Influenza D Virus Circulation Among Bovines, Swine, Equines, and Wild Boars in Italy: A Sero-Epidemiological Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Samples
2.2. Influenza Viruses
2.3. Haemagglutination Inhibition Assay
2.4. Virus Neutralization Assay
2.5. Data Analysis
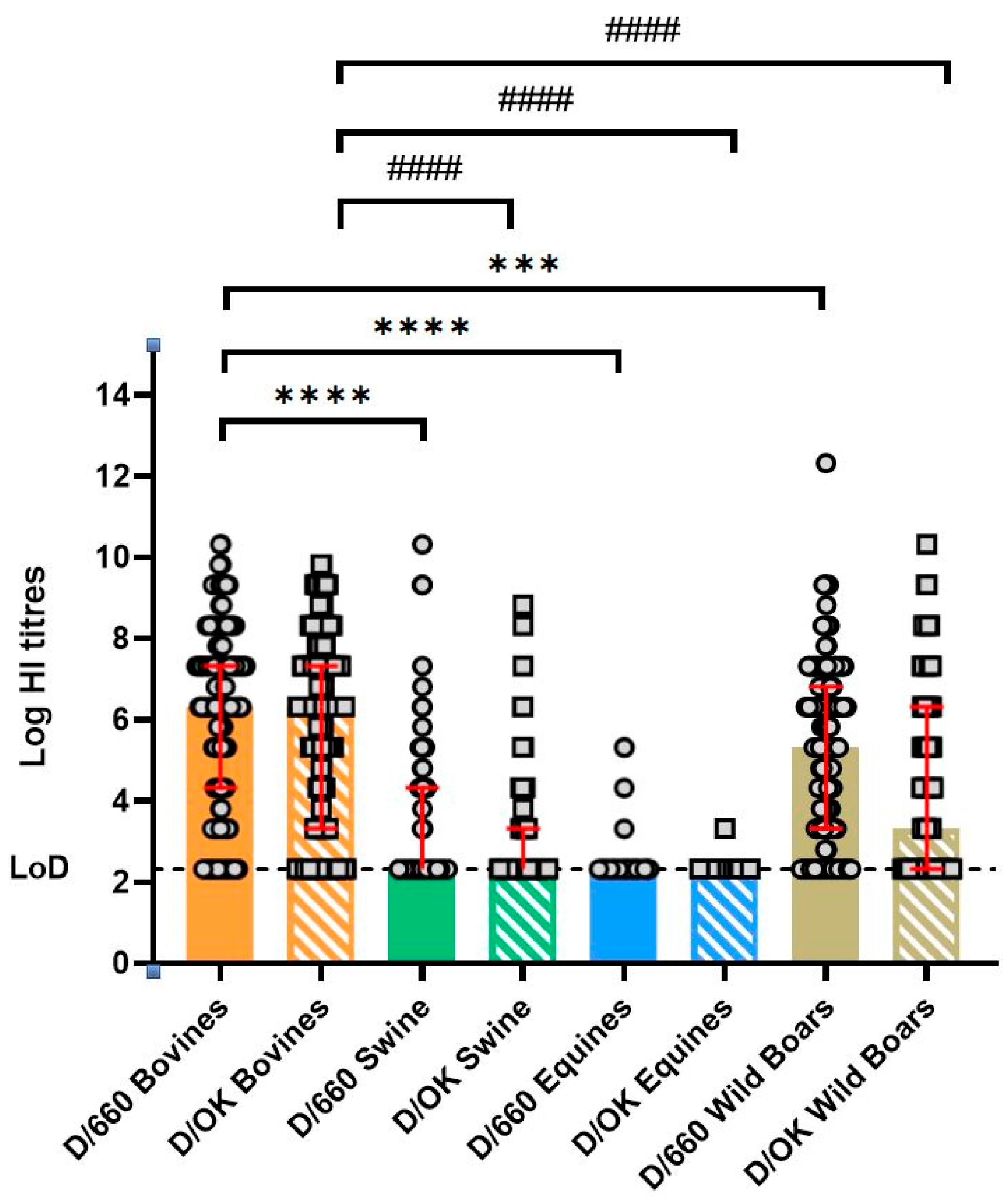
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a Novel Swine Influenza Virus from Oklahoma in 2011 Which Is Distantly Related to Human Influenza C Viruses. PLoS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.L.; Zhang, H.; Chen, S.N.; Zhou, X.; Lin, T.; Liu, R.; Lv, D.-H.; Wen, X.-H.; Wei, W.-K.; Wang, D.; et al. Influenza D Virus in Animal Species in Guangdong Province, Southern China. Emerg. Infect. Dis. 2017, 23, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.; Eckard, L.; Epperson, W.B.; Long, L.-P.; Smith, D.; Huston, C.; Genova, S.; Webby, R.; Wan, X.-F. Influenza D Virus Infection in Mississippi Beef Cattle. Virology 2016, 486, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Endoh, M.; Kobayashi, T.; Takenaka-Uema, A.; Chambers, J.K.; Uchida, K.; Nishihara, M.; Hause, B.; Horimoto, T. Influenza D Virus Infection in Herd of Cattle, Japan. Emerg. Infect. Dis. 2016, 22, 1517–1519. [Google Scholar] [CrossRef]
- Mazzetto, E.; Bortolami, A.; Fusaro, A.; Mazzacan, E.; Maniero, S.; Vascellari, M.; Beato, M.S.; Schiavon, E.; Chiapponi, C.; Terregino, C.; et al. Replication of Influenza D Viruses of Bovine and Swine Origin in Ovine Respiratory Explants and Their Attachment to the Respiratory Tract of Bovine, Sheep, Goat, Horse, and Swine. Front. Microbiol. 2020, 11, 1136. [Google Scholar] [CrossRef]
- Trombetta, C.M.; Marchi, S.; Manini, I.; Kistner, O.; Li, F.; Piu, P.; Manenti, A.; Biuso, F.; Sreenivasan, C.; Druce, J.; et al. Influenza D Virus: Serological Evidence in the Italian Population from 2005 to 2017. Viruses 2019, 12, 30. [Google Scholar] [CrossRef]
