1. Introduction
Malaria remains a major global health challenge, with over 263 million cases reported annually and continued transmission despite widespread deployment of vector control, chemoprevention, and artemisinin-based combination therapies [
1]. The pre-erythrocytic liver stage represents a critical bottleneck in the
Plasmodium life cycle, during which a small number of sporozoites infect hepatocytes and undergo extensive replication before initiating symptomatic blood-stage infection [
2,
3]. Because parasite biomass is low and genetically non-diverse at this stage, the liver stage offers an attractive target for both prophylactic vaccines and host-directed therapies aimed at preventing progression to clinical disease [
4,
5].
Once inside hepatocytes,
Plasmodium develops within a modified parasitophorous vacuole membrane (PVM), which acts as the primary interface for host–pathogen interactions [
6,
7,
8]. Host cells mount a rapid cytosolic defense response upon parasite invasion, including interferon-driven immune pathways and endolysosomal remodeling [
9,
10,
11]. A key component of this response is the recruitment of LC3 (microtubule-associated protein 1 light chain 3), a member of the ATG8 (autophagy-related protein 8) family, to the PVM [
7,
12,
13]; however, studies primarily using
Plasmodium berghei liver-stage infection models have demonstrated that this process does not reflect classical xenophagy but instead represents a form of noncanonical autophagy [
13,
14,
15].
This pathway, known as conjugation of ATG8 to single membranes (CASM), a form of noncanonical autophagy distinct from classical double-membrane autophagosome formation, is initiated by V-ATPase-dependent recruitment of ATG16L1 and leads to LC3 lipidation on the single-membrane PVM, forming the
Plasmodium-associated autophagy-related (PAAR) response [
15]. Most mechanistic insights into CASM/PAAR during malaria have been derived from liver-stage experimental models, and the degree of conservation across
Plasmodium species and clinical malaria settings remains to be fully established. PAAR represents an early cell-intrinsic defense that limits parasite survival and developmental progression within hepatocytes—a process referred to here as parasite restriction, defined as the suppression of parasite growth, maturation, or persistence through host-mediated intracellular mechanisms rather than direct parasite killing [
16,
17].
To counteract this response,
Plasmodium expresses PVM-resident proteins such as UIS3 and UIS4, which interfere with host autophagy by sequestering LC3 through direct binding and functional immobilization at the parasitophorous vacuole membrane and by driving perivacuolar actin remodeling, thereby limiting lysosomal access [
12,
18,
19]. At later stages, parasites exploit host autophagy machinery by selectively recruiting GABARAP (γ-aminobutyric acid receptor-associated protein) paralogs, a subset of ATG8-family proteins involved in vesicular trafficking and autophagy regulation, and activating TFEB (transcription factor EB), a master regulator of lysosomal biogenesis, thereby enhancing access to lipids and nutrients required for intracellular development [
20,
21,
22,
23].
Together, these findings reveal that autophagy plays a dual role in liver-stage malaria—both restricting and supporting parasite development depending on the molecular context. Importantly, these interactions reveal an inherently asymmetric landscape in which hepatocyte-intrinsic restriction and parasite-driven exploitation can operate in parallel, often drawing on overlapping autophagy machinery but yielding opposite outcomes. A comprehensive understanding of CASM activation, parasite evasion mechanisms, and autophagy-mediated metabolic remodeling—defined here as parasite-driven reprogramming of hepatocyte lysosomal biogenesis, membrane trafficking, and lipid mobilization through selective engagement of autophagy signaling pathways—is therefore crucial for designing selective, host-directed strategies to prevent liver-stage progression and reduce the emergence of antimalarial resistance. This review synthesizes current advances in the mechanistic interplay between Plasmodium liver stages and hepatocyte autophagy. Unless otherwise specified, the majority of mechanistic insights discussed here are derived from studies using the rodent malaria parasite Plasmodium berghei, which remains the dominant experimental model for liver-stage autophagy research. Where available, evidence from Plasmodium falciparum or human-relevant systems is explicitly indicated.
2. Mechanistic Architecture of Autophagy During Plasmodium Liver-Stage Infection
The liver stage of
Plasmodium infection begins with sporozoite entry into hepatocytes, a process traditionally viewed as parasite-driven but now understood to involve substantial host contributions. Live-cell imaging studies demonstrate that the initial contact between motile sporozoites and hepatocytes induces pronounced plasma membrane ruffling and filopodia extension, driven in part by Rho GTPase signaling, which increases host cell susceptibility to productive invasion [
24]. These host-directed membrane dynamics facilitate plasma membrane invagination, giving rise to the single-membrane Parasitophorous vacuole membrane (PVM) that envelops the intracellular parasite and provides the structural platform for subsequent liver-stage development [
25,
26]. Once formed, the PVM becomes the central interface for host–parasite interaction and a focal point for a specialized form of noncanonical autophagy, CASM. Recent work has shown that CASM—rather than canonical xenophagy—governs LC3 recruitment to the PVM through V-ATPase–ATG16L1-dependent lipidation, constituting a major hepatocyte-intrinsic defense mechanism during early liver-stage infection [
14,
15].
