Transcriptomic Analysis Reveals Selective Metabolic Adaptation of Streptococcus suis to Porcine Blood and Cerebrospinal Fluid
Abstract
:1. Introduction
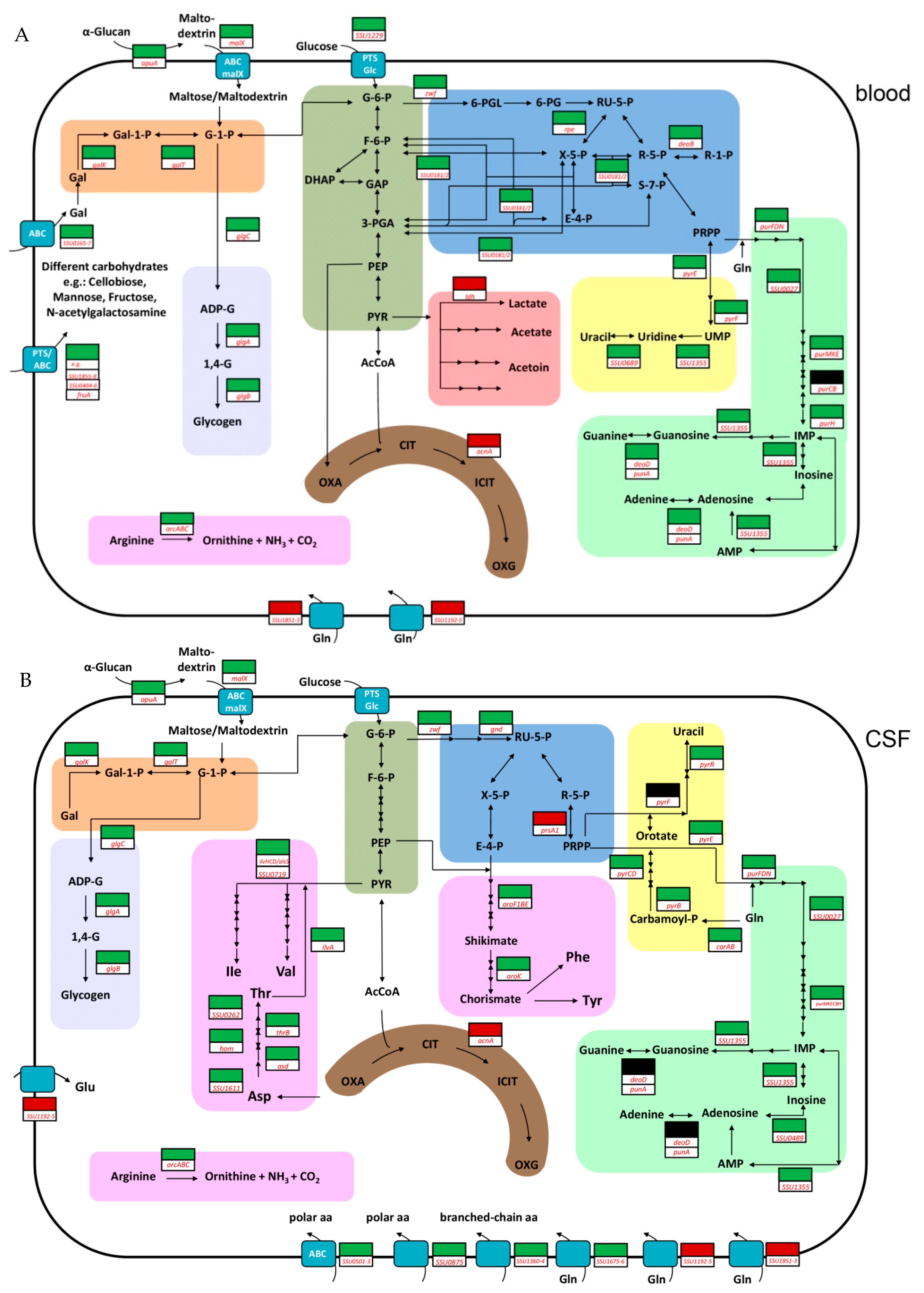
2. Results and Discussion
3. Materials and Methods
3.1. Bacteria and Growth Conditions
3.2. RNA Extraction and RNA Deep Sequencing (RNA-seq)
3.3. Reverse Transcriptase Quantitative-PCR (RT-qPCR)
3.4. Determination of Amino Acid Concentrations in Porcine Serum and CSF
3.5. Data Accession Number
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gottschalk, M.; Xu, J.; Calzas, C.; Segura, M. Streptococcus suis: A new emerging or an old neglected zoonotic pathogen? Future Microbiol. 2010, 5, 371–391. [Google Scholar] [CrossRef] [PubMed]
- Huong, V.T.; Ha, N.; Huy, N.T.; Horby, P.; Nghia, H.D.; Thiem, V.D.; Zhu, X.; Hoa, N.T.; Hien, T.T.; Zamora, J.; et al. Epidemiology, Clinical Manifestations, and Outcomes of Streptococcus suis Infection in Humans. Emerg. Infect. Dis. 2014, 20, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.; Calzas, C.; Grenier, D.; Gottschalk, M. Initial steps of the pathogenesis of the infection caused by Streptococcus suis: Fighting against nonspecific defenses. FEBS Lett. 2016, 590, 3772–3799. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Segura, M. The pathogenesis of the meningitis caused by Streptococcus suis: The unresolved questions. Vet. Microbiol. 2000, 76, 259–272. [Google Scholar] [CrossRef]
- Fittipaldi, N.; Segura, M.; Grenier, D.; Gottschalk, M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 2012, 7, 259–279. [Google Scholar] [CrossRef] [PubMed]
- Doran, K.S.; Fulde, M.; Gratz, N.; Kim, B.J.; Nau, R.; Prasadarao, N.; Schubert-Unkmeir, A.; Tuomanen, E.I.; Valentin-Weigand, P. Host-pathogen interactions in bacterial meningitis. Acta Neuropathol. 2016, 131, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Willenborg, J.; Goethe, R. Metabolic traits of pathogenic streptococci. FEBS Lett. 2016, 590, 3905–3919. [Google Scholar] [CrossRef] [PubMed]
- Richards, V.P.; Choi, S.C.; Pavinski Bitar, P.D.; Gurjar, A.A.; Stanhope, M.J. Transcriptomic and genomic evidence for Streptococcus agalactiae adaptation to the bovine environment. BMC Genom. 2013, 14, 920. [Google Scholar] [CrossRef] [PubMed]
- Szafranska, A.K.; Oxley, A.P.; Chaves-Moreno, D.; Horst, S.A.; Rosslenbroich, S.; Peters, G.; Goldmann, O.; Rohde, M.; Sinha, B.; Pieper, D.H.; et al. High-Resolution Transcriptomic Analysis of the Adaptive Response of Staphylococcus aureus during Acute and Chronic Phases of Osteomyelitis. MBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Mereghetti, L.; Sitkiewicz, I.; Green, N.M.; Musser, J.M. Extensive Adaptive Changes Occur in the Transcriptome of Streptococcus agalactiae (Group B Streptococcus) in Response to Incubation with Human Blood. PLoS ONE 2008, 3, e3143. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wu, C.; Shao, J.; Zhu, Z.; Wang, W.; Zhang, W.; Tang, M.; Pei, N.; Fan, H.; Li, J.; et al. The Streptococcus suis transcriptional landscape reveals adaptation mechanisms in pig blood and cerebrospinal fluid. RNA 2014, 20, 882–898. [Google Scholar] [CrossRef] [PubMed]
