Genotype-Specific HPV mRNA Triage Improves CIN2+ Detection Efficiency Compared to Cytology: A Population-Based Study of HPV DNA-Positive Women
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.1.1. Primary HPV DNA Screening
2.1.2. Triage Procedures
2.2. Follow-Up and Outcome Ascertainment
2.3. Statistical Analysis
2.4. Ethical Approval
3. Results
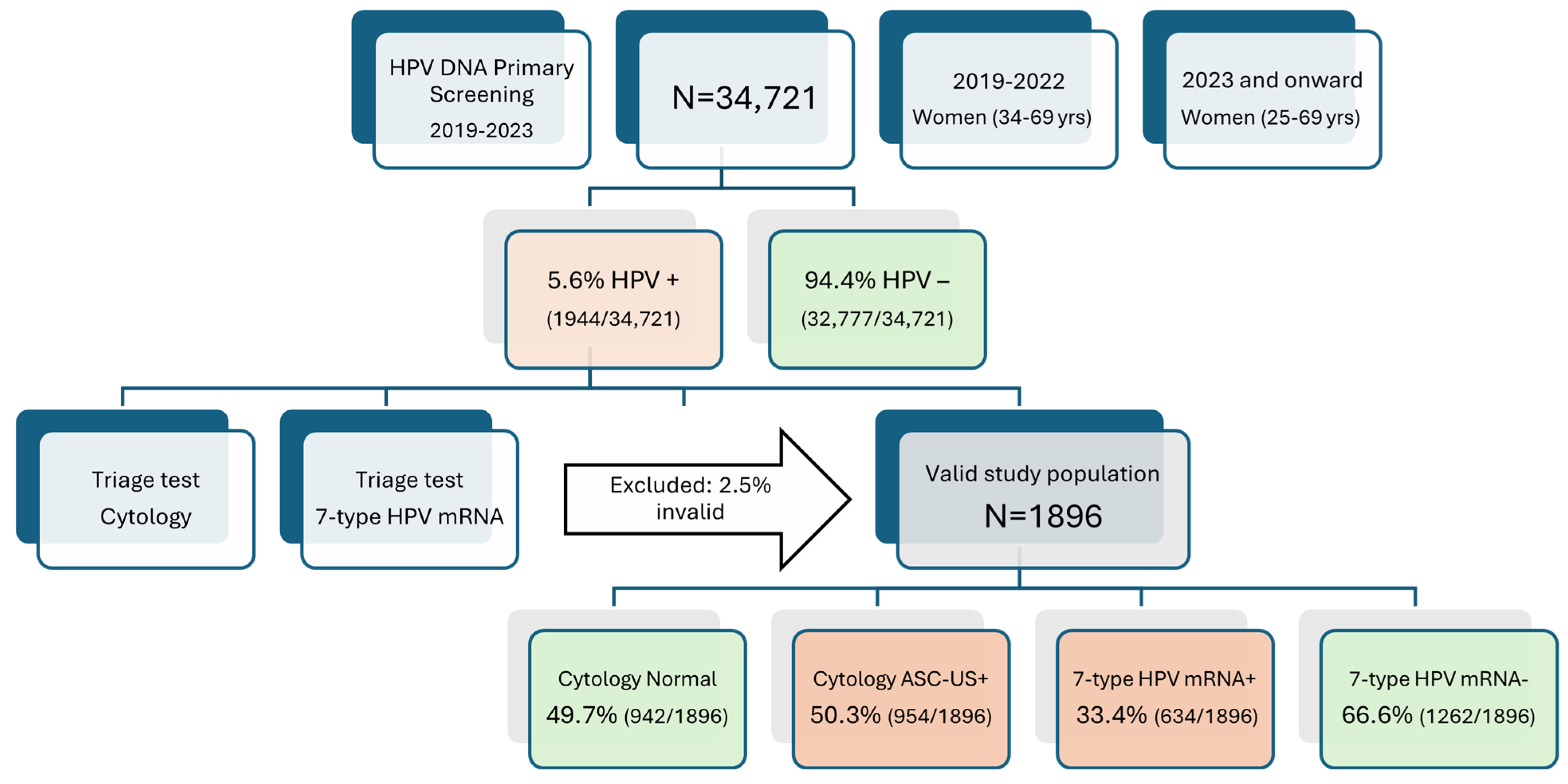
3.1. Primary Screening Outcomes and Triage Cohort Formation
3.2. Comparative Triage Positivity Rates and Impact on Referral Burden
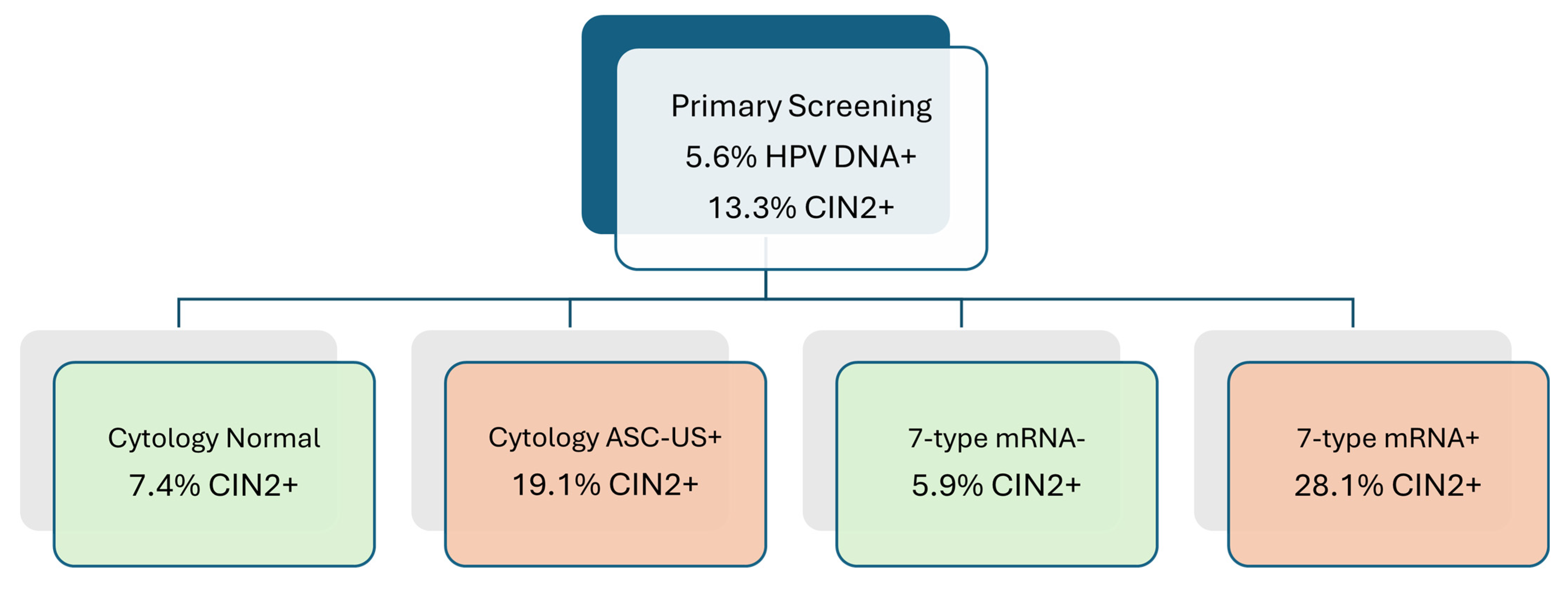
3.3. CIN2+ Detection Rates and Risk Stratification by Triage Modality
3.4. Comparative Diagnostic Accuracy of Cytology and HPV mRNA Triage for CIN2+ Detection
3.5. Colposcopy Efficiency: Procedures Required per CIN2+ Case Detected
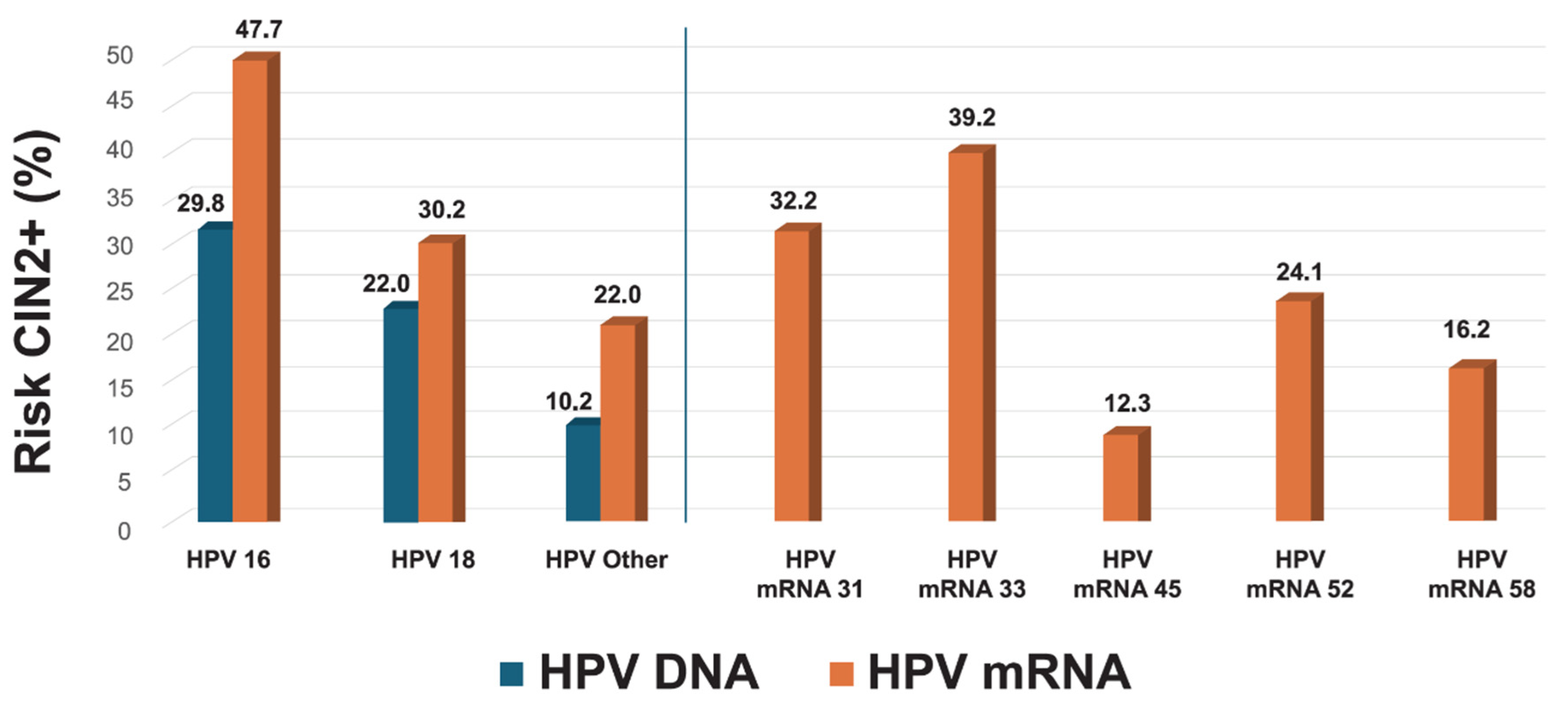
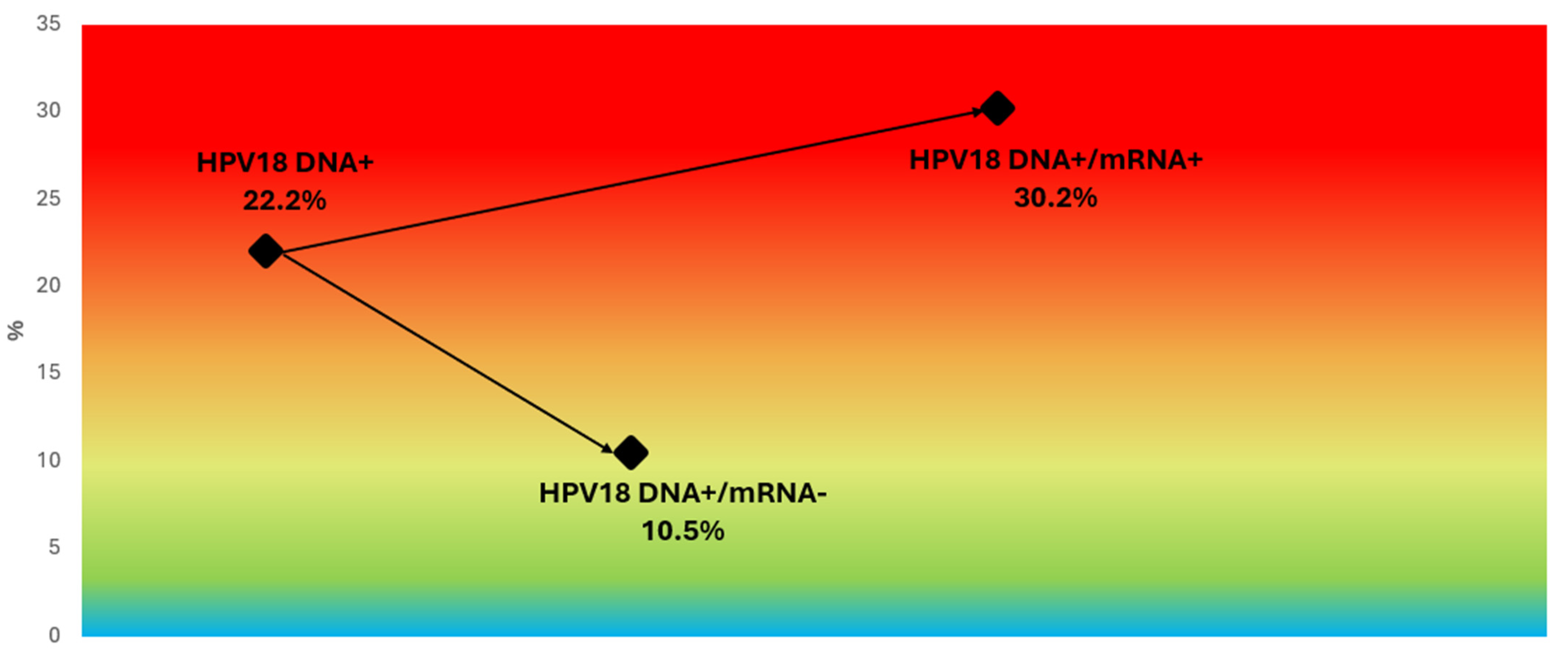
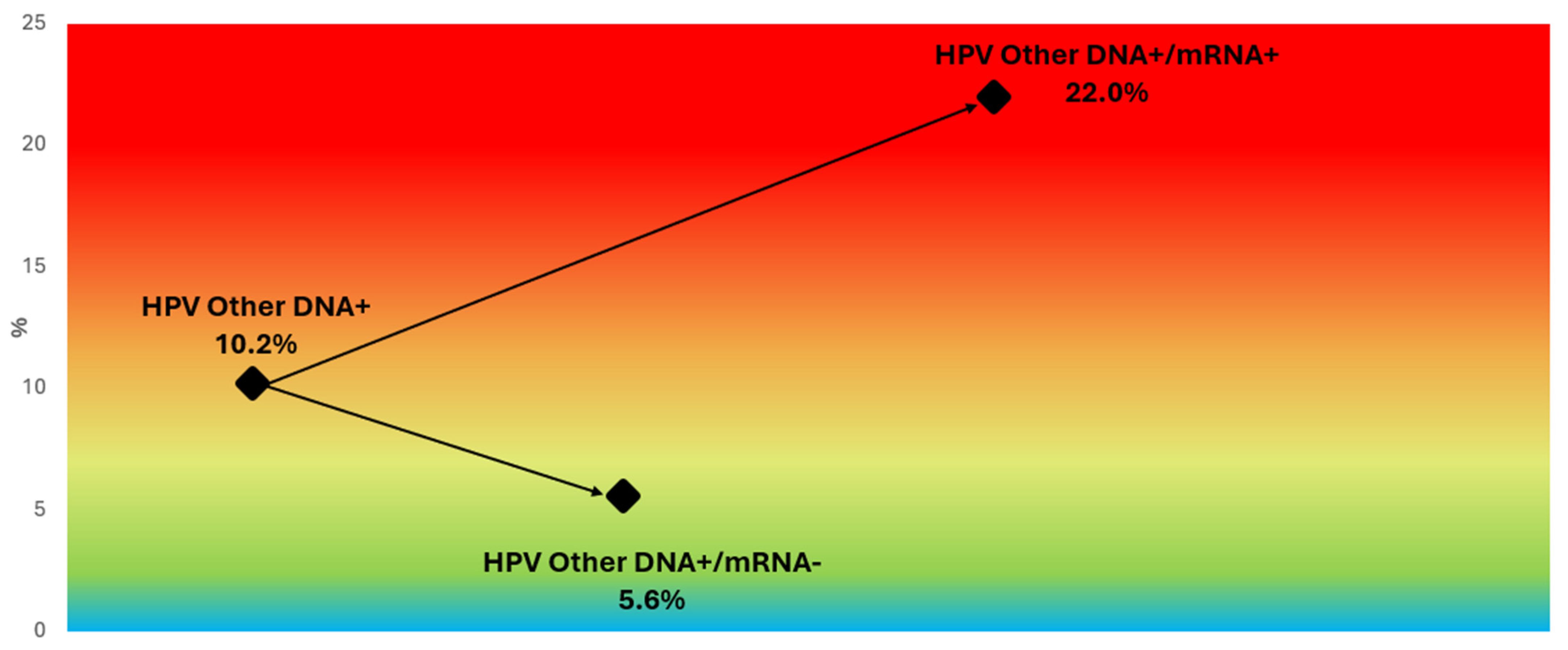
3.6. Genotype-Specific Predictive Values
3.7. Refining Risk Stratification in HPV16/18 DNA-Positive Women and Other Genotype Subgroups
4. Discussion
4.1. Clinical Utility of Genotype-Specific HPV mRNA Triage
4.2. Enhanced Stratification Within HPV DNA 16/18
4.3. Negative Predictive Value and Long-Term Safety of mRNA Triage
4.4. HPV mRNA Versus Other Molecular Triage Strategies
4.5. Colposcopy Efficiency and Health System Impact
4.6. Alignment with National Data on HPV Genotype Risk Stratification
4.7. Study Limitations and Need for Further Validation