2.1. Activation of Noncanonical Autophagy (CASM) at the PVM
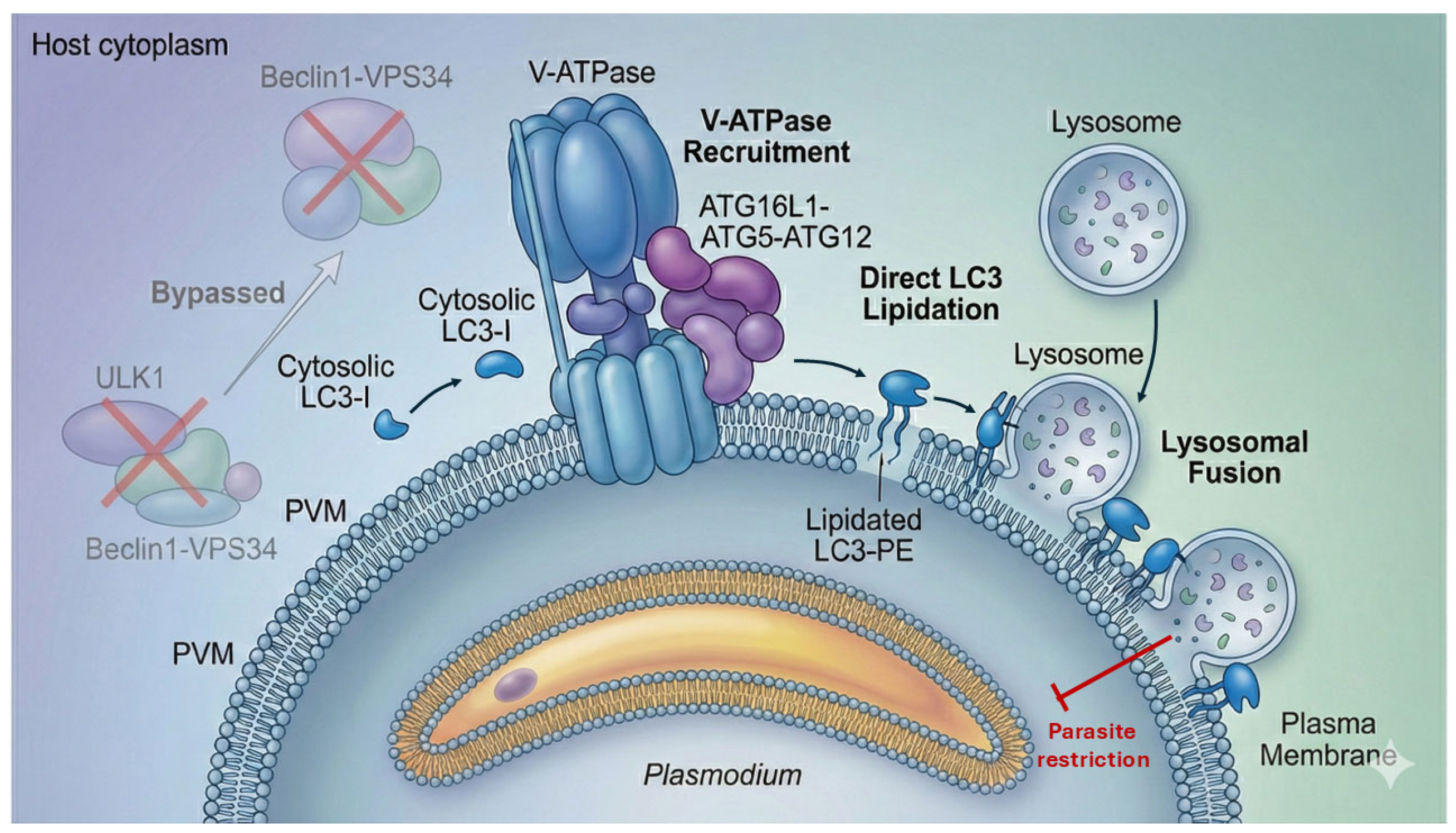
Upon productive invasion, the PVM undergoes rapid biophysical and ionic perturbations—including changes in membrane tension, lipid organization, and local ion fluxes such as proton and calcium gradients—that serve as upstream signals for the recruitment of the V-ATPase complex and the ATG16L1–ATG5–ATG12 machinery to the vacuole surface [
27,
28]. Unlike canonical autophagy, which is a degradative pathway characterized by the formation of double-membrane autophagosomes initiated by the ULK1 and Beclin1–VPS34 complexes, these initiation pathways instead enable the direct conjugation of LC3 to the single-membrane PVM bilayer [
29,
30].
This modification defines the PAAR—a hallmark of hepatocyte defense—characterized by long-lasting LC3B decoration of the PVM [
15,
27]. LC3 lipidation marks the PVM for interaction with endolysosomal machinery, enabling downstream tethering events required for parasite restriction (
Figure 1).
2.2. LC3-Dependent Restriction and Lysosomal Engagement
LC3-decorated PVMs can recruit ubiquitin and autophagy adaptors, enhancing the recognition of the Parasitophorous compartment as damaged or foreign [
9,
12]. Subsequent fusion with host lysosomes leads to parasite degradation through a nitric oxide-independent mechanism, redefining the centrality of noncanonical autophagy in parasite elimination [
10,
16]. These observations collectively establish that CASM-driven LC3 lipidation, not classical xenophagy, is the predominant autophagy pathway engaged by hepatocytes during early liver-stage infection. Disruption of V-ATPase or ATG16L1 significantly impairs LC3 recruitment and compromises parasite clearance, confirming CASM as a core arm of hepatocyte cell-autonomous immunity [
15].
2.3. Parasite Interference with CASM Execution
To survive within the hepatocyte,
Plasmodium deploys specialized PVM-resident proteins that interfere with the execution—but not necessarily the initiation—of the PAAR response. The best-characterized factor, UIS3, identified primarily in
Plasmodium berghei, binds directly to LC3B, acting as a molecular sink that prevents LC3-engaged PVMs from progressing toward lysosomal fusion [
12,
31]. In parallel, UIS4, characterized in rodent malaria models, organizes actin cytoskeleton remodeling around the PVM, creating a dense perivacuolar barrier that restricts access of lysosomes, ubiquitin, and autophagy adaptors [
18,
19]. These evasion mechanisms do not abolish LC3 lipidation but instead derail the maturation of CASM into a fully restrictive pathway, allowing the parasite to remain protected despite early detection by hepatocyte surveillance systems. It is important to note, however, that the molecular mechanisms underlying UIS3-mediated LC3 sequestration have been primarily delineated using in vitro hepatocyte models, including transformed hepatoma cell lines and primary hepatocytes, often under highly synchronized infection conditions [
12]. While complementary rodent in vivo studies, such as UIS3 depletion models, confirm the essential role of UIS3 for liver-stage development [
31], these systems do not fully recapitulate the metabolic heterogeneity, cellular architecture, and immune complexity of the human liver. Consequently, the quantitative contribution and regulatory dynamics of UIS3-dependent autophagy evasion in human
Plasmodium infections remain to be fully resolved.
2.4. Divergent ATG8 Paralog Engagement
While LC3B is associated primarily with parasite restriction, liver-stage parasites selectively recruit GABARAP paralogs at later developmental stages [
32]. Recent work shows that GABARAP enrichment at the PVM promotes TFEB activation, the master regulator of lysosomal biogenesis and autophagy gene expression [
20]. Unlike LC3-driven PAAR, this pathway enhances intracellular trafficking capacity and lipid mobilization—processes that support parasite replication and metabolic expansion [
21]. This differential ATG8 engagement underscores a fundamental asymmetry in the autophagy landscape:
LC3 → Restrictive, CASM-dependent defense;
GABARAP → Supportive, TFEB-dependent exploitation.
Understanding the determinants of this paralog specificity remains a key unresolved question.
Together, these mechanistic insights define the molecular architecture of noncanonical autophagy at the
Plasmodium PVM. CASM-driven LC3 lipidation, lysosomal tethering, and the counteracting roles of UIS3 and UIS4, along with selective ATG8 recruitment, illustrate that the hepatocyte–parasite interface is shaped by tightly regulated, molecule-specific interactions rather than a uniform autophagic program. However, these mechanisms do not operate in isolation. Instead, they form a dynamic and competitive landscape in which hepatocyte-intrinsic defense and parasite-driven exploitation unfold simultaneously and often asymmetrically. To understand how these discrete molecular events translate into infection outcomes, it is necessary to move beyond individual pathways and examine how host and parasite autophagy programs intersect, oppose, or reinforce each other.
Section 3 synthesizes these interactions into an integrated conceptual framework that captures the emergent behavior of the autophagy interface during liver-stage malaria.
3. Integrated Host–Parasite Autophagy Dynamics During Liver-Stage Malaria
The molecular events described in
Section 2 reveal that hepatocytes initiate a CASM-driven PAAR response that decorates the PVM with LC3 and promotes lysosomal engagement. However, the liver stage of
Plasmodium infection is not defined solely by isolated autophagy pathways. Instead, it reflects a dynamic and asymmetric interplay in which hepatocyte restriction mechanisms and parasite survival strategies operate simultaneously, often competing for the same autophagy machinery. This section synthesizes these interactions into a unified conceptual framework that captures how noncanonical autophagy shapes liver-stage infection outcomes.