- Echenique-Rivera, H.; Muzzi, A.; Del Tordello, E.; Seib, K.L.; Francois, P.; Rappuoli, R.; Pizza, M.; Serruto, D. Transcriptome Analysis of Neisseria meningitidis in Human Whole Blood and Mutagenesis Studies Identify Virulence Factors Involved in Blood Survival. PLoS Pathog. 2011, 7, e1002027. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.R.; Virtaneva, K.; Porcella, S.F.; Barry, W.T.; Gowen, B.B.; Johnson, C.R.; Wright, F.A.; Musser, J.M. Group A Streptococcus Transcriptome Dynamics during Growth in Human Blood Reveals Bacterial Adaptive and Survival Strategies. Am. J. Pathol. 2005, 166, 455–465. [Google Scholar] [CrossRef]
- Brinsmade, S.R. CodY, a master integrator of metabolism and virulence in Gram-positive bacteria. Curr. Genet. 2016. [Google Scholar] [CrossRef] [PubMed]
- Willenborg, J.; Huber, C.; Koczula, A.; Lange, B.; Eisenreich, W.; Valentin-Weigand, P.; Goethe, R. Characterization of the Pivotal Carbon Metabolism of Streptococcus suis serotype 2 under ex Vivo and Chemically Defined in Vitro Conditions by Isotopologue Profiling. J. Biol. Chem. 2015, 290, 5840–5854. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.F.; Gao, T.; Liu, W.Q.; Zhang, C.Y.; Yang, X.; Zhu, J.W.; Teng, M.Y.; Li, L.; Zhou, R. MsmK, an ATPase, Contributes to Utilization of Multiple Carbohydrates and Host Colonization of Streptococcus suis. PLoS ONE 2015, 10, e0130792. [Google Scholar] [CrossRef] [PubMed]
- Bidossi, A.; Mulas, L.; Decorosi, F.; Colomba, L.; Ricci, S.; Pozzi, G.; Deutscher, J.; Viti, C.; Oggioni, M.R. A Functional Genomics Approach to Establish the Complement of Carbohydrate Transporters in Streptococcus pneumoniae. PLoS ONE 2012, 7, e33320. [Google Scholar] [CrossRef] [PubMed]
- Ferrando, M.L.; Fuentes, S.; de Greeff, A.; Smith, H.; Wells, J.M. ApuA, a multifunctional α-glucan-degrading enzyme of Streptococcus suis, mediates adhesion to porcine epithelium and mucus. Microbiology 2010, 156, 2818–2828. [Google Scholar] [CrossRef] [PubMed]
- Willenborg, J.; Fulde, M.; de Greeff, A.; Rohde, M.; Smith, H.E.; Valentin-Weigand, P.; Goethe, R. Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis. Microbiology 2011, 157, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Willenborg, J.; de Greeff, A.; Jarek, M.; Valentin-Weigand, P.; Goethe, R. The CcpA regulon of Streptococcus suis reveals novel insights into the regulation of the streptococcal central carbon metabolism by binding of CcpA to two distinct binding motifs. Mol. Microbiol. 2014, 92, 61–83. [Google Scholar] [CrossRef] [PubMed]
- Rosenling, T.; Slim, C.L.; Christin, C.; Coulier, L.; Shi, S.; Stoop, M.P.; Bosman, J.; Suits, F.; Horvatovich, P.L.; Stockhofe-Zurwieden, N.; et al. The Effect of Preanalytical Factors on Stability of the Proteome and Selected Metabolites in Cerebrospinal Fluid (CSF). J. Proteome Res. 2009, 8, 5511–5522. [Google Scholar] [CrossRef] [PubMed]
- Hickman, R.; Alp, M.H. A predictable pathophysiological model of porcine hepatic failure. Eur. Surg. Res. 1986, 18, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Mesavage, W.C.; Suchy, S.F.; Weiner, D.L.; Nance, C.S.; Flannery, D.B.; Wolf, B. Amino acids in amniotic fluid in the second trimester of gestation. Pediatr. Res. 1985, 19, 1021–1024. [Google Scholar] [CrossRef] [PubMed]