4.8. Interpretation of Diagnostic Differences and Clinical Relevance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Castellsagué, X. Natural History and Epidemiology of HPV Infection and Cervical Cancer. Gynecol. Oncol. 2008, 110 (Suppl. S2), S4–S7. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, M.G.; Snijders, P.J.; Arbyn, M.; Rijkaart, D.C.; Berkhof, J.; Meijer, C.J. Cervical Cancer Screening: On the Way to a Shift from Cytology to Full Molecular Screening. Ann. Oncol. 2014, 25, 927–935. [Google Scholar] [CrossRef]
- Cuschieri, K.; Cubie, H.; Graham, C.; Rowan, J.; Hardie, A.; Horne, A.; Earle, C.B.; Bailey, A.; Crosbie, E.J.; Kitchener, H. Clinical Performance of RNA and DNA Based HPV Testing in a Colposcopy Setting: Influence of Assay Target, Cut-Off and Age. J. Clin. Virol. 2014, 59, 104–108. [Google Scholar] [CrossRef]
- Tjalma, W.A.A. Diagnostic Performance of Dual-Staining Cytology for Cervical Cancer Screening: A Systematic Literature Review. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017, 210, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Verhoef, L.; Bleeker, M.C.G.; Polman, N.; Steenbergen, R.D.M.; Meijer, C.J.L.M.; Melchers, W.J.G.; Bekkers, R.L.; Molijn, A.C.; Quint, W.G.; van Kemenade, F.J.; et al. Performance of DNA Methylation Analysis of ASCL1, LHX8, ST6GALNAC5, GHSR, ZIC1 and SST for the Triage of HPV-Positive Women: Results from a Dutch Primary HPV-Based Screening Cohort. Int. J. Cancer 2021, 149, 2136–2144. [Google Scholar] [CrossRef]
- Wentzensen, N.; Silver, M.I. Biomarkers for Cervical Cancer Prevention Programs: The Long and Winding Road From Discovery to Clinical Use. J. Low. Genit. Tract Dis. 2016, 20, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Cuschieri, K.; Ronco, G.; Lorincz, A.; Smith, L.; Ogilvie, G.; Mirabello, L.; Carozzi, F.; Cubie, H.; Wentzensen, N.; Snijders, P.; et al. Eurogin Roadmap 2017: Triage Strategies for the Management of HPV-Positive Women in Cervical Screening Programs. Int. J. Cancer 2018, 143, 735–745. [Google Scholar] [CrossRef]
- Cattani, P.; Zannoni, G.F.; Ricci, C.; D’Onghia, S.; Trivellizzi, I.N.; Di Franco, A.; Vellone, V.G.; Durante, M.; Fadda, G.; Scambia, G.; et al. Clinical Performance of Human Papillomavirus E6 and E7 mRNA Testing for High-Grade Lesions of the Cervix. J. Clin. Microbiol. 2009, 47, 3895–3901. [Google Scholar] [CrossRef]
- Derbie, A.; Mekonnen, D.; Woldeamanuel, Y.; Van Ostade, X.; Abebe, T. HPV E6/E7 mRNA Test for the Detection of High Grade Cervical Intraepithelial Neoplasia (CIN2+): A Systematic Review. Infect. Agents Cancer 2020, 15, 9. [Google Scholar] [CrossRef]
- Benevolo, M.; Vocaturo, A.; Caraceni, D.; French, D.; Rosini, S.; Zappacosta, R.; Terrenato, I.; Ciccocioppo, L.; Frega, A.; Giorgi Rossi, P. Sensitivity, Specificity, and Clinical Value of Human Papillomavirus (HPV) E6/E7 mRNA Assay as a Triage Test for Cervical Cytology and HPV DNA Test. J. Clin. Microbiol. 2011, 49, 2643–2650. [Google Scholar] [CrossRef]
- Origoni, M.; Cristoforoni, P.; Carminati, G.; Stefani, C.; Costa, S.; Sandri, M.T.; Mariani, L.; Preti, M. E6/E7 mRNA Testing for Human Papilloma Virus-Induced High-Grade Cervical Intraepithelial Disease (CIN2/CIN3): A Promising Perspective. Ecancermedicalscience 2015, 9, 533. [Google Scholar] [CrossRef]