3.1. Host PAAR/CASM as a Restrictive Autophagy Pathway
Hepatocytes deploy PAAR as a form of cell-autonomous immunity, characterized by persistent LC3 lipidation onto the single-membrane PVM through V-ATPase-ATG16L1-dependent CASM activation [
14,
15]. This LC3 enrichment enables the PVM to be recognized by ubiquitin and autophagy adaptors, allowing it to be tethered to lysosomes, which culminates in parasite degradation independent of nitric oxide [
10,
16]. At the systems level, PAAR represents an early restriction axis that acts during the narrow temporal window immediately following sporozoite invasion, when parasite numbers are low and host defense is at its peak.
To consolidate the mechanistic insights outlined above, we provide a structured summary of the key components involved in hepatocyte CASM/PAAR activation.
Table 1 synthesizes the major molecular triggers, autophagy machinery, and downstream restrictive processes that shape the early host response to
Plasmodium liver-stage infection. This table provides a concise reference to the core mechanisms underlying noncanonical autophagy at the PVM.
3.2. Parasite Evasion of PAAR Execution
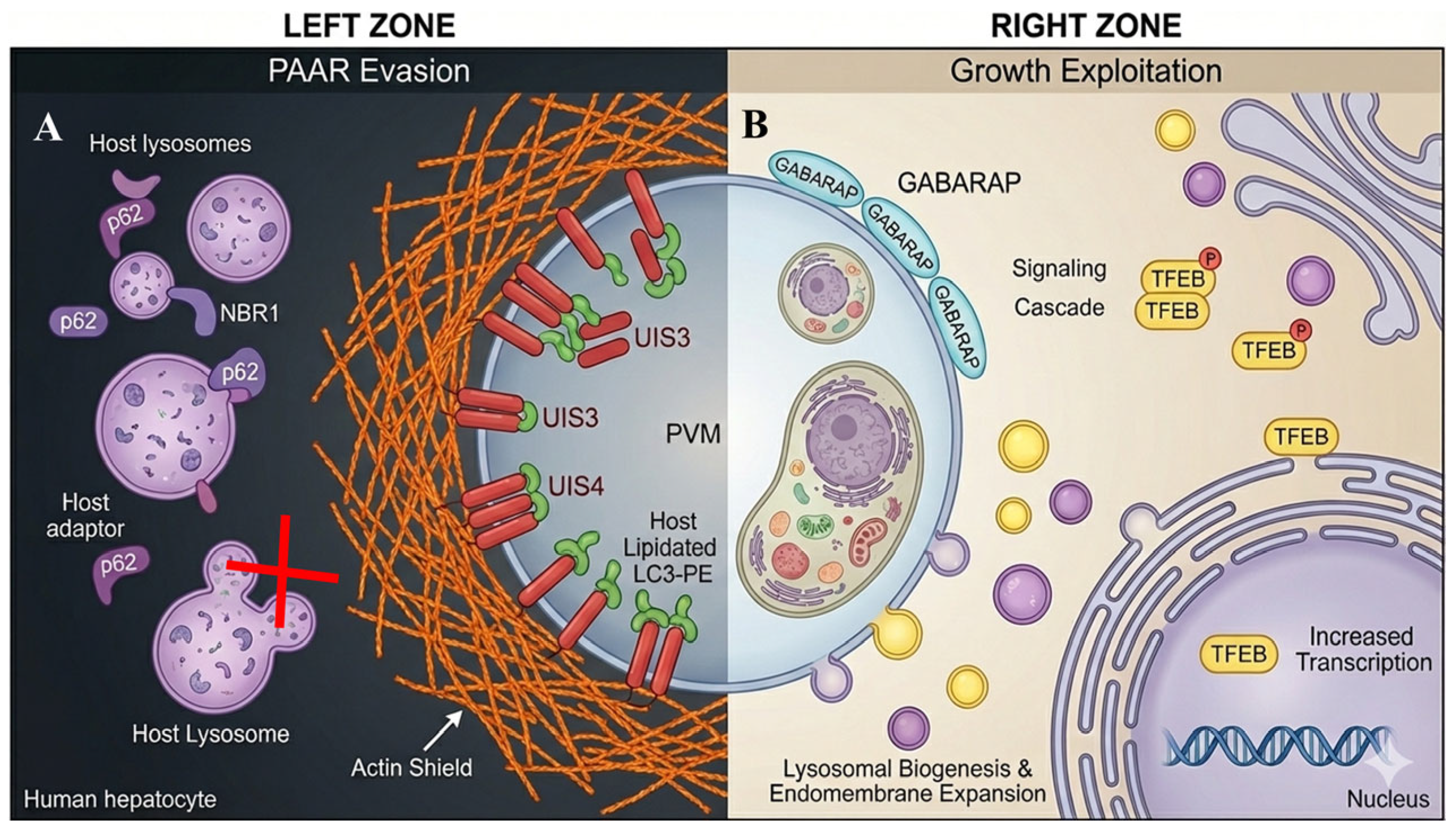
To withstand PAAR-mediated restriction,
Plasmodium deploys targeted interference strategies that divert or block the downstream maturation of LC3-decorated PVMs. UIS3 binds LC3 directly, preventing its recruitment of degradative adaptors and effectively trapping LC3 in a non-restrictive configuration [
12,
31]. In parallel, UIS4 remodels the perivacuolar actin network to create a dense physical barrier that limits access of lysosomes and ubiquitin (
Figure 2) [
18,
19]. These mechanisms do not inhibit CASM initiation; instead, they selectively target the execution phase of PAAR. As a result, hepatocytes continue to recognize and label the PVM with LC3, but parasites prevent the transition from LC3 decoration to effective lysosomal killing. This creates a biological stalemate in which LC3 recruitment becomes necessary but insufficient for eliminating the parasite.
3.3. Parasite Exploitation of Autophagy Components for Intracellular Growth
Beyond PAAR evasion, liver-stage parasites co-opt host autophagy machinery to support their metabolic expansion. Recent work using
Plasmodium berghei liver-stage parasites shows selective recruitment of GABARAP paralogs—distinct members of the ATG8 family with trafficking and signaling functions—to the PVM at later stages [
20,
32]. Unlike LC3, which drives restriction, GABARAP promotes activation of TFEB, the master transcriptional regulator of lysosomal biogenesis and autophagy gene expression (
Figure 2) [
23].
TFEB activation expands the hepatocyte’s lysosomal network and increases endomembrane trafficking capacity, generating a nutrient-rich and membrane-rich environment that accelerates parasite replication [
20,
21]. Through this strategy, the parasite effectively repurposes host autophagy infrastructure to meet its metabolic and organellar demands during schizogony.
Given that hepatocyte CASM/PAAR activity is counterbalanced by parasite-driven evasion and exploitation mechanisms, it is essential to contextualize these opposing processes within a unified framework.
Table 2 summarizes the principal strategies
Plasmodium uses to interfere with LC3-mediated restriction and to co-opt autophagy components for selective metabolic advantage. This overview highlights how molecular evasion (UIS3 and UIS4) and metabolic exploitation (GABARAP-TFEB signaling) collectively reshape the autophagy landscape at the PVM.
