A Look Under the Carpet of a Successful Eradication Campaign Against Small Ruminant Lentiviruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Diagnostic Tests
2.2.1. Serology
2.2.2. Real-Time PCR
2.3. Clinical Examination
2.4. Interviews and Questionnaire
3. Results
3.1. Analysis of Historical Laboratory Data
3.2. Serology and Real-Time PCR
3.3. Clinical Examination
3.4. Animal Movements and Potential Transmission Routes
4. Discussion
5. Conclusions and Recommendations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SRLV | small ruminant lentiviruses |
| CAEV | caprine arthritis encephalitis virus |
| MVV | Maedi-visna virus |
| qPCR | nested real-time PCR |
References
- Leroux, C.; Chastang, J.; Greenland, T.; Mornex, J.F. Genomic heterogeneity of small ruminant lentiviruses: Existence of heterogeneous populations in sheep and of the same lentiviral genotypes in sheep and goats. Arch. Virol. 1997, 142, 1125–1137. [Google Scholar] [CrossRef]
- Leroux, C.; Cruz, J.C.; Mornex, J.F. Srlvs: A genetic continuum of lentiviral species in sheep and goats with cumulative evidence of cross species transmission. Curr. HIV Res. 2010, 8, 94–100. [Google Scholar] [CrossRef]
- Olech, M.; Kuzmak, J. Molecular characterization of small ruminant lentiviruses in polish mixed flocks supports evidence of cross species transmission, dual infection, a recombination event, and reveals the existence of new subtypes within group a. Viruses 2021, 13, 2529. [Google Scholar] [CrossRef] [PubMed]
- Grego, E.; Bertolotti, L.; Quasso, A.; Profiti, M.; Lacerenza, D.; Muz, D.; Rosati, S. Genetic characterization of small ruminant lentivirus in italian mixed flocks: Evidence for a novel genotype circulating in a local goat population. J. Gen. Virol. 2007, 88, 3423–3427. [Google Scholar] [CrossRef] [PubMed]
- Bouzalas, I.; Apostolidi, E.D.; Scalas, D.; Davidopoulou, E.; Chassalevris, T.; Rosati, S.; Colitti, B. A combined approach for the characterization of small ruminant lentivirus strains circulating in the islands and mainland of greece. Animals 2024, 14, 1119. [Google Scholar] [CrossRef] [PubMed]
- De Martin, E.; Golomingi, A.; Zahno, M.; Cachim, J.; Di Labio, E.; Perler, L.; Abril, C.; Zanoni, R.; Bertoni, G. Diagnostic response to a cross-border challenge for the swiss caprine arthritis encephalitis virus eradication program. Schweiz. Arch. Tierheilkd. 2019, 161, 93–104. [Google Scholar] [CrossRef]
- Crawford, T.B.; Adams, D.S.; Cheevers, W.P.; Cork, L.C. Chronic arthritis in goats caused by a retrovirus. Science 1980, 207, 997–999. [Google Scholar] [CrossRef]
- Thomann, B.; Falzon, L.C.; Bertoni, G.; Vogt, H.R.; Schupbach-Regula, G.; Magouras, I. A census to determine the prevalence and risk factors for caprine arthritis-encephalitis virus and visna/maedi virus in the swiss goat population. Prev. Vet. Med. 2017, 137, 52–58. [Google Scholar] [CrossRef]
- Deubelbeiss, M.; Blatti-Cardinaux, L.; Zahno, M.L.; Zanoni, R.; Vogt, H.R.; Posthaus, H.; Bertoni, G. Characterization of small ruminant lentivirus a4 subtype isolates and assessment of their pathogenic potential in naturally infected goats. Virol. J. 2014, 11, 65–75. [Google Scholar] [CrossRef]
- Cardinaux, L.; Zahno, M.L.; Deubelbeiss, M.; Zanoni, R.; Vogt, H.R.; Bertoni, G. Virological and phylogenetic characterization of attenuated small ruminant lentivirus isolates eluding efficient serological detection. Vet. Microbiol 2013, 162, 572–581. [Google Scholar] [CrossRef]
- Swiss Federal Food Safety and Veterinary Office (FSVO): 2015–2022. «Tiergesundheitsstatistik». Available online: https://www.blv.admin.ch/blv/de/home/tiere/publikationen/statistiken-berichte-tiere.html (accessed on 28 April 2023).