- Frega, A.; Lorenzon, L.; Giovagnoli, M.R.; De Sanctis, L.; Fabiano, V.; Lukic, A.; Moscarini, M.; Torrisi, M.R.; French, D. Prognostic Implication of High-Risk Human Papillomavirus E6 and E7 mRNA in Patients with Intraepithelial Lesions of the Cervix in Relationship to Age. Int. J. Immunopathol. Pharmacol. 2011, 24, 461–470. [Google Scholar] [CrossRef]
- Ratnam, S.; Coutlee, F.; Fontaine, D.; Bentley, J.; Escott, N.; Ghatage, P.; Gadag, V.; Holloway, G.; Bartellas, E.; Kum, N.; et al. Clinical Performance of the PreTect HPV-Proofer E6/E7 mRNA Assay in Comparison with That of the Hybrid Capture 2 Test for Identification of Women at Risk of Cervical Cancer. J. Clin. Microbiol. 2010, 48, 2779–2785. [Google Scholar] [CrossRef]
- Sørbye, S.W.; Falang, B.M.; Antonsen, M. Performance of a 7-Type HPV mRNA Test in Triage of HPV DNA Primary Screen Positive Women Compared to Liquid-Based Cytology. J. Mol. Pathol. 2023, 4, 69–80. [Google Scholar] [CrossRef]
- Sørbye, S.W.; Fismen, S.; Gutteberg, T.; Mortensen, E.S. Triage of Women with Minor Cervical Lesions: Data Suggesting a “Test and Treat” Approach for HPV E6/E7 mRNA Testing. PLoS ONE 2010, 5, e12724. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Flores, C.E.A.; Falang, B.M.; Gómez-Laguna, L.; Gutiérrez, G.G.; León, J.M.O.; Uribe, M.; Cruz, O.; Sørbye, S.W. Enhancing Cervical Cancer Screening with 7-Type HPV mRNA E6/E7 Testing on Self-Collected Samples: Multicentric Insights from Mexico. Cancers 2024, 16, 2485. [Google Scholar] [CrossRef]
- Aranda Flores, C.E.; Gomez Gutierrez, G.; Ortiz Leon, J.M.; Cruz Rodriguez, D.; Sørbye, S.W. Self-collected versus clinician-collected cervical samples for the detection of HPV infections by 14-type DNA and 7-type mRNA tests. BMC Infect. Dis. 2021, 21, 504. [Google Scholar] [CrossRef] [PubMed]