- Sitkiewicz, I.; Green, N.M.; Guo, N.; Bongiovanni, A.M.; Witkin, S.S.; Musser, J.M. Transcriptome Adaptation of Group B Streptococcus to Growth in Human Amniotic Fluid. PLoS ONE 2009, 4, e6114. [Google Scholar] [CrossRef] [PubMed]
- Molzen, T.E.; Burghout, P.; Bootsma, H.J.; Brandt, C.T.; van der Gaast-de Jongh, C.E.; Eleveld, M.J.; Verbeek, M.M.; Frimodt-Moller, N.; Ostergaard, C.; Hermans, P.W. Genome-Wide Identification of Streptococcus pneumoniae Genes Essential for bacterial Replication during Experimental Meningitis. Infect. Immun. 2011, 79, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Fittipaldi, N.; Harel, J.; D’Amours, B.; Lacouture, S.; Kobisch, M.; Gottschalk, M. Potential use of an unencapsulated and aromatic amino acid-auxotrophic Streptococcus suis mutant as a live attenuated vaccine in swine. Vaccine 2007, 25, 3524–3535. [Google Scholar] [CrossRef] [PubMed]
- Fulde, M.; Willenborg, J.; de Greeff, A.; Benga, L.; Smith, H.E.; Valentin-Weigand, P.; Goethe, R. ArgR is an essential local transcriptional regulator of the arcABC operon in Streptococcus suis and is crucial for biological fitness in an acidic environment. Microbiology 2011, 157, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Aronesty, E. Comparison of sequencing utility programs. Open Bioinform. J. 2013, 7, 1–8. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McClure, R.; Balasubramanian, D.; Sun, Y.; Bobrovskyy, M.; Sumby, P.; Genco, C.A.; Vanderpool, C.K.; Tjaden, B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013, 41, e140. [Google Scholar] [CrossRef] [PubMed]
- Vervuert, I.; Coenen, M.; Watermulder, E. Metabolic responses to oral tryptophan supplementation before exercise in horses. J. Anim. Physiol. Anim. Nutr. 2005, 89, 140–145. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koczula, A.; Jarek, M.; Visscher, C.; Valentin-Weigand, P.; Goethe, R.; Willenborg, J. Transcriptomic Analysis Reveals Selective Metabolic Adaptation of Streptococcus suis to Porcine Blood and Cerebrospinal Fluid. Pathogens 2017, 6, 7. https://doi.org/10.3390/pathogens6010007
Koczula A, Jarek M, Visscher C, Valentin-Weigand P, Goethe R, Willenborg J. Transcriptomic Analysis Reveals Selective Metabolic Adaptation of Streptococcus suis to Porcine Blood and Cerebrospinal Fluid. Pathogens. 2017; 6(1):7. https://doi.org/10.3390/pathogens6010007
Chicago/Turabian StyleKoczula, Anna, Michael Jarek, Christian Visscher, Peter Valentin-Weigand, Ralph Goethe, and Jörg Willenborg. 2017. "Transcriptomic Analysis Reveals Selective Metabolic Adaptation of Streptococcus suis to Porcine Blood and Cerebrospinal Fluid" Pathogens 6, no. 1: 7. https://doi.org/10.3390/pathogens6010007
APA StyleKoczula, A., Jarek, M., Visscher, C., Valentin-Weigand, P., Goethe, R., & Willenborg, J. (2017). Transcriptomic Analysis Reveals Selective Metabolic Adaptation of Streptococcus suis to Porcine Blood and Cerebrospinal Fluid. Pathogens, 6(1), 7. https://doi.org/10.3390/pathogens6010007