- Trombetta, C.M.; Montomoli, E.; Di Bartolo, I.; Ostanello, F.; Chiapponi, C.; Marchi, S. Detection of Antibodies against Influenza D Virus in Swine Veterinarians in Italy in 2004. J. Med. Virol. 2022, 94, 2855–2859. [Google Scholar] [CrossRef]
- White, S.K.; Ma, W.; McDaniel, C.J.; Gray, C.J.; Lednicky, J.A. Serologic Evidence of Exposure to Influenza D Virus among Persons with Occupational Contact with Cattle. J. Clin. Virol. 2016, 81, 31–33. [Google Scholar] [CrossRef]
- Chiapponi, C.; Faccini, S.; Fusaro, A.; Moreno, A.; Prosperi, A.; Merenda, M.; Baioni, L.; Gabbi, V.; Rosignoli, C.; Alborali, G.L.; et al. Detection of a New Genetic Cluster of Influenza D Virus in Italian Cattle. Viruses 2019, 11, 1110. [Google Scholar] [CrossRef]
- Weklak, D.; Pembaur, D.; Koukou, G.; Jönsson, F.; Hagedorn, C.; Kreppel, F. Genetic and Chemical Capsid Modifications of Adenovirus Vectors to Modulate Vector–Host Interactions. Viruses 2021, 13, 1300. [Google Scholar] [CrossRef]
- Nemanichvili, N.; Berends, A.J.; Tomris, I.; Barnard, K.N.; Parrish, C.R.; Gröne, A.; Rijks, J.M.; Verheije, M.H.; de Vries, R.P. Influenza D Binding Properties Vary amongst the Two Major Virus Clades and Wildlife Species. Vet. Microbiol. 2022, 264, 109298. [Google Scholar] [CrossRef] [PubMed]
- Quast, M.; Sreenivasan, C.; Sexton, G.; Nedland, H.; Singrey, A.; Fawcett, L.; Miller, G.; Lauer, D.; Voss, S.; Pollock, S.; et al. Serological evidence for the presence of influenza D virus in small ruminants. Vet. Microbiol. 2015, 180, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Salem, E.; Cook, E.A.J.; Lbacha, H.A.; Oliva, J.; Awoume, F.; Aplogan, G.L.; Hymann, E.C.; Muloi, D.; Deem, S.L.; Alali, S.; et al. Serologic Evidence for Influenza C and D Virus among Ruminants and Camelids, Africa, 1991–2015. Emerg. Infect. Dis. 2019, 25, 1229–1237. [Google Scholar] [CrossRef]
- Holwerda, M.; Kelly, J.; Laloli, L.; Stürmer, I.; Portmann, J.; Stalder, H.; Dijkman, R. Determining the Replication Kinetics and Cellular Tropism of Influenza D Virus on Primary Well-Differentiated Human Airway Epithelial Cells. Viruses 2019, 11, 377. [Google Scholar] [CrossRef]