- Schaer, J.; Cvetnic, Z.; Sukalic, T.; Dorig, S.; Grisiger, M.; Iscaro, C.; Feliziani, F.; Pfeifer, F.; Origgi, F.; Zanoni, R.G.; et al. Evaluation of serological methods and a new real-time nested pcr for small ruminant lentiviruses. Pathogens 2022, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, G.; Cardinaux, L.; Deubelbeiss, M.; Zahno, M.-L.; Vogt, H.-R. Su5 serology as a novel tool to support a challenging caprine arthritis encephalitis virus (caev) eradication campaign. In Lbh: 7. Leipziger Tierärztekongress; Rackwitz, R., Pees, M., Aschenbach, J.R., Gäbel, G., Eds.; University of Leipzig: Leipzig, Germany, 2014; pp. 229–232. [Google Scholar]
- Mordasini, F.; Vogt, H.R.; Zahno, M.L.; Maeschli, A.; Nenci, C.; Zanoni, R.; Peterhans, E.; Bertoni, G. Analysis of the antibody response to an immunodominant epitope of the envelope glycoprotein of a lentivirus and its diagnostic potential. J. Clin. Microbiol. 2006, 44, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Brülisauer, F.; Vogt, H.-R.; Perler, L.; Rüfenacht, J. Risk factors for the infection of swiss goat herds with small ruminant lentivirus: A case-control study. Vet. Rec. 2005, 157, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Kaba, J.; Czopowicz, M.; Witkowski, L.; Szalus-Jordanow, O.; Mickiewicz, M.; Markowska-Daniel, I.; Puchala, R.; Bagnicka, E. Longitudinal study on seroreactivity of goats exposed to colostrum and milk of small ruminant lentivirus-infected dams. J. Vet. Res. 2022, 66, 511–521. [Google Scholar] [CrossRef]
- Pisoni, G.; Bertoni, G.; Manarolla, G.; Vogt, H.R.; Scaccabarozzi, L.; Locatelli, C.; Moroni, P. Genetic analysis of small ruminant lentiviruses following lactogenic transmission. Virology 2010, 407, 91–99. [Google Scholar] [CrossRef]
- Illius, A.W.; Savill, N.J. Maternal transmission of small ruminant lentivirus has no epidemiological importance. Prev. Vet. Med. 2024, 230, 106297. [Google Scholar] [CrossRef]
- Clawson, M.L.; Redden, R.; Schuller, G.; Heaton, M.P.; Workman, A.; Chitko-McKown, C.G.; Smith, T.P.; Leymaster, K.A. Genetic subgroup of small ruminant lentiviruses that infects sheep homozygous for tmem154 frameshift deletion mutation a4delta53. Vet. Res. 2015, 46, 22. [Google Scholar] [CrossRef]
- L’Homme, Y.; Ouardani, M.; Levesque, V.; Bertoni, G.; Simard, C.; Pisoni, G. Molecular characterization and phylogenetic analysis of small ruminant lentiviruses isolated from canadian sheep and goats. Virol. J. 2011, 8, 271. [Google Scholar] [CrossRef]
