Induce Differential Cytokine and Chemokine Production by Human Neutrophils. Pathogens. 2019; 8(2):59. https://doi.org/10.3390/pathogens8020059
Chicago/Turabian StyleVashishta, Aruna, Emeri Jimenez-Flores, Christopher K. Klaes, Shifu Tian, Irina Miralda, Richard J. Lamont, and Silvia M. Uriarte. 2019. "Putative Periodontal Pathogens, Filifactor alocis and Peptoanaerobacter stomatis, Induce Differential Cytokine and Chemokine Production by Human Neutrophils" Pathogens 8, no. 2: 59. https://doi.org/10.3390/pathogens8020059
APA StyleVashishta, A., Jimenez-Flores, E., Klaes, C. K., Tian, S., Miralda, I., Lamont, R. J., & Uriarte, S. M. (2019). Putative Periodontal Pathogens, Filifactor alocis and Peptoanaerobacter stomatis, Induce Differential Cytokine and Chemokine Production by Human Neutrophils. Pathogens, 8(2), 59. https://doi.org/10.3390/pathogens8020059






