Introduction and Spatial–Temporal Distribution of Oropouche Virus in Rio de Janeiro State, Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Study Population and Ethics Committee
2.3. Detection of Arboviruses by Real-Time RT-PCR and Nucleotide Sequencing
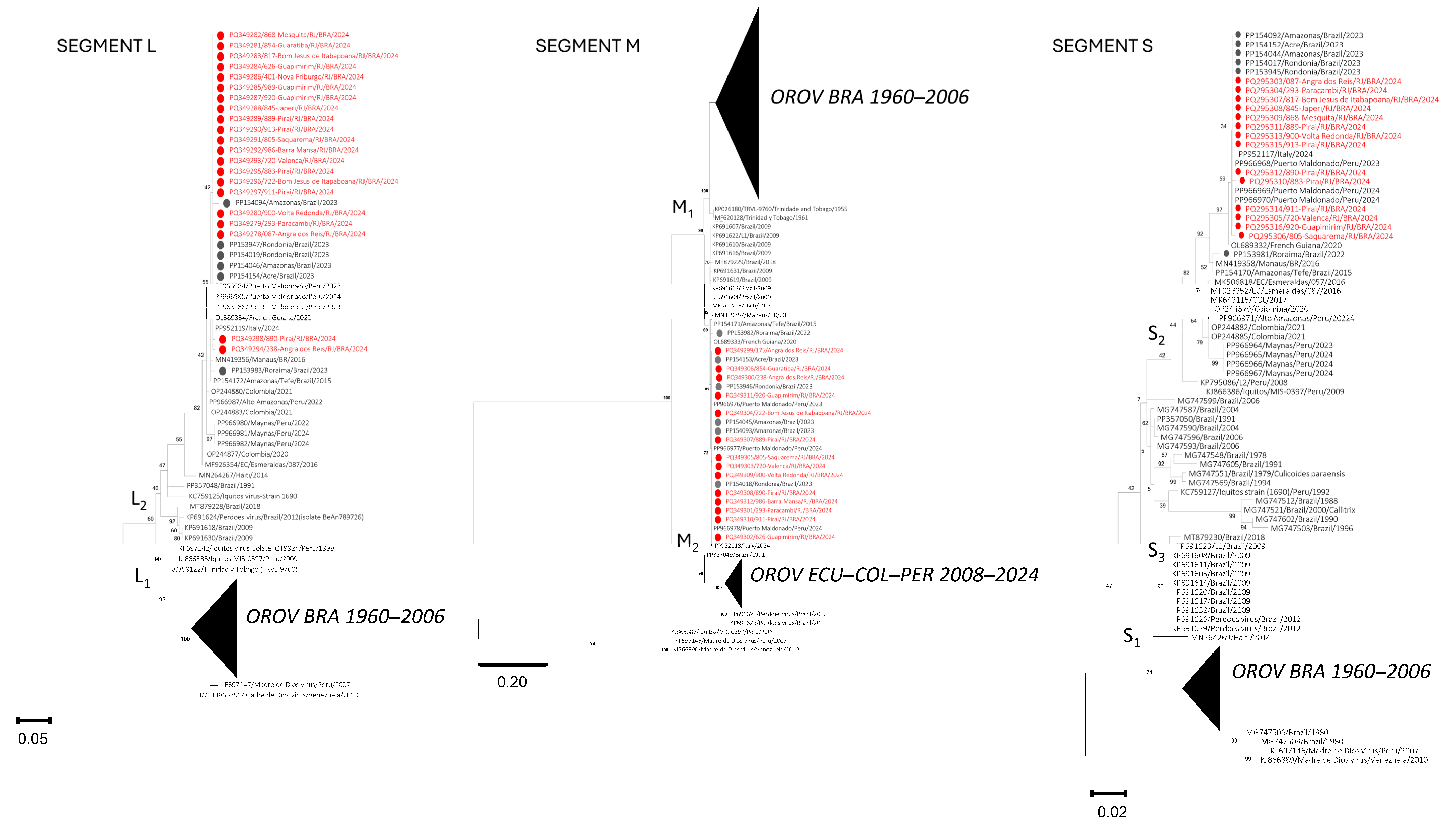
2.4. Phylogenetic Analysis
2.5. Statistical Analysis
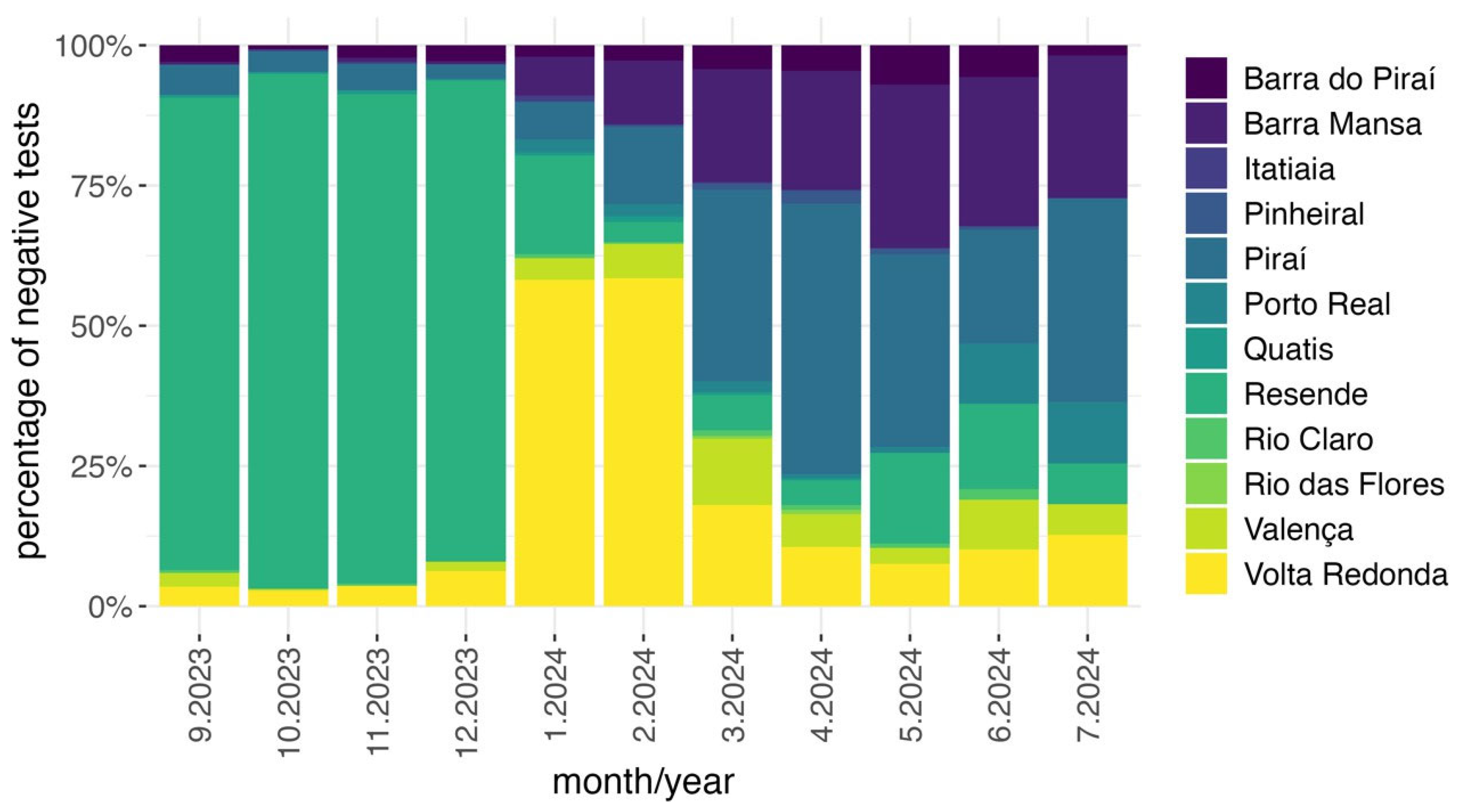
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Da Rosa, J.F.T.; De Souza, W.M.; De Paula Pinheiro, F.; Figueiredo, M.L.; Cardoso, J.F.; Acrani, G.O.; Nunes, M.R.T. Oropouche Virus: Clinical, Epidemiological, and Molecular Aspects of a Neglected Orthobunyavirus. Am. J. Trop. Med. Hyg. 2017, 96, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.R.; Spence, L.; Downs, W.G.; Aitken, T.H.G. Oropouche Virus: A New Human Disease Agent from Trinidad, West Indies. Am. J. Trop. Med. Hyg. 1961, 10, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.E.; López, K.; Eastwood, G.; Guzman, H.; Carrington, C.V.F.; Tesh, R.B.; Auguste, A.J. Phylogenetic Characterization of Orthobunyaviruses Isolated from Trinidad Shows Evidence of Natural Reassortment. Virus Genes 2023, 59, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Sakkas, H.; Bozidis, P.; Franks, A.; Papadopoulou, C. Oropouche Fever: A Review. Viruses 2018, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Naveca, F.G.; de Almeida, T.A.P.; Souza, V.; Nascimento, V.; Silva, D.; Nascimento, F.; Mejía, M.; de Oliveira, Y.S.; Rocha, L.; Xavier, N.; et al. Human Outbreaks of a Novel Reassortant Oropouche Virus in the Brazilian Amazon Region. Nat. Med. 2024, 30, 3509–3521. [Google Scholar] [CrossRef] [PubMed]
- Mourão, M.P.G.; Bastos, M.S.; Gimaque, J.B.L.; Mota, B.R.; Souza, G.S.; Grimmer, G.H.N.; Galusso, E.S.; Arruda, E.; Figueiredo, L.T.M. Oropouche Fever Outbreak, Manaus, Brazil, 2007–2008. Emerg. Infect. Dis. 2009, 15, 2063–2064. [Google Scholar] [CrossRef] [PubMed]
- Sciancalepore, S.; Schneider, M.C.; Kim, J.; Galan, D.I.; Riviere-Cinnamond, A. Presence and Multi-Species Spatial Distribution of Oropouche Virus in Brazil within the One Health Framework. Trop. Med. Infect. Dis. 2022, 7, 111. [Google Scholar] [CrossRef] [PubMed]
- Centro de Inteligência em Saúde, Painel de Monitoramento das Arboviroses. 2024. Available online: https://cisshiny.saude.rj.gov.br/_d/arboviroses/ (accessed on 18 August 2024).