3.4. An Asymmetric Autophagy Landscape: Restriction vs. Exploitation
Taken together, the host and parasite simultaneously engage autophagy pathways in a fundamentally asymmetric manner. LC3-driven CASM/PAAR represents a restrictive pathway that hepatocytes deploy to eliminate intracellular parasites. GABARAP-dependent TFEB activation, by contrast, represents an exploitative pathway hijacked by the parasite to enhance intracellular growth. These pathways use overlapping autophagy components but yield opposite biological outcomes. This asymmetry yields three key conceptual insights (
Figure 3):
Autophagy is not uniformly protective: its impact depends on which ATG8 paralog predominates at the PVM.
Host defense and parasite survival occur in parallel: evasion mechanisms (UIS3 and UIS4) prevent the host from converting LC3 labeling into effective killing.
The parasite dynamically transitions its strategy across developmental time: early evasion of LC3-dependent restriction precedes late-stage exploitation of GABARAP–TFEB signaling.
This integrated framework redefines the liver stage as a competitive interface between noncanonical autophagy-mediated restriction and autophagy-driven metabolic exploitation, laying the foundation for the therapeutic approaches discussed in
Section 4.
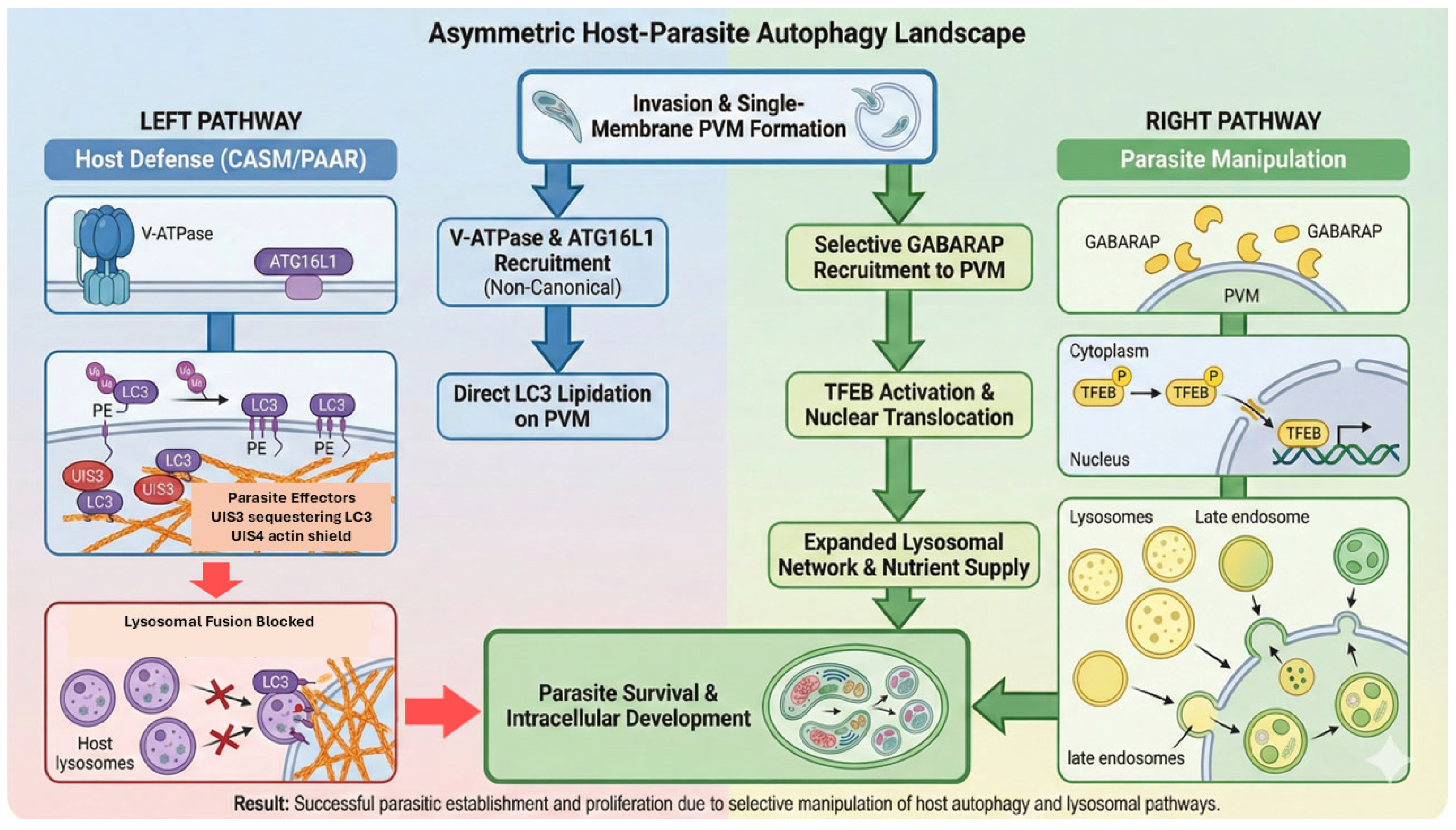
Figure 3.
The asymmetric host–parasite autophagy landscape: defense vs. exploitation. This schematic summarizes the divergent autophagy pathways occurring at the PVM interface. (Left) Host defense: the host initiates cell-autonomous immunity (CASM/PAAR) by recruiting V-ATPase and ATG16L1 to facilitate direct LC3 lipidation on the PVM, thereby driving lysosomal fusion. However, this restriction mechanism is counteracted by parasite effectors (UIS3 sequestration and the UIS4-mediated actin shield), which effectively block lysosomal attack. (Right) Parasite manipulation: concurrently, the parasite exploits the pathway by selectively recruiting the host ATG8 paralog, GABARAP. This recruitment triggers a signaling cascade leading to the activation and nuclear translocation of TFEB. This results in upregulated lysosomal biogenesis and endomembrane expansion, providing an expanded nutrient supply essential for parasite survival and development. Colors and symbols are used for clarity, and arrows indicate the direction of host–parasite interactions and pathway progression at the PVM.
Figure 3.
The asymmetric host–parasite autophagy landscape: defense vs. exploitation. This schematic summarizes the divergent autophagy pathways occurring at the PVM interface. (Left) Host defense: the host initiates cell-autonomous immunity (CASM/PAAR) by recruiting V-ATPase and ATG16L1 to facilitate direct LC3 lipidation on the PVM, thereby driving lysosomal fusion. However, this restriction mechanism is counteracted by parasite effectors (UIS3 sequestration and the UIS4-mediated actin shield), which effectively block lysosomal attack. (Right) Parasite manipulation: concurrently, the parasite exploits the pathway by selectively recruiting the host ATG8 paralog, GABARAP. This recruitment triggers a signaling cascade leading to the activation and nuclear translocation of TFEB. This results in upregulated lysosomal biogenesis and endomembrane expansion, providing an expanded nutrient supply essential for parasite survival and development. Colors and symbols are used for clarity, and arrows indicate the direction of host–parasite interactions and pathway progression at the PVM.