- Lanave, G.; Camero, M.; Coppola, C.; Marchi, S.; Cascone, G.; Salina, F.; Coltraro, M.; Odigie, A.E.; Montomoli, E.; Chiapponi, C.; et al. Serological Evidence for Circulation of Influenza D Virus in the Ovine Population in Italy. Pathogens 2024, 13, 162. [Google Scholar] [CrossRef]
- Ruiz, M.; Puig, A.; Bassols, M.; Fraile, L.; Armengol, R. Influenza D Virus: A Review and Update of Its Role in Bovine Respiratory Syndrome. Viruses 2022, 14, 2717. [Google Scholar] [CrossRef]
- Limaye, S.; Lohar, T.; Dube, H.; Ramasamy, S.; Kale, M.; Kulkarni-Kale, U.; Kuchipudi, S.V. Rapid Evolution Leads to Extensive Genetic Diversification of Cattle Flu Influenza D Virus. Commun. Biol. 2024, 7, 1276. [Google Scholar] [CrossRef]
- Su, S.; Fu, X.; Li, G.; Kerlin, F.; Veit, M. Novel Influenza D Virus: Epidemiology, Pathology, Evolution and Biological Characteristics. Virulence 2017, 8, 1580–1591. [Google Scholar] [CrossRef]
- Ferguson, L.; Luo, K.; Olivier, A.K.; Cunningham, F.L.; Blackmon, S.; Hanson-Dorr, K.; Sun, H.; Baroch, J.; Lutman, M.W.; Quade, B.; et al. Influenza D Virus Infection in Feral Swine Populations, United States. Emerg. Infect. Dis. 2018, 24, 1020–1028. [Google Scholar] [CrossRef] [PubMed]
- O’Donovan, T.; Donohoe, L.; Ducatez, M.F.; Meyer, G.; Ryan, E. Seroprevalence of Influenza D Virus in Selected Sample Groups of Irish Cattle, Sheep and Pigs. Ir. Vet. J. 2019, 72, 11. [Google Scholar] [CrossRef]
- Gorin, S.; Fablet, C.; Quéguiner, S.; Barbier, N.; Paboeuf, F.; Hervé, S.; Rose, N.; Simon, G. Assessment of Influenza D Virus in Domestic Pigs and Wild Boars in France. Viruses 2019, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Kwasnik, M.; Rola, J.; Rozek, W. Influenza D in Domestic and Wild Animals. Viruses 2023, 15, 2433. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, M.; Moreno, A.; Snoeck, C.J.; Zohari, S.; Saegerman, C.; O’dOnovan, T.; Ryan, E.; Zanni, I.; Foni, E.; Sausy, A.; et al. Emerging Influenza D Virus Infection in European Livestock as Determined in Serology Studies. Transbound. Emerg. Dis. 2021, 68, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, A.; Pellegrini, F.; Camero, M.; Catella, C.; Buonavoglia, D.; Fusco, G.; Martella, V.; Lanave, G. Genetic Diversity of Porcine Circovirus Types 2 and 3 in Wild Boar in Italy. Animals 2022, 12, 953. [Google Scholar] [CrossRef]
- Sreenivasan, C.C.; Uprety, T.; Reedy, S.E.; Temeeyasen, G.; Hause, B.M.; Wang, D.; Li, F.; Chambers, T.M. Experimental Infection of Horses with Influenza D Virus. Viruses 2022, 14, 661. [Google Scholar] [CrossRef] [PubMed]
- Nedland, H.; Wollman, J.; Sreenivasan, C.; Quast, M.; Singrey, A.; Fawcett, L.; Christopher-Hennings, J.; Nelson, E.; Kaushik, R.S.; Wang, D.; et al. Serological evidence for the co-circulation of two lineages of influenza D viruses in equine populations of the Midwest United States. Zoonoses Public Health 2018, 65, e148–e154. [Google Scholar] [CrossRef]
- Bailey, E.S.; Choi, J.Y.; Zemke, J.; Yondon, M.; Gray, G.C. Molecular Surveillance of Respiratory Viruses with Bioaerosol Sampling in an Airport. Trop. Dis. Travel Med. Vaccines 2018, 4, 11. [Google Scholar] [CrossRef]
- Choi, J.Y.; Zemke, J.; Philo, S.E.; Bailey, E.S.; Yondon, M.; Gray, G.C. Aerosol Sampling in a Hospital Emergency Room Setting: A Complementary Surveillance Method for the Detection of Respiratory Viruses. Front. Public Health 2018, 6, 174. [Google Scholar] [CrossRef]
- Borkenhagen, L.K.; Mallinson, K.A.; Tsao, R.W.; Ha, S.-J.; Lim, W.-H.; Toh, T.-H.; Anderson, B.D.; Fieldhouse, J.K.; Philo, S.E.; Chong, K.-S.; et al. Surveillance for Respiratory and Diarrheal Pathogens at the Human-Pig Interface in Sarawak, Malaysia. PLoS ONE 2018, 13, e0201295. [Google Scholar] [CrossRef]
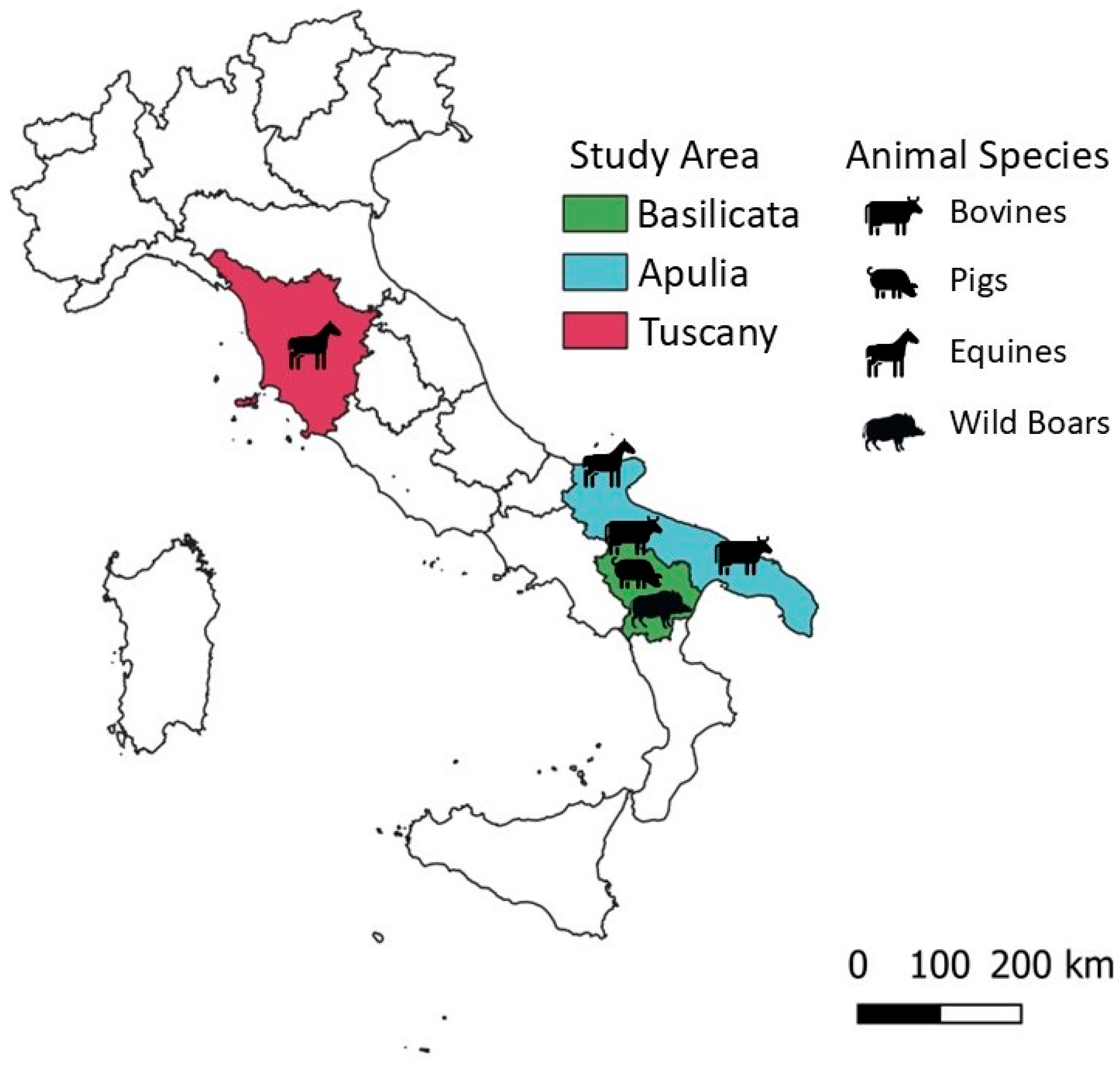
| Animal Species | Number of Samples | Area | Year |
|---|---|---|---|
| Bovine | 246 | Apulia and Basilicata | 2014–2015 |
| Swine | 249 | Basilicata | 2021 |
| Equine | 98 | Tuscany and Apulia | 2001 and 2021 |