- IBGE–Instituto Brasileiro de Geografia e Estatística. Censo 2022; IBGE: Rio de Janeiro, Brazil, 2022. Available online: https://censo2022.ibge.gov.br/panorama/ (accessed on 18 August 2024).
- Naveca, F.G.; Nascimento, V.A.D.; Souza, V.C.; Nunes, B.T.D.; Rodrigues, D.S.G.; Vasconcelos, P. Multiplexed Reverse Transcription Real-Time Polymerase Chain Reaction for Simultaneous Detection of Mayaro, Oropouche, and Oropouche-like Viruses. Memórias Inst. Oswaldo Cruz 2017, 112, 510–513. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, V.A.D.; Santos, J.H.A.; Monteiro, D.C.D.S.; Pessoa, K.P.; Cardoso, A.J.L.; de Souza, V.C.; Abdalla, L.F.; Naveca, F.G. Oropouche Virus Detection in Saliva and Urine. Memórias Inst. Oswaldo Cruz 2020, 115. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000; ISBN 9780195350517. [Google Scholar]
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023.
- Delatorre, E.; de Mendonça, G.C.; Gatti, F.D.; Có, A.C.G.; Pereira, J.d.P.; Tavares, E.A.; Nodari, J.Z.; Rossi, A.; de Azevedo, S.S.D.; Sacchi, C.T.; et al. Emergence of Oropouche Virus in Espírito Santo State, Brazil, 2024. Emerg. Infect. Dis. 2025, 31, 1178–1188. [Google Scholar] [CrossRef] [PubMed]
- Gräf, T.; Delatorre, E.; do Nascimento Ferreira, C.; Rossi, A.; Santos, H.G.G.; Pizzato, B.R.; Nascimento, V.; Souza, V.; de Lima, G.B.; Dezordi, F.Z.; et al. Expansion of Oropouche Virus in Non-Endemic Brazilian Regions: Analysis of Genomic Characterisation and Ecological Drivers. Lancet Infect. Dis. 2025, 25, 379–389. [Google Scholar] [CrossRef] [PubMed]
- IBGE–Instituto Brasileiro de Geografia e Estatística. Censo Agropecuário 2017; IBGE: Rio de Janeiro, Brazil, 2017. Available online: https://censoagro2017.ibge.gov.br/templates/censo_agro/resultadosagro/index.html (accessed on 18 August 2024).