![Pathogens 15 00070 g003 Pathogens 15 00070 g003]()
4. Therapeutic Modulation of Autophagy
Given the dual and highly specialized roles of autophagy during Plasmodium liver-stage infection—where CASM-driven PAAR restricts parasites while GABARAP–TFEB signaling supports parasite growth—the therapeutic potential of autophagy modulation depends on selectively enhancing host-protective pathways while avoiding reinforcement of parasite-beneficial mechanisms. This emerging paradigm underpins host-directed therapy (HDT), which aims to manipulate hepatocyte pathways that govern parasite fate while reducing the selective pressure imposed by parasite-targeted drugs.
4.1. Enhancing PAAR/CASM-Mediated Parasite Clearance
Recent studies demonstrate that liver-stage parasite elimination is mediated by noncanonical autophagy, reliant on the V-ATPase-ATG16L1-LC3 axis [
16]. CASM activation begins at invasion, when membrane tension and ionic perturbation recruit the ATG16L1 WD40-domain to the PVM [
15]. Agents capable of stabilizing LC3 lipidation or disrupting UIS3-LC3B interaction—a central parasite evasion mechanism [
12]—represent promising HDT candidates. Strengthening PAAR execution without stimulating canonical autophagy may enhance parasite elimination while minimizing off-target metabolic effects.
4.2. Preventing Parasite Exploitation of Host Autophagy
While LC3-driven PAAR restricts parasites, liver-stage
Plasmodium can exploit GABARAP paralogs to activate TFEB, the master regulator of lysosomal biogenesis [
20]. This creates a nutrient-rich intracellular environment favorable for growth. Thus, autophagy activators that globally elevate ATG8-family activity could inadvertently enhance parasite survival. Selective inhibitors that disrupt GABARAP–TFEB coupling while preserving LC3-dependent PAAR represent a more precise HDT strategy.
4.3. Repurposing Existing Autophagy Modulators
Several autophagy-modulating drugs offer repurposing potential:
Rapamycin enhances autophagy through mTOR inhibition and reduces the liver-stage burden, although immunosuppressive effects limit its translational viability [
33].
Chloroquine (CQ)/hydroxychloroquine (HCQ) inhibit lysosomal acidification and may disrupt parasite access to nutrients, but resistance and toxicity constrain their use [
9].
Carbamazepine (CBZ) induces mTOR-independent autophagy, potentially enhancing hepatocyte resilience [
34].
Metformin activates AMPK and reprograms hepatocyte metabolism, reducing liver-stage replication [
18].
These observations demonstrate that manipulating autophagy yields context-dependent effects, underscoring the need for pathway-selective approaches.
4.4. Integrating Host Autophagy Modulators with Artemisinin-Based Therapies
Because artemisinin resistance is associated with enhanced stress tolerance and metabolic dormancy, combining ACTs with PAAR-enhancing therapies may accelerate parasite elimination and limit the emergence of resistant blood-stage clones [
35]. Strengthening hepatocyte-driven clearance before parasites reach the bloodstream reduces parasite biomass and restricts the evolutionary space available for resistance selection [
36].
4.5. Challenges and Prospects for Autophagy-Targeted Host-Directed Therapies
To complement the mechanistic and strategic framework presented above,
Table 3 summarizes candidate host-directed therapeutics (HDTs) that target autophagy-related pathways in the liver stage. The table highlights both clinically available drugs and experimental molecules, together with their mechanistic basis and translational potential.
Challenges for autophagy-targeted therapies include hepatotoxicity risk, inter-individual variation in autophagy gene polymorphisms, and the difficulty of manipulating CASM without perturbing essential hepatocyte physiology. Advances in ATG8-selective chemistry, TFEB inhibitors, and humanized liver models may enable the development of precise autophagy modulators that enhance PAAR while preventing parasite exploitation.
Collectively, these insights position CASM-targeted HDT as a promising strategy with the potential to block liver-to-blood stage transition, reduce artemisinin resistance, and create evolution-resilient therapeutic combinations.
5. Knowledge Gaps and Future Perspectives
Despite major advances in understanding the interaction between Plasmodium liver-stage parasites and hepatocyte autophagy, several key gaps remain unresolved, limiting our ability to design targeted HDTs and to predict how different autophagy pathways influence infection outcome.
5.1. Unresolved Molecular Triggers of PAAR/CASM Activation
Although recent studies demonstrate that noncanonical autophagy is initiated during productive invasion via a V-ATPase-ATG16L1-dependent mechanism [
15], the upstream sensing events that distinguish productive invasion from abortive entry or phagocytic uptake remain unclear. The precise membrane tension, ionic perturbations, and lipid compositions that activate CASM in hepatocytes have not been fully defined. Moreover, the influence of hepatocyte heterogeneity on PAAR initiation has not been examined. A deeper molecular and biophysical understanding of CASM activation at the PVM is needed to harness this pathway therapeutically.
5.2. Incomplete Understanding of Parasite Evasion Pathways
While UIS3-LC3 binding and UIS4-dependent actin remodeling are established as major evasion strategies [
12,
18], the extent to which these pathways act redundantly or synergistically remains unknown. It also remains unclear whether
Plasmodium expresses additional PVM proteins that antagonize PAAR or prevent endolysosomal tethering. The structural basis of the UIS3-LC3 interaction is only partially characterized, and high-resolution structural studies could enable the rational design of UIS3 inhibitors. Furthermore, the dynamics of the perivacuolar actin network have never been visualized in vivo, representing a major gap in our understanding of evasion mechanisms.
5.3. Uncharacterized Functions of ATG8 Paralog Selectivity
Emerging evidence shows that
Plasmodium can differentially recruit LC3 versus GABARAP paralogs to the PVM, enabling the parasite to block PAAR while simultaneously exploiting host TFEB-driven lysosomal expansion [
20]. However, the determinants of this paralog selectivity are unknown. Whether ATG4 isoforms, lipid microdomains, or parasite-derived lipids shape ATG8 recruitment remains to be elucidated. Since ATG8 proteins have distinct roles in autophagy, membrane dynamics, and vesicular trafficking, understanding paralog-specific interactions is essential for developing selective HDTs.
5.4. Limited Translational Tools for Targeting CASM and PAAR
Although recent work shows that hepatocyte killing of liver-stage parasites depends on noncanonical autophagy [
16], no selective CASM activators or ATG16L1 modulators are currently available. Most known autophagy drugs broadly target mTOR, AMPK, or lysosomes and therefore risk enhancing parasite exploitation pathways (e.g., the GABARAP-TFEB axis) [
37,
38]. Likewise, no pharmacological inhibitors of UIS3-LC3 interaction or UIS4-actin remodeling currently exist. Thus, there is an urgent need for chemical probes that selectively modulate LC3 lipidation, ATG16L1 localization, or TFEB activity in infected hepatocytes.
5.5. Modeling Barriers and Inter-Individual Variability
The lack of physiologically relevant models constrains progress in the field. Most mechanistic studies use transformed hepatoma lines, which differ substantially from primary hepatocytes in lipid metabolism, autophagy flux, and endolysosomal dynamics [
39,
40]. Humanized liver mouse models address some limitations but remain costly and low-throughput [
41,
42]. Although these mechanisms are robustly supported in
Plasmodium berghei models, their conservation and quantitative contribution during
Plasmodium falciparum liver-stage infection in humans remain incompletely defined.