- Zahno, M.L.; Bertoni, G. An immunodominant region of the envelope glycoprotein of small ruminant lentiviruses may function as decoy antigen. Viruses 2018, 10, 231. [Google Scholar] [CrossRef]
- Herrmann-Hoesing, L.M.; Palmer, G.H.; Knowles, D.P. Evidence of proviral clearance following postpartum transmission of an ovine lentivirus. Virology 2007, 362, 226–234. [Google Scholar] [CrossRef]
- Bertolotti, L.; Reina, R.; Mazzei, M.; Preziuso, S.; Camero, M.; Carrozza, M.L.; Cavalli, A.; Juganaru, M.; Profiti, M.; De, M.D.; et al. Small ruminant lentivirus genotype b and e interaction: Evidences on the role of roccaverano strain on reducing proviral load of the challenging caev strain. Vet. Microbiol. 2012, 163, 33–41. [Google Scholar] [CrossRef]
- Venturino, E.; Collino, S.; Ferreri, L.; Bertolotti, L.; Rosati, S.; Giacobini, M. An effective management strategy for the control of two lentiviruses in goat breedings. J. Theor. Biol. 2019, 469, 96–106. [Google Scholar] [CrossRef]
- Brotto Rebuli, K.; Giacobini, M.; Bertolotti, L. Caprine arthritis encephalitis virus disease modelling review. Animals 2021, 11, 1457. [Google Scholar] [CrossRef]
- Samoilenko, M.; Nedosekov, V.; Bertoni, G. Testing the tenacity of small ruminant lentiviruses in vitro to assess the potential risk of indirect fomites’ transmission. Viruses 2025, 17, 419. [Google Scholar] [CrossRef] [PubMed]
- Olech, M.; Osinski, Z.; Kuzmak, J. Seroprevalence of small ruminant lentivirus (srlv) infection in wild cervids in poland. Prev. Vet. Med. 2020, 176, 104905. [Google Scholar] [CrossRef] [PubMed]
- Patton, K.M.; Bildfell, R.J.; Anderson, M.L.; Cebra, C.K.; Valentine, B.A. Fatal caprine arthritis encephalitis virus-like infection in 4 rocky mountain goats (Oreamnos americanus). J. Vet. Diagn. Investig. 2012, 24, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Erhouma, E.; Guiguen, F.; Chebloune, Y.; Gauthier, D.; Lakhal, L.M.; Greenland, T.; Mornex, J.F.; Leroux, C.; Alogninouwa, T. Small ruminant lentivirus proviral sequences from wild ibexes in contact with domestic goats. J. Gen. Virol. 2008, 89, 1478–1484. [Google Scholar] [CrossRef]
- Molaee, V.; Bazzucchi, M.; De Mia, G.M.; Otarod, V.; Abdollahi, D.; Rosati, S.; Luhken, G. Phylogenetic analysis of small ruminant lentiviruses in germany and iran suggests their expansion with domestic sheep. Sci. Rep. 2020, 10, 2243. [Google Scholar] [CrossRef]
- Shah, C.; Huder, J.B.; Boni, J.; Schonmann, M.; Muhlherr, J.; Lutz, H.; Schupbach, J. Direct evidence for natural transmission of small-ruminant lentiviruses of subtype a4 from goats to sheep and vice versa. J. Virol. 2004, 78, 7518–7522. [Google Scholar] [CrossRef]