- Tegally, H.; Dellicour, S.; Poongavanan, J.; Mavian, C.; Dor, G.; Fonseca, V.; Tagliamonte, M.S.; Dunaiski, M.; Moir, M.; Wilkinson, E.; et al. Dynamics and Ecology of a Multi-Stage Expansion of Oropouche Virus in Brazil. medRxiv 2024. [Google Scholar] [CrossRef]
- Brugger, K.; Rubel, F. Characterizing the Species Composition of European Culicoides Vectors by Means of the Köppen-Geiger Climate Classification. Parasites Vectors 2013, 6, 333. [Google Scholar] [CrossRef] [PubMed]
- Benitez, A.J.; Alvarez, M.; Perez, L.; Gravier, R.; Serrano, S.; Hernandez, D.M.; Perez, M.M.; Gutierrez-Bugallo, G.; Martinez, Y.; Companioni, A.; et al. Oropouche Fever, Cuba, May 2024. Emerg. Infect. Dis. 2024, 30, 2155–2159. [Google Scholar] [CrossRef] [PubMed]
- Castilletti, C.; Mori, A.; Matucci, A.; Ronzoni, N.; Van Duffel, L.; Rossini, G.; Sponga, P.; D’Errico, M.L.; Rodari, P.; Cristini, F.; et al. Oropouche Fever Cases Diagnosed in Italy in Two Epidemiologically Non-Related Travellers from Cuba, Late May to Early June 2024. Eurosurveillance 2024, 29, 2400362. [Google Scholar] [CrossRef] [PubMed]
- Scachetti, G.C.; Forato, J.; Claro, I.M.; Hua, X.; Salgado, B.B.; Vieira, A.; Simeoni, C.L.; Barbosa, A.R.C.; Rosa, I.L.; de Souza, G.F.; et al. Reemergence of Oropouche Virus between 2023 and 2024 in Brazil. medRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, E.d.A.N.; da Silva, A.F.; da Silva, V.G.; Machado, L.C.; de Lima, G.B.; Ishigami, B.I.M.; e Silva, K.M.P.; Costa, M.M.d.O.M.d.; Falcão, D.A.; Vasconcelos, A.P.; et al. Genomic and Phenotypic Characterization of the Oropouche Virus Strain Implicated in the 2022–24 Large-scale Outbreak in Brazil. J. Med. Virol. 2024, 96, e70012. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.R.R.; Dutra, J.V.R.; de Carvalho, A.H.B.; Reis, C.R.; Rios, J.S.H.; Ribeiro, M. de O.; Arruda, M.B.; Alvarez, P.; Souza, R.P.; Voloch, C.; et al. Oropouche Virus Genomic Surveillance in Brazil. Lancet Infect. Dis. 2024, 24, e664–e666. [Google Scholar] [CrossRef] [PubMed]

| Region | Suspected Cases (September 2023 to July 2024) | Suspected Cases (March 2024 to July 2024) | Positive Cases |
|---|---|---|---|
| Coastal Lowland | 172 | 159 | 2 |
| Ilha Grande Bay | 77 | 75 | 4 |
| South Center | 165 | 156 | 2 |
| Metropolitan I | 878 | 804 | 14 |
| Metropolitan II | 240 | 233 | 3 |
| Middle Paraíba | 1049 | 448 | 76 |
| Northwest | 190 | 183 | 7 |
| North | 82 | 77 | 0 |
| Mountain | 301 | 293 | 9 |
| Other state | 56 | 47 | 1 |
| Total | 3210 | 2475 | 118 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Costa, F.B.; Cavalcanti, A.C.; Erbisti, R.S.; Dias, V.Z.; Moreira, C.G.d.C.; Grifo, M.M.; Vaz, M.C.d.S.; Camargo, A.C.; de Souza, L.M.; dos Santos, F.B.; et al. Introduction and Spatial–Temporal Distribution of Oropouche Virus in Rio de Janeiro State, Brazil. Pathogens 2025, 14, 833. https://doi.org/10.3390/pathogens14080833
da Costa FB, Cavalcanti AC, Erbisti RS, Dias VZ, Moreira CGdC, Grifo MM, Vaz MCdS, Camargo AC, de Souza LM, dos Santos FB, et al. Introduction and Spatial–Temporal Distribution of Oropouche Virus in Rio de Janeiro State, Brazil. Pathogens. 2025; 14(8):833. https://doi.org/10.3390/pathogens14080833
Chicago/Turabian Styleda Costa, Fábio Burack, Andrea Cony Cavalcanti, Rafael Santos Erbisti, Vanessa Zaquieu Dias, Cristiane Gomes de Castro Moreira, Mateus Marques Grifo, Maria Carmelita dos Santos Vaz, Adriana Cardoso Camargo, Leandro Magalhães de Souza, Flávia Barreto dos Santos, and et al. 2025. "Introduction and Spatial–Temporal Distribution of Oropouche Virus in Rio de Janeiro State, Brazil" Pathogens 14, no. 8: 833. https://doi.org/10.3390/pathogens14080833
APA Styleda Costa, F. B., Cavalcanti, A. C., Erbisti, R. S., Dias, V. Z., Moreira, C. G. d. C., Grifo, M. M., Vaz, M. C. d. S., Camargo, A. C., de Souza, L. M., dos Santos, F. B., Ribeiro, M. S., Malirat, V., Alves Honório, N., & Azevedo, R. C. (2025). Introduction and Spatial–Temporal Distribution of Oropouche Virus in Rio de Janeiro State, Brazil. Pathogens, 14(8), 833. https://doi.org/10.3390/pathogens14080833