Additionally, common human polymorphisms in autophagy genes—such as ATG16L1 T300A—may influence host susceptibility to liver-stage infection yet remain unexplored in malaria [
35]. Understanding host genetic variability will be crucial for predicting patient-specific responses to HDTs.
5.6. Future Directions
Future research should prioritize dissecting CASM activation mechanisms, resolving the structural interactions between UIS3/UIS4, characterizing ATG8 paralog selectivity, and developing selective autophagy modulators compatible with HDT. Although this review focuses on Plasmodium liver stages, related apicomplexan parasites also interface with host autophagy and endolysosomal pathways. Comparative work across these intracellular vacuolar pathogens may help identify conserved versus Plasmodium-specific principles of autophagy engagement and immune evasion. Integrating single-cell transcriptomics, CRISPR perturbation screens, humanized liver platforms, and high-resolution imaging of the PVM will be key to generating a comprehensive map of hepatocyte defense pathways. Ultimately, defining the autophagy landscape of the infected hepatocyte—and how parasites rewire it—will accelerate the development of evolutionary-resilient interventions capable of blocking liver-stage progression and reducing the emergence of artemisinin resistance.
Together, these unresolved questions highlight that although substantial progress has been made in defining the molecular architecture of hepatocyte–Plasmodium interactions, our understanding of liver-stage malaria remains incomplete. The outstanding gaps—ranging from the upstream triggers of CASM activation to the structural basis of UIS3–LC3 sequestration to the determinants of ATG8 paralog selectivity and the limitations of current hepatocyte model systems—underscore the complexity of the autophagy landscape that shapes parasite fate. Addressing these knowledge gaps is not only critical for clarifying the fundamental biology of liver-stage infection but also essential for enabling the design of next-generation host-directed therapies that selectively reinforce protective autophagy while preventing parasite exploitation of those same pathways.
In light of these scientific and translational challenges, it becomes increasingly important to synthesize the emerging conceptual framework and articulate its implications for malaria biology and intervention. The following section integrates these mechanistic insights and outlines the broader significance of autophagy—both as a determinant of hepatocyte-intrinsic immunity and as a target of parasite manipulation—providing a unified perspective on how these processes can be leveraged for more effective malaria control strategies.
6. Conclusions
Recent advances have substantially refined our understanding of how autophagy influences Plasmodium liver-stage infection. Current evidence indicates that hepatocytes do not rely on canonical xenophagy; instead, they activate a noncanonical, CASM-dependent pathway that promotes LC3 lipidation on the parasitophorous vacuole membrane through a V-ATPase-ATG16L1 mechanism. This PAAR response represents a critical component of cell-intrinsic immunity and contributes to early parasite restriction.
At the same time, Plasmodium liver-stage parasites employ dedicated strategies to counteract this defense. UIS3-mediated sequestration of LC3 and UIS4-driven actin remodeling limit the execution of PAAR and maintain PVM integrity. Moreover, parasites can exploit host autophagy components—particularly GABARAP paralogs and TFEB-associated lysosomal remodeling—to enhance their access to nutrients and support intracellular development. These findings demonstrate that autophagy serves both protective and permissive functions, depending on the specific molecular interactions occurring at the host–parasite interface.
This duality underscores the necessity for therapeutic approaches that differentiate between beneficial and parasite-supportive autophagy pathways. Strategies that selectively reinforce CASM-mediated LC3 lipidation, while avoiding activation of GABARAP–TFEB signaling, may offer a targeted route for host-directed intervention. Such approaches have the potential to limit liver-to-blood stage progression and reduce selective pressures that facilitate antimalarial resistance.
Despite this progress, several challenges remain. Key unresolved questions include the upstream triggers of CASM activation, the breadth of parasite effectors contributing to PAAR evasion, and the determinants of ATG8-paralog specificity at the PVM. In addition, current hepatocyte models do not fully reproduce the metabolic and immunological complexity of human liver tissue. Advances in single-cell analysis, gene perturbation technologies, and humanized liver platforms will be instrumental for addressing these gaps.
Beyond their mechanistic implications, these insights have direct relevance for global malaria control efforts. A deeper understanding of hepatocyte-intrinsic immunity and parasite evasion pathways supports the development of innovative host-directed strategies aligned with the United Nations Sustainable Development Goal (SDG) 3—particularly Target 3.3, which aims to end malaria as a public health threat by 2030. By informing therapeutic approaches that reduce liver-stage progression and limit the emergence of drug resistance, research in this area contributes not only to basic science but also to long-term global health priorities.
Author Contributions
Conceptualization, A.B.; methodology, A.B. and S.K.; validation, A.B., S.K. and A.K.; investigation, A.B. and S.K.; writing—original draft preparation, A.B. and S.K.; supervision, E.Y.S. and A.K.; funding acquisition, A.B. and EYS. All authors have read and agreed to the published version of the manuscript.
Funding
The publication fee was supported by Universitas Padjadjaran through the Indonesian Endowment Fund for Education (LPDP) of the Ministry of Higher Education, Science, and Technology, under the EQUITY Program (Contract No. 4303/B3/DT.03.08/2025 and 3927/UN6.RKT/HK.07.00/2025).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
This article is a narrative review, and no new data were generated or analyzed. Therefore, data sharing is not applicable.
Acknowledgments
The authors sincerely acknowledge the Directorate of Research, Downstream, and Community Engagement at Universitas Padjadjaran for institutional support throughout the preparation of this article. Artificial intelligence–based tools, including ChatGPT version 5.0 and Google Gemini 3 Pro (Nano Banana Pro), were used under the full supervision of the authors to support language editing and the development of original schematic figures. The authors assume full responsibility for the content, accuracy, and originality of all figures. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ACTs | Artemisinin-based Combination Therapies |
| AMPK | AMP-Activated Protein Kinase |
| ATG | Autophagy-Related Gene |
| ATG4 | Autophagy-Related Protein 4 |
| ATG5 | Autophagy-Related Protein 5 |
| ATG8 | Autophagy-Related Protein 8 |
| ATG12 | Autophagy-Related Protein 12 |
| ATG16L1 | Autophagy-Related Protein 16 Like 1 |
| CASM | Conjugation of ATG8 to single Membrane |
| CBZ | Carbamazepine |
| CQ | Chloroquine |
| GABARAP | Gaba Type A Receptor-Associated Protein |
| HCQ | Hydroxychloroquine |
| HDT | Host-directed Therapy |
| LC3/LC3B | Microtubule-Associated Protein 1A/1B Light Chain 3 |
| mTOR | Mechanistic Target of Rapamycin |
| NO | Nitric Oxide |
| PAAR | Plasmodium-associated autophagy-related |
| PV | Parasitophorous vacuole |
| PVM | Parasitophorous Vacuole Membrane |
| SDGs | Sustainable Development Goals |
| TFEB | Transcription Factor EB |
| UIS3 | Upregulated in Infective Sporozoites 3 |
| UIS4 | Upregulated in Infective Sporozoites 4 |
References
- World Health Organization. World Malaria Report 2024: Addressing Inequity in the Global Malaria Response; World Health Organization: Geneva, Switzerland, 2024; pp. 6–26. Available online: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2024 (accessed on 1 November 2025).