- Alvarez, V.; Arranz, J.; Daltabuit-Test, M.; Leginagoikoa, I.; Juste, R.A.; Amorena, B.; de Andres, D.; Lujan, L.L.; Badiola, J.J.; Berriatua, E. Relative contribution of colostrum from maedi-visna virus (mvv) infected ewes to mvv-seroprevalence in lambs. Res. Vet. Sci. 2005, 78, 237–243. [Google Scholar] [CrossRef]
- De Boer, G.F.; Terpstra, C.; Houwers, D.J.; Hendriks, J. Studies in epidemiology of maedi/visna in sheep. Res. Vet. Sci. 1979, 26, 202–208. [Google Scholar] [CrossRef]
- Minguijon, E.; Reina, R.; Perez, M.; Polledo, L.; Villoria, M.; Ramirez, H.; Leginagoikoa, I.; Badiola, J.J.; Garcia-Marin, J.F.; de Andres, D.; et al. Small ruminant lentivirus infections and diseases. Vet. Microbiol. 2015, 181, 75–89. [Google Scholar] [CrossRef]
- Olech, M. The genetic variability of small-ruminant lentiviruses and its impact on tropism, the development of diagnostic tests and vaccines and the effectiveness of control programmes. J. Vet. Res. 2023, 67, 479–502. [Google Scholar] [CrossRef]
- Leginagoikoa, I.; Juste, R.A.; Barandika, J.; Amorena, B.; de Andres, D.; Lujan, L.; Badiola, J.; Berriatua, E. Extensive rearing hinders maedi-visna virus (mvv) infection in sheep. Vet. Res. 2006, 37, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Gjerset, B.; Jonassen, C.M.; Rimstad, E. Natural transmission and comparative analysis of small ruminant lentiviruses in the norwegian sheep and goat populations. Virus Res. 2007, 125, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Pisoni, G.; Quasso, A.; Moroni, P. Phylogenetic analysis of small-ruminant lentivirus subtype b1 in mixed flocks: Evidence for natural transmission from goats to sheep. Virology 2005, 339, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Larruskain, A.; Jugo, B.M. Retroviral infections in sheep and goats: Small ruminant lentiviruses and host interaction. Viruses 2013, 5, 2043–2061. [Google Scholar] [CrossRef]
- Heaton, M.P.; Clawson, M.L.; Chitko-McKown, C.G.; Leymaster, K.A.; Smith, T.P.; Harhay, G.P.; White, S.N.; Herrmann-Hoesing, L.M.; Mousel, M.R.; Lewis, G.S.; et al. Reduced lentivirus susceptibility in sheep with tmem154 mutations. PLoS Genet. 2012, 8, e1002467. [Google Scholar] [CrossRef]
- Rachid, A.; Croise, B.; Russo, P.; Vignoni, M.; Lacerenza, D.; Rosati, S.; Kuzmak, J.; Valas, S. Diverse host-virus interactions following caprine arthritis-encephalitis virus infection in sheep and goats. J. Gen. Virol. 2013, 94, 634–642. [Google Scholar] [CrossRef]