- Reinholdt, K.; Juul, K.E.; Dehlendorff, C.; Munk, C.; Kjaer, S.K.; Thomsen, L.T. Triage of low-grade squamous intraepithelial lesions using human papillomavirus messenger ribonucleic acid tests—A prospective population-based register study. Acta Obstet. Gynecol. Scand. 2019, 98, 1374–1381. [Google Scholar] [CrossRef]
- Alaghehbandan, R.; Fontaine, D.; Bentley, J.; Escott, N.; Ghatage, P.; Lear, A.; Coutlee, F.; Ratnam, S. Performance of ProEx C and PreTect HPV-Proofer E6/E7 mRNA tests in comparison with the Hybrid Capture 2 HPV DNA test for triaging ASCUS and LSIL cytology. Diagn. Cytopathol. 2013, 41, 767–775. [Google Scholar] [CrossRef]
- Verdoodt, F.; Szarewski, A.; Halfon, P.; Cuschieri, K.; Arbyn, M. Triage of women with minor abnormal cervical cytology: Meta-analysis of the accuracy of an assay targeting messenger ribonucleic acid of 5 high-risk human papillomavirus types. Cancer Cytopathol. 2013, 121, 675–687. [Google Scholar] [CrossRef]
- Tainio, K.; Athanasiou, A.; Tikkinen, K.A.O.; Aaltonen, R.; Cárdenas Hernándes, J.; Glazer-Livson, S.; Jakobsson, M.; Joronen, K.; Kiviharju, M.; Louvanto, K.; et al. Clinical Course of Untreated Cervical Intraepithelial Neoplasia Grade 2 under Active Surveillance: Systematic Review and Meta-Analysis. BMJ 2018, 360, k499. [Google Scholar] [CrossRef] [PubMed]
- St-Martin, G.; Holst Thamsborg, L.; Andersen, B.; Christensen, J.; Ejersbo, D.; Jochumsen, K.; Lynge, E. Management of Low-Grade Cervical Cytology in Young Women: Cohort Study from Denmark. Acta Oncol. 2021, 60, 444–451. [Google Scholar] [CrossRef]