- Zhu, C.; Jiao, S.; Xu, W. CD8+ Trms against malaria liver-stage: Prospects and challenges. Front. Immunol. 2024, 15, 1344941. [Google Scholar] [CrossRef] [PubMed]
- Goswami, D.; Kumar, S.; Betz, W.; Armstrong, J.M.; Haile, M.T.; Camargo, N.; Parthiban, C.; Seilie, A.M.; Murphy, S.C.; Vaughan, A.M.; et al. A Plasmodium falciparum ATP-binding cassette transporter is essential for liver stage entry into schizogony. iScience 2022, 25, 104224. [Google Scholar] [CrossRef] [PubMed]
- Roques, M.; Bindschedler, A.; Beyeler, R.; Heussler, V.T. Same, same but different: Exploring Plasmodium cell division during liver stage development. PLoS Pathog. 2023, 19, e1011210. [Google Scholar] [CrossRef] [PubMed]
- Soulard, V.; Bosson-Vanga, H.; Lorthiois, A.; Roucher, C.; Franetich, J.F.; Zanghi, G.; Bordessoulles, M.; Tefit, M.; Thellier, M.; Morosan, S.; et al. Plasmodium falciparum full life cycle and Plasmodium ovale liver stages in humanized mice. Nat. Commun. 2015, 6, 7690. [Google Scholar] [CrossRef]
- Goldberg, D.E.; Zimmerberg, J. Hardly Vacuous: The Parasitophorous Vacuolar Membrane of Malaria Parasites. Trends Parasitol. 2019, 36, 138–146. [Google Scholar] [CrossRef]
- Posfai, D.; Sylvester, K.; Reddy, A.; Ganley, J.G.; Wirth, J.; Cullen, Q.E.; Dave, T.; Kato, N.; Dave, S.S.; Derbyshire, E.R. Plasmodium Parasite Exploits Host Aquaporin-3 During Liver Stage Malaria Infection. PLoS Pathog. 2018, 14, e1007057. [Google Scholar] [CrossRef]
- Burda, P.-C.; Caldelari, R.; Heussler, V. Manipulation of the Host Cell Membrane during Plasmodium Liver Stage Egress. mBio 2017, 8, e00139-17. [Google Scholar] [CrossRef]
- Agop-Nersesian, C.; Niklaus, L.; Wacker, R.; Heussler, V. Host cell cytosolic immune response during Plasmodium liver stage development. FEMS Microbiol. Rev. 2018, 42, 324–334. [Google Scholar] [CrossRef]
- Niklaus, L.; Agop-Nersesian, C.; Schmuckli-Maurer, J.; Wacker, R.; Grünig, V.; Heussler, V.T. Deciphering host lysosome-mediated elimination of Plasmodium berghei liver stage parasites. Sci. Rep. 2019, 9, 7967. [Google Scholar] [CrossRef]
- Boonhok, R.; Rachaphaew, N.; Duangmanee, A.; Chobson, P.; Pattaradilokrat, S.; Utaisincharoen, P.; Sattabongkot, J.; Ponpuak, M. LAP-like Process as an Immune Mechanism Downstream of IFN-γ in Control of the Human Malaria Plasmodium vivax liver Stage. Proc. Natl. Acad. Sci. USA 2016, 113, E3519–E3528. [Google Scholar] [CrossRef]
- Real, E.; Rodrigues, L.; Cabal, G.G.; Enguita, F.J.; Mancio-Silva, L.; Mello-Vieira, J.; Beatty, W.; Vera, I.M.; Zuzarte-Luís, V.; Figueira, T.N.; et al. Plasmodium UIS3 sequesters host LC3 to avoid elimination by autophagy in hepatocytes. Nat. Microbiol. 2018, 3, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Setua, S.; Enguita, F.J.; Chora, Â.; Ranga-prasad, H.; Lahree, A.; Marques, S.; Sundaramurthy, V.; Mota, M.M. Disrupting Plasmodium UIS3–host LC3 interaction with a small molecule causes parasite elimination from host cells. Commun. Biol. 2020, 3, 688. [Google Scholar] [CrossRef] [PubMed]
- Wacker, R.; Eickel, N.; Schmuckli-Maurer, J.; Annoura, T.; Niklaus, L.; Khan, S.M.; Guan, J.L.; Heussler, V.T. LC3-association with the parasitophorous vacuole membrane of Plasmodium berghei liver stages follows a noncanonical autophagy pathway. Cell Microbiol. 2017, 19, e12754. [Google Scholar] [CrossRef] [PubMed]
- Bindschedler, A.; Schmuckli-Maurer, J.; Buchser, S.; Fischer, T.D.; Wacker, R.; Davalan, T.; Brunner, J.; Heussler, V.T. LC3B labeling of the parasitophorous vacuole membrane of Plasmodium berghei liver stage parasites depends on the V-ATPase and ATG16L1. Mol. Microbiol. 2024, 121, 1095–1111. [Google Scholar] [CrossRef]
- Schepis, A.; Mertens, J.E.; Lewis, P.; Patel, H.; Stegman, N.; Reynolds, L.; Minkah, N.K.; Kappe, S.H.I. Elimination of intra-hepatocytic malaria parasites is driven by non-canonical autophagy but not nitric oxide production. iScience 2025, 28, 112052. [Google Scholar] [CrossRef]
- Mishra, A.; Rajput, S.; Srivastava, P.N.; Ali, H.S.; Mishra, S. Autophagy protein Atg7 is essential for maintaining malaria parasite cellular homeostasis and organelle biogenesis. mBio 2025, 16, e0273524. [Google Scholar] [CrossRef]
- Gomes-Santos, C.S.S.; Itoe, M.A.; Afonso, C.; Henriques, R.; Gardner, R.; Sepúlveda, N.; Simões, P.D.; Raquel, H.; Almeida, A.P.; Moita, L.F.; et al. Highly dynamic host actin reorganization around developing Plasmodium inside hepatocytes. PLoS ONE 2012, 7, e29408. [Google Scholar] [CrossRef]
- Petersen, W.; Petersen, W.; Stenzel, W.; Silvie, O.; Blanz, J.; Saftig, P.; Matuschewski, K.; Ingmundson, A. Sequestration of cholesterol within the host late endocytic pathway restricts liver-stage Plasmodium development. Mol. Biol. Cell. 2017, 28, 726–735. [Google Scholar] [CrossRef]
- Schmuckli-Maurer, J.; Bindschedler, A.F.; Wacker, R.; Würgler, O.M.; Rehmann, R.; Lehmberg, T.; Murphy, L.O.; Nguyen, T.N.; Lazarou, M.; Monfregola, J.; et al. Plasmodium berghei liver stage parasites exploit host GABARAP proteins for TFEB activation. Commun. Biol. 2024, 7, 1554. [Google Scholar] [CrossRef]
- Leleu, I.; Alloo, J.; Cazenave, P.-A.; Roland, J.; Pied, S. Autophagy Pathways in the Genesis of Plasmodium-Derived Microvesicles: A Double-Edged Sword? Life 2022, 12, 415. [Google Scholar] [CrossRef]