| Wild Boar | 445 | Basilicata | 2020–2021 |
| Animal Species | HI D/660 | HI D/OK | HI D/660–D/OK | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ≤5 | ≥10–<40 | ≥40 | Overall seroprevalence ≥10 | ≤5 | ≥10–<40 | ≥40 | Overall seroprevalence ≥10 | D/660 and D/OK overall seroprevalence ≥10 | |
| Bovine | 14% (34/246) 95% CI 9.76–18.7% | 13% (31/246) 95% CI 8.73–17.4% | 74% (181/246) 95% CI 67.60–78.98% | 87% (212/246) 95% CI 81.22–90.24% | 20% (48/246) 95% CI 14.75–25.02% | 10% (25/246) 95% CI 64.19–75.96% | 70% (173/246) 95% CI 64.19–75.96% | 80% (198/246) 95% CI 74.98–85.25% | 80% (196/246) 95% CI 74.10–84.52% |
| Swine | 69% (172/249) 95% CI 62.93–74.76% | 18% (44/249) 95% CI 13.14–22.99% | 13% (33/249) 95% CI 9.30–18.11% | 31% (77/249) 95% CI 25.24–37.07% | 71% (178/249) 95% CI 65.44–77.01% | 21% (52/249) 95% CI 16.01–26.47% | 8% (19/249) 95% CI 4.66–11.66% | 29% (71/249) 95% CI 22.99–34.56% | 20% (20/249) 95% CI 4.98–12.13% |
| Equine | 95% (93/98) 95% CI 88.49–98.32% | 3% (3/98) 95% CI 0.64–8.68% | 2% (2/98) 95% CI 0.25–7.18% | 5% (5/98) 95% CI 1.68–11.51% | 99% (97/98) 95% CI 94.45–99.97% | 1% (1/98) 95% CI 0.03–5.55% | 0% (0/98) 95% CI 0.00–3.69% | 1% (1/98) 95% CI 0.03–5.55 | 1% (1/98) 95% CI 0.03–5.55 |
| Wild Boar | 19% (86/445) 95% CI 15.76–23.31% | 23% (102/445) 95% CI 19.09–27.11% | 57% (252/445) 95% CI 51.88–61.29% | 80% (354/445) 95% CI 75.50–83.20% | 44% (198/445) 95% CI 39.81–49.25% | 20% (87/445) 95% CI 15.97–23.55% | 36% (160/445) 95% CI 31.49–40.61% | 56% (247/445) 95% CI 50.75–60.19% | 51% (225/445) 95% CI 45.781–55.30% |
| Animal Groups | VN D/660 | VN D/OK | ||||||
|---|---|---|---|---|---|---|---|---|
| ≤5 | ≥10–<40 | ≥40 | Overall seroprevalence ≥ 10 | ≤5 | ≥10–<40 | ≥40 | Overall seroprevalence ≥ 10 | |
| Bovine | 13% (33/246) 95% CI 9.42–18.32% | 10% (24/246) 95% CI 6.35–18.32% | 77% (189/246) 95% CI 71.05–81.95% | 87% (213/246) 95% CI 81.68–90.58% | 29% (43/246) 95% CI 12.95–22.81% | 6% (14/246) 95% CI 3.15–9.36% | 77% (189/246) 95% CI 71.05–81.95% | 83% (203/246) 95% CI 77.19–87.05% |
| Swine | 79% (197/249) 95% CI 73.53–83.99% | 11% (28/249) 95% CI 7.60–15.84% | 10% (24/249) 95% CI 6.27–14.00% | 21% (52/249) 95% CI 16.01–26.47% | 76% (190/249) 95% CI 70.53–81.45% | 14% (34/249) 95% CI 9.64–18.55% | 10% (25/249) 95% CI 6.60–14.46% | 24% (59/249) 95% CI 18.55–29.47% |
| Equine | 98% (96/98) 95% CI 92.82–99.75% | 2% (2/98) 95% CI 0.25–7.18% | 0% (0/98) 95% CI 0–3.69% | 2% (2/98) 95% CI 0.25–7.18% | 98% (96/98) 95% CI 92.82–99.75% | 2% (2/98) 95% CI 0.25–7.18% | 0% (0/98) 95% CI 0–3.69% | 2% (2/98) 95% CI 0.25–7.18% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falsini, A.; Coppola, C.; Fiori, A.; Buonavoglia, D.; Marchi, S.; Montomoli, E.; Pellegrini, F.; Lanave, G.; Martella, V.; Camero, M.; et al. Influenza D Virus Circulation Among Bovines, Swine, Equines, and Wild Boars in Italy: A Sero-Epidemiological Study. Pathogens 2025, 14, 891. https://doi.org/10.3390/pathogens14090891
Falsini A, Coppola C, Fiori A, Buonavoglia D, Marchi S, Montomoli E, Pellegrini F, Lanave G, Martella V, Camero M, et al. Influenza D Virus Circulation Among Bovines, Swine, Equines, and Wild Boars in Italy: A Sero-Epidemiological Study. Pathogens. 2025; 14(9):891. https://doi.org/10.3390/pathogens14090891
Chicago/Turabian StyleFalsini, Alessandro, Chiara Coppola, Aurora Fiori, Domenico Buonavoglia, Serena Marchi, Emanuele Montomoli, Francesco Pellegrini, Gianvito Lanave, Vito Martella, Michele Camero, and et al. 2025. "Influenza D Virus Circulation Among Bovines, Swine, Equines, and Wild Boars in Italy: A Sero-Epidemiological Study" Pathogens 14, no. 9: 891. https://doi.org/10.3390/pathogens14090891
APA StyleFalsini, A., Coppola, C., Fiori, A., Buonavoglia, D., Marchi, S., Montomoli, E., Pellegrini, F., Lanave, G., Martella, V., Camero, M., & Trombetta, C. M. (2025). Influenza D Virus Circulation Among Bovines, Swine, Equines, and Wild Boars in Italy: A Sero-Epidemiological Study. Pathogens, 14(9), 891. https://doi.org/10.3390/pathogens14090891