- Rad, A.; Sørbye, S.W.; Tiwari, S.; Løchen, M.-L.; Skjeldestad, F.E. Risk of Intraepithelial Neoplasia Grade 3 or Worse (CIN3+) among Women Examined by a 5-Type HPV mRNA Test during 2003 and 2004, Followed through 2015. Cancers 2023, 15, 3106. [Google Scholar] [CrossRef]
- Wentzensen, N.; Clarke, M.A.; Bremer, R.; Poitras, N.; Tokugawa, D.; Goldhoff, P.E.; Castle, P.E.; Schiffman, M.; Kingery, J.D.; Grewal, K.K.; et al. Clinical Evaluation of Human Papillomavirus Screening with p16/Ki-67 Dual Stain Triage in a Large Organized Cervical Cancer Screening Program. JAMA Intern. Med. 2019, 179, 881–888. [Google Scholar] [CrossRef]
- De Strooper, L.M.; Meijer, C.J.; Berkhof, J.; Hesselink, A.T.; Snijders, P.J.; Steenbergen, R.D.; Heideman, D.A. Methylation Analysis of the FAM19A4 Gene in Cervical Scrapes Is Highly Efficient in Detecting Cervical Carcinomas and Advanced CIN2/3 Lesions. Cancer Prev. Res. 2014, 7, 1251–1257. [Google Scholar] [CrossRef] [PubMed]
- Carcea, F.; Vavoulidis, E.; Petousis, S.; Papandreou, P.; Margioula Siarkou, C.; Nasioutziki, M.; Papanikolaou, A.; Dinas, K.; Daniilidis, A. Diagnostic Performance of HPV E6/E7 mRNA Testing Towards HPV-DNA Testing and p16/Ki67 Immunostaining as a Biomarker of High-Risk HPV Recurrence in Greek Women Surgically Treated for Their Cervical Lesions. J. Obstet. Gynaecol. Res. 2021, 47, 3082–3090. [Google Scholar] [CrossRef]
- Wright, T.C., Jr.; Behrens, C.M.; Ranger-Moore, J.; Rehm, S.; Sharma, A.; Stoler, M.H.; Ridder, R. Triaging HPV-Positive Women with p16/Ki-67 Dual-Stained Cytology: Results from a Sub-Study Nested into the ATHENA Trial. Gynecol. Oncol. 2017, 144, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Wentzensen, N.; Fetterman, B.; Castle, P.E.; Schiffman, M.; Wood, S.N.; Stiemerling, E.; Tokugawa, D.; Bodelon, C.; Poitras, N.; Lorey, T.; et al. p16/Ki-67 Dual Stain Cytology for Detection of Cervical Precancer in HPV-Positive Women. J. Natl. Cancer Inst. 2015, 107, djv257. [Google Scholar] [CrossRef]
- McMenamin, M.; McKenna, M.; McDowell, A. Clinical Utility of CINtec PLUS Triage in Equivocal Cervical Cytology and Human Papillomavirus Primary Screening. Am. J. Clin. Pathol. 2018, 150, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Øvestad, I.T.; Dalen, I.; Andersland, M.S.; Vintermyr, O.K.; Moltu, P.; Berland, J.M.; Janssen, E.A.M.; Haugland, H.K. Triaging HPV-Positive Cervical Samples with p16 and Ki-67 Dual Stained Cytology within an Organized Screening Program—A Prospective Observational Study from Western Norway. Int. J. Mol. Sci. 2023, 24, 7158. [Google Scholar] [CrossRef]
- Giorgi Rossi, P.; Carozzi, F.; Ronco, G.; Allia, E.; Bisanzi, S.; Gillio-Tos, A.; De Marco, L.; Rizzolo, R.; Gustinucci, D.; Del Mistro, A.; et al. New Technology for Cervical Cancer 2 Working Group. p16/ki67 and E6/E7 mRNA Accuracy and Prognostic Value in Triaging HPV DNA-Positive Women. J. Natl. Cancer Inst. 2021, 113, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Hashim, D.; Engesæter, B.; Baadstrand Skare, G.; Castle, P.E.; Bjørge, T.; Tropé, A.; Nygård, M. Real-World Data on Cervical Cancer Risk Stratification by Cytology and HPV Genotype to Inform the Management of HPV-Positive Women in Routine Cervical Screening. Br. J. Cancer 2020, 122, 1715–1723. [Google Scholar] [CrossRef] [PubMed]
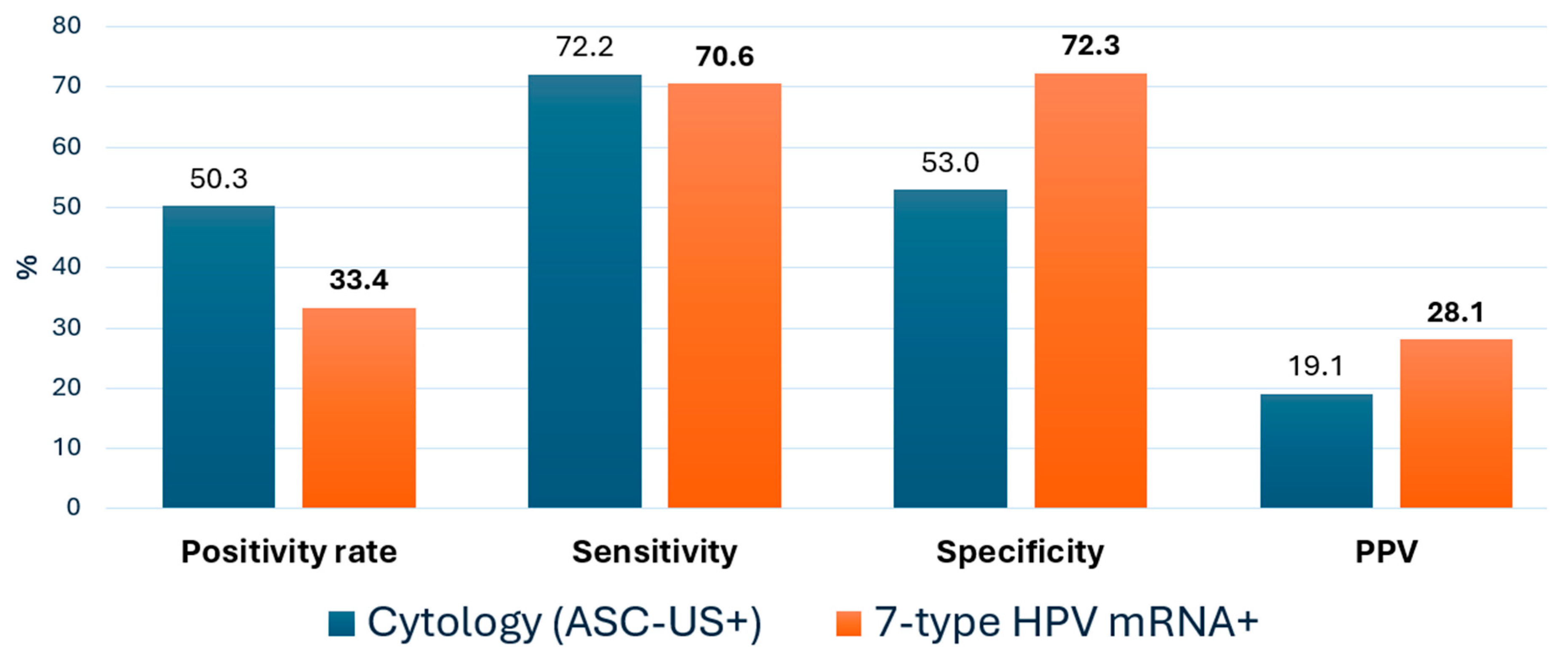

| Metric Value | Cytology ≥ ASC-US % (95% CI) | 7-Type HPV mRNA% (95% CI) | Δ (mRNA-Cyt) pp * |
|---|---|---|---|
| Sensitivity | 72.2 (66.3–77.5) | 70.6 (64.6–75.9) | −1.6 |
| Specificity | 53.0 (50.3–55.7) | 72.3 (69.8–74.7) | +19.3 |
| PPV | 19.1 (16.4–22.0) | 28.1 (24.9–31.6) | +9.0 |
| NPV | 93.0 (91.1–94.6) | 94.6 (92.8–96.0) | +1.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sørbye, S.; Falang, B.M.; Antonsen, M.; Mortensen, E. Genotype-Specific HPV mRNA Triage Improves CIN2+ Detection Efficiency Compared to Cytology: A Population-Based Study of HPV DNA-Positive Women. Pathogens 2025, 14, 749. https://doi.org/10.3390/pathogens14080749
Sørbye S, Falang BM, Antonsen M, Mortensen E. Genotype-Specific HPV mRNA Triage Improves CIN2+ Detection Efficiency Compared to Cytology: A Population-Based Study of HPV DNA-Positive Women. Pathogens. 2025; 14(8):749. https://doi.org/10.3390/pathogens14080749
Chicago/Turabian StyleSørbye, S., B. M. Falang, M. Antonsen, and E. Mortensen. 2025. "Genotype-Specific HPV mRNA Triage Improves CIN2+ Detection Efficiency Compared to Cytology: A Population-Based Study of HPV DNA-Positive Women" Pathogens 14, no. 8: 749. https://doi.org/10.3390/pathogens14080749
APA StyleSørbye, S., Falang, B. M., Antonsen, M., & Mortensen, E. (2025). Genotype-Specific HPV mRNA Triage Improves CIN2+ Detection Efficiency Compared to Cytology: A Population-Based Study of HPV DNA-Positive Women. Pathogens, 14(8), 749. https://doi.org/10.3390/pathogens14080749