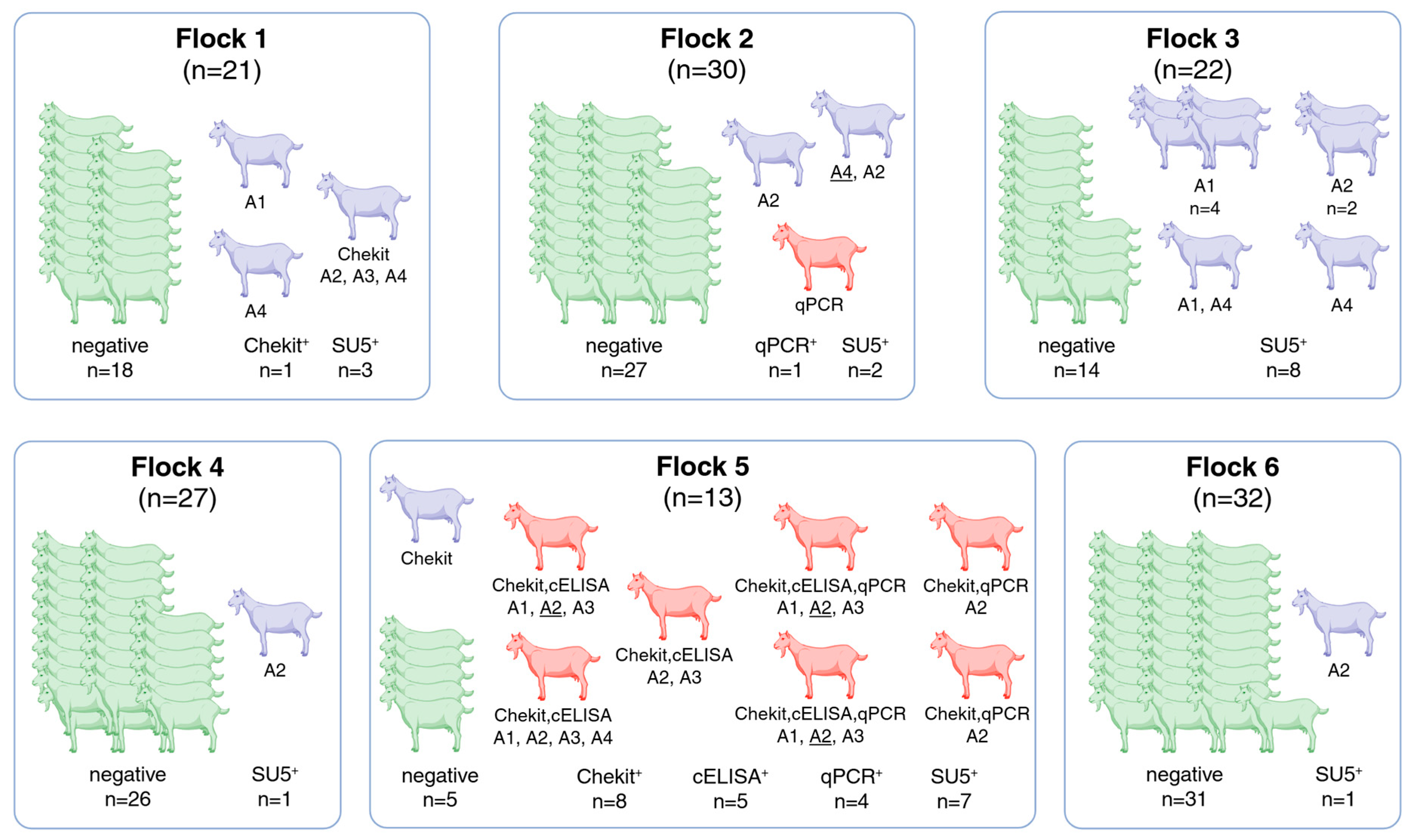
| Flock 1 | Flock 2 | Flock 3 | Flock 4 | Flock 5 | Flock 6 | Total | |
|---|---|---|---|---|---|---|---|
| No. goats | 21 | 30 | 22 | 27 | 13 | 32 | 145 |
| Tested negative | 18 | 27 | 14 | 26 | 5 | 31 | 122 (84%) |
| SRLV-positive by reference lab definition (red in Figure 1) | 0 | 1 | 0 | 0 | 7 | 0 | 8 (5.5%) |
| SRLV-positive due to positive test results, but officially SRLV negative (yellow in Figure 1) | 3 | 2 | 8 | 1 | 1 | 1 | 15 (10.3%) |
| Chekit | 1 | 7 | 8 (5.5%) | ||||
| cELISA | 5 | 5 (3.4%) | |||||
| qPCR | 1 | 4 | 4 (2.8%) | ||||
| SU5 A1 | 1 | 5 | 4 | 10 (6.9%) | |||
| SU5 A2 | 1 | 2 | 2 | 1 | 7 | 1 | 13 (9.0%) |
| SU5 A3 | 1 | 5 | 6 (4.1%) | ||||
| SU5 A4 | 2 | 1 | 2 | 1 | 6 (4.1%) |
| Flock 1 | Flock 2 | Flock 3 | Flock 4 | Flock 5 | Flock 6 | |
|---|---|---|---|---|---|---|
| No. goats | 21 | 30 | 22 | 27 | 13 | 32 |
| No. females | 20 | 21 | 21 | 26 | 13 | 32 |
| No. males | 1 | 9 | 1 | 1 | 0 | 0 |
| Production type | dairy | suckler | dairy | dairy | pasture care | dairy |
| Member SZZV * | yes | no | no | yes | yes | yes |
| Contact with sheep or goats from other flocks | yes (sheep sporadic) | no | no | no | Yes (>300 sheep) | unknown |
| Buy-in of animals | yes | yes | no | yes | unknown | yes |
| SRLV-positive animals or offsprings remained in herd | yes | no | yes | yes (offspring) | yes | unknown |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vincenz, F.; Samoilenko, M.; Abril, C.E.; Zanolari, P.; Bertoni, G.; Thomann, B. A Look Under the Carpet of a Successful Eradication Campaign Against Small Ruminant Lentiviruses. Pathogens 2025, 14, 719. https://doi.org/10.3390/pathogens14070719
Vincenz F, Samoilenko M, Abril CE, Zanolari P, Bertoni G, Thomann B. A Look Under the Carpet of a Successful Eradication Campaign Against Small Ruminant Lentiviruses. Pathogens. 2025; 14(7):719. https://doi.org/10.3390/pathogens14070719
Chicago/Turabian StyleVincenz, Fadri, Maksym Samoilenko, Carlos Eduardo Abril, Patrik Zanolari, Giuseppe Bertoni, and Beat Thomann. 2025. "A Look Under the Carpet of a Successful Eradication Campaign Against Small Ruminant Lentiviruses" Pathogens 14, no. 7: 719. https://doi.org/10.3390/pathogens14070719
APA StyleVincenz, F., Samoilenko, M., Abril, C. E., Zanolari, P., Bertoni, G., & Thomann, B. (2025). A Look Under the Carpet of a Successful Eradication Campaign Against Small Ruminant Lentiviruses. Pathogens, 14(7), 719. https://doi.org/10.3390/pathogens14070719