- Byun, S.; Kim, Y.; Zhang, Y.; Kong, B.; Guo, G.L.; Sadoshima, J.; Ma, J.; Kemper, B.; Kemper, J.K. A postprandial FGF19-SHP-LSD1 regulatory axis mediates epigenetic repression of hepatic autophagy. EMBO J. 2017, 36, 1755–1769. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Lee, H.M.; Kim, J.K.; Yang, C.; Kim, T.S.; Jung, M.; Jin, H.S.; Kim, S.; Jang, J.; Oh, G.T.; et al. PPAR-α Activation Mediates Innate Host Defense through Induction of TFEB and Lipid Catabolism. J. Immunol. 2017, 198, 3283–3295. [Google Scholar] [CrossRef] [PubMed]
- Schepis, A.; Kumar, S.; Kappe, S.H.I. Malaria parasites harness Rho GTPase signaling and host cell membrane ruffling for productive invasion of hepatocytes. Cell Rep. 2023, 42, 111927. [Google Scholar] [CrossRef] [PubMed]
- Sturm, A.; Amino, R.; van de Sand, C.; Regen, T.; Retzlaff, S.; Rennenberg, A.; Krueger, A.; Pollok, J.M.; Menard, R.; Heussler, V.T. Manipulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science 2006, 313, 1287–1290. [Google Scholar] [CrossRef]
- Risco-Castillo, V.; Topçu, S.; Marinach, C.; Manzoni, G.; Bigorgne, A.E.; Briquet, S.; Baudin, X.; Lebrun, M.; Dubremetz, J.F.; Silvie, O. Malaria Sporozoites Traverse Host Cells within Transient Vacuoles. Cell Host Microbe 2015, 18, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Schmuckli-Maurer, J.; Reber, V.; Wacker, R.; Bindschedler, A.; Zakher, A.; Heussler, V.T. Inverted recruitment of autophagy proteins to the Plasmodium berghei parasitophorous vacuole membrane. PLoS ONE 2017, 12, e0183797. [Google Scholar] [CrossRef]
- Durgan, J.; Florey, O. Many roads lead to CASM: Diverse stimuli of noncanonical autophagy share a unifying molecular mechanism. Sci. Adv. 2022, 8, eabo1274. [Google Scholar] [CrossRef]
- Hooper, K.M.; Jacquin, E.; Li, T.; Goodwin, J.M.; Brumell, J.H.; Durgan, J.; Florey, O. V-ATPase is a universal regulator of LC3 associated phagocytosis and non-canonical autophagy. bioRxiv 2021. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; Zaretski, S.; Liu, T.; Adams, P.D.; Hansen, M. Post-translational modifications of ATG8 proteins—An emerging mechanism of autophagy control. J. Cell Sci. 2023, 136, jcs259725. [Google Scholar] [CrossRef]
- Kumar, H.; Sattler, J.M.; Singer, M.; Heiss, K.; Reinig, M.; Hammerschmidt-Kamper, C.; Heussler, V.; Mueller, A.K.; Frischknecht, F. Protective efficacy and safety of liver stage attenuated malaria parasites. Sci. Rep. 2016, 6, 26824. [Google Scholar] [CrossRef]
- Wang, Q.; Chang, C.; Gu, N.; Pan, C.; Xu, C. Effect of autophagy on liver regeneration. Hereditas 2015, 37, 1116–1124. [Google Scholar] [PubMed]
- Prado, M.; Eickel, N.; De Niz, M.; Heitmann, A.; Agop-Nersesian, C.; Wacker, R.; Schmuckli-Maurer, J.; Caldelari, R.; Janse, C.J.; Khan, S.M.; et al. Long-term live imaging reveals cytosolic immune responses of host hepatocytes against Plasmodium infection and parasite escape mechanisms. Autophagy 2015, 11, 1561–1579. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, S.; Zhang, X.; Li, T.; Tang, Y.; Liu, H.; Yang, W.; Le, W. Autophagy Enhancer Carbamazepine Alleviates Memory Deficits and Cerebral Amyloid-β Pathology in a Mouse Model of Alzheimer’s Disease. Curr. Alzheimer Res. 2013, 10, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Marques-da-Silva, C.; Schmidt-Silva, C.; Kurup, S.P. Hepatocytes and the art of killing Plasmodium softly. Trends Parasitol. 2024, 40, 466–476. [Google Scholar] [CrossRef]
- Sá e Cunha, C.; Nyboer, B.; Heiss, K.; Sanches-Vaz, M.; Fontinha, D.; Wiedtke, E.; Grimm, D.; Przyborski, J.M.; Mota, M.M.; Prudêncio, M.; et al. Plasmodium berghei EXP-1 interacts with host Apolipoprotein H during Plasmodium liver-stage development. Proc. Natl. Acad. Sci. USA 2017, 114, E1138–E1147. [Google Scholar] [CrossRef]
- Rebecca, V.W.; Nicastri, M.C.; McLaughlin, N.; Fennelly, C.; McAfee, Q.; Ronghe, A.; Nofal, M.; Lim, C.Y.; Witze, E.; Chude, C.I.; et al. A unified approach to targeting the lysosome’s degradative and growth signaling roles. Cancer Discov. 2017, 7, 1266–1283. [Google Scholar] [CrossRef]
- Capurro, M.I.; Prashar, A.; Gao, X.; Jones, N.L. Survival of intracellular pathogens in response to mTORC1- or TRPML1-TFEB-induced xenophagy. Autophagy Rep. 2023, 2, 2191918. [Google Scholar] [CrossRef]
- Portugal, S.; Rodriguez, A.; Prudêncio, M.; Maria, M. Mota: Bringing Plasmodium Liver Infection to the Centre Stage of Malaria Research. Front. Cell Infect. Microbiol. 2022, 12, 851484. [Google Scholar] [CrossRef]
- Yang, A.S.P.; Dutta, D.; Kretzschmar, K.; Hendriks, D.; Puschhof, J.; Hu, H.; Boonekamp, K.E.; van Waardenburg, Y.; de Sousa Lopes, S.M.C.; van Gemert, G.-J.; et al. Development of Plasmodium falciparum liver-stages in hepatocytes derived from human fetal liver organoid cultures. Nat. Commun. 2023, 14, 4631. [Google Scholar] [CrossRef]
- Ng, S.; March, S.; Galstian, A.; Gural, N.; Stevens, K.R.; Mota, M.M.; Bhatia, S.N. Towards a Humanized Mouse Model of Liver Stage Malaria Using Ectopic Artificial Livers. Sci. Rep. 2017, 7, 45424. [Google Scholar] [CrossRef]
- Colón-Thillet, R.; Stone, D.; Loprieno, M.A.; Klouser, L.; Roychoudhury, P.; Santo, T.K.; Xie, H.; Stensland, L.; Upham, S.L.; Pepper, G.; et al. Liver humanized NSG-PiZ mice support the study of chronic hepatitis B virus infection and antiviral therapies. Microbiol. Spectr. 2023, 11, e0517622. [Google Scholar] [CrossRef]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |