Emergence of Dengue Virus Serotypes 1 and 3 in Mahottari and Adjacent Areas of Southern Nepal
Abstract
1. Introduction
2. Materials and Methods
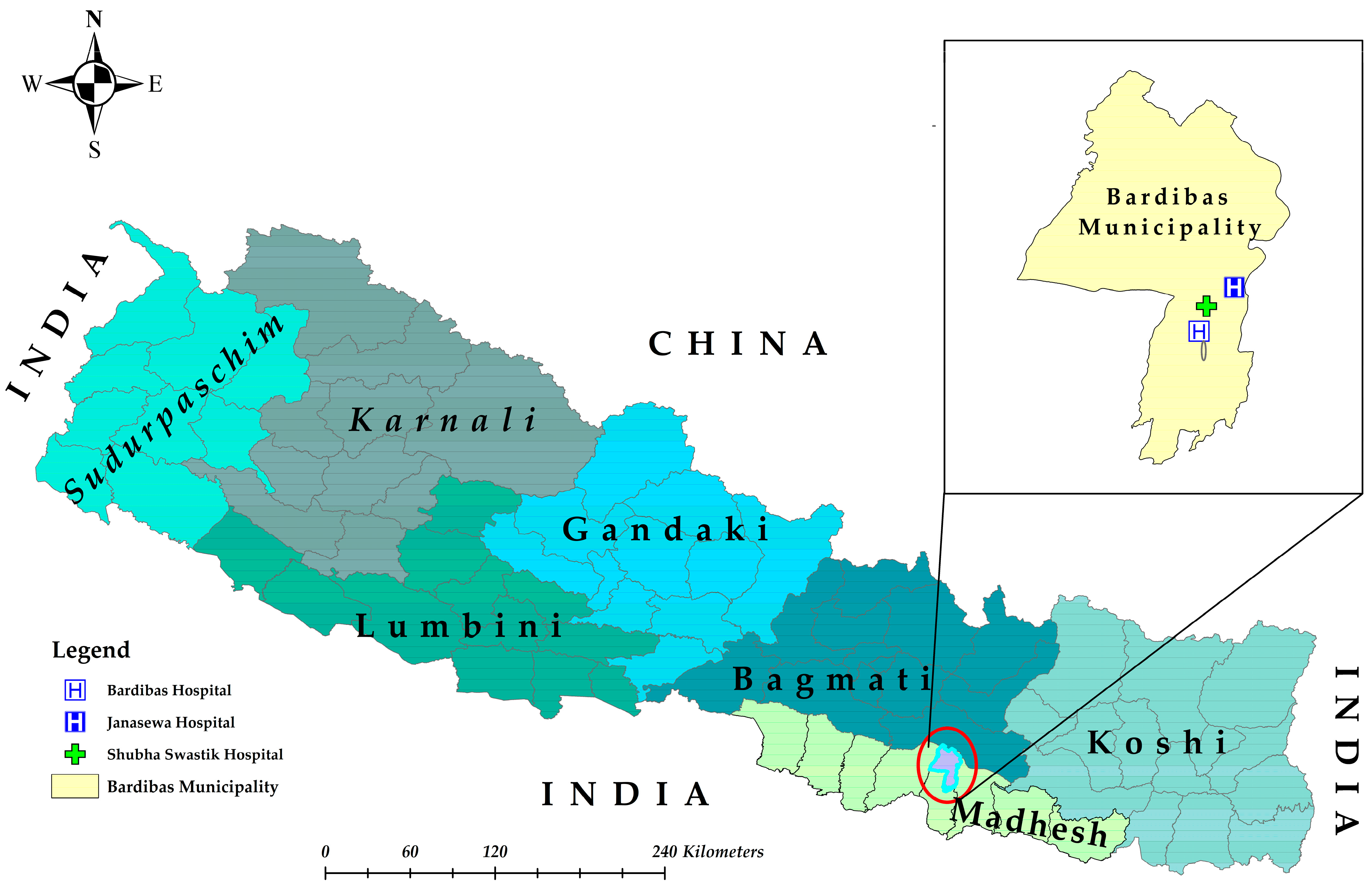
2.1. Study Design and Sites
2.2. Patient Enrollment, Questionnaire, and Sample Collection
2.3. Dengue Diagnosis
2.4. Molecular Investigation
2.4.1. Viral RNA Extraction and Quality and Quantity Assessment
2.4.2. One-Step Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.4.3. Quantification of DENV Genome Levels
2.5. Data Analysis
3. Results
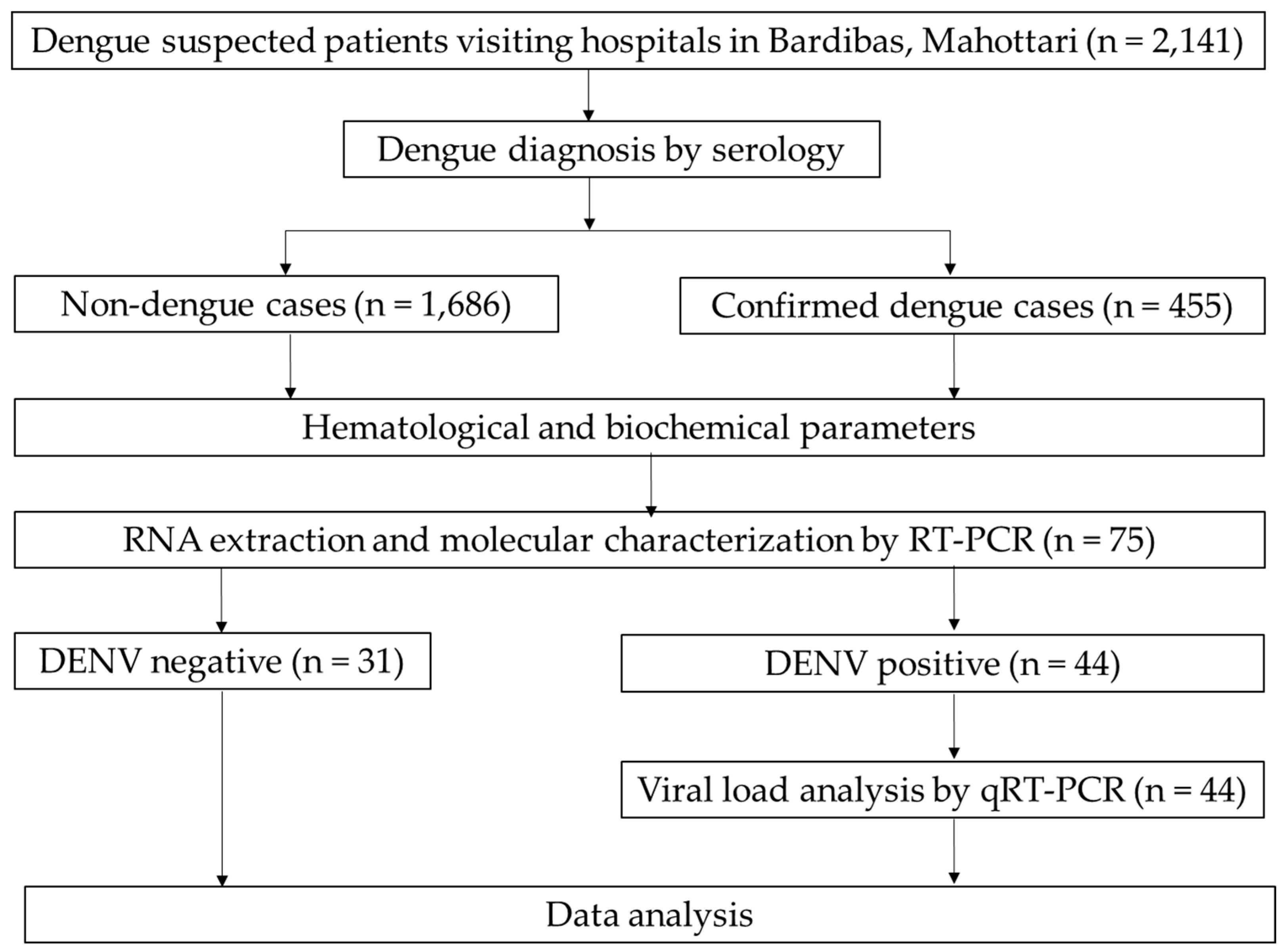
3.1. General Profiles of the Study Participants
3.2. Seasonal Trends of Dengue Cases in Bardibas, Mahottari
3.3. Blood Parameters of Dengue and Non-Dengue Patients
3.4. Blood Parameters of Dengue Patients by Admission Status
3.5. Blood Parameters of Dengue Patients in Primary and Secondary Infection
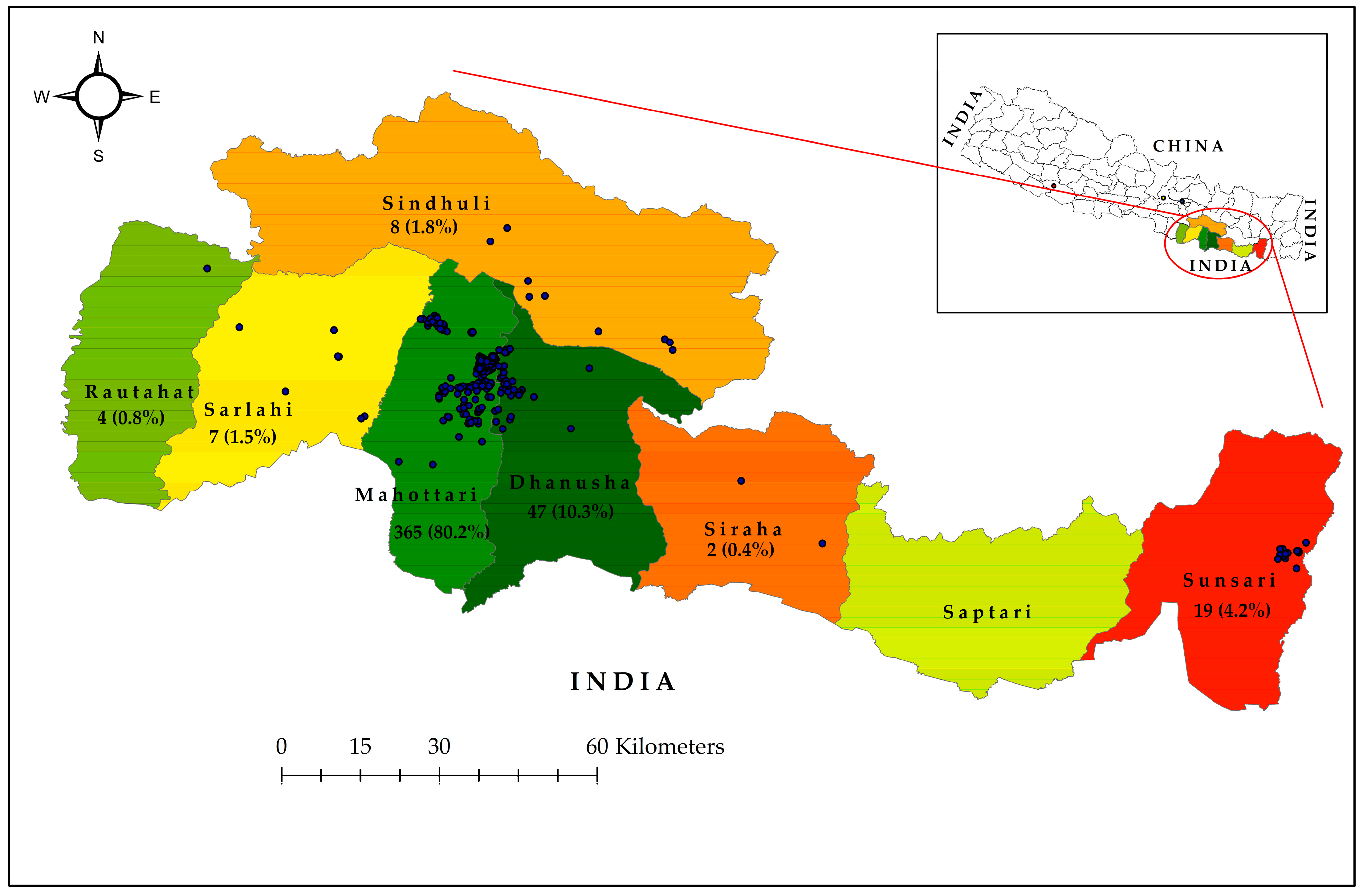
3.6. DENV Serotypes 1, 2, and 3 Detected During 2023 Dengue Outbreak in Southern Nepal
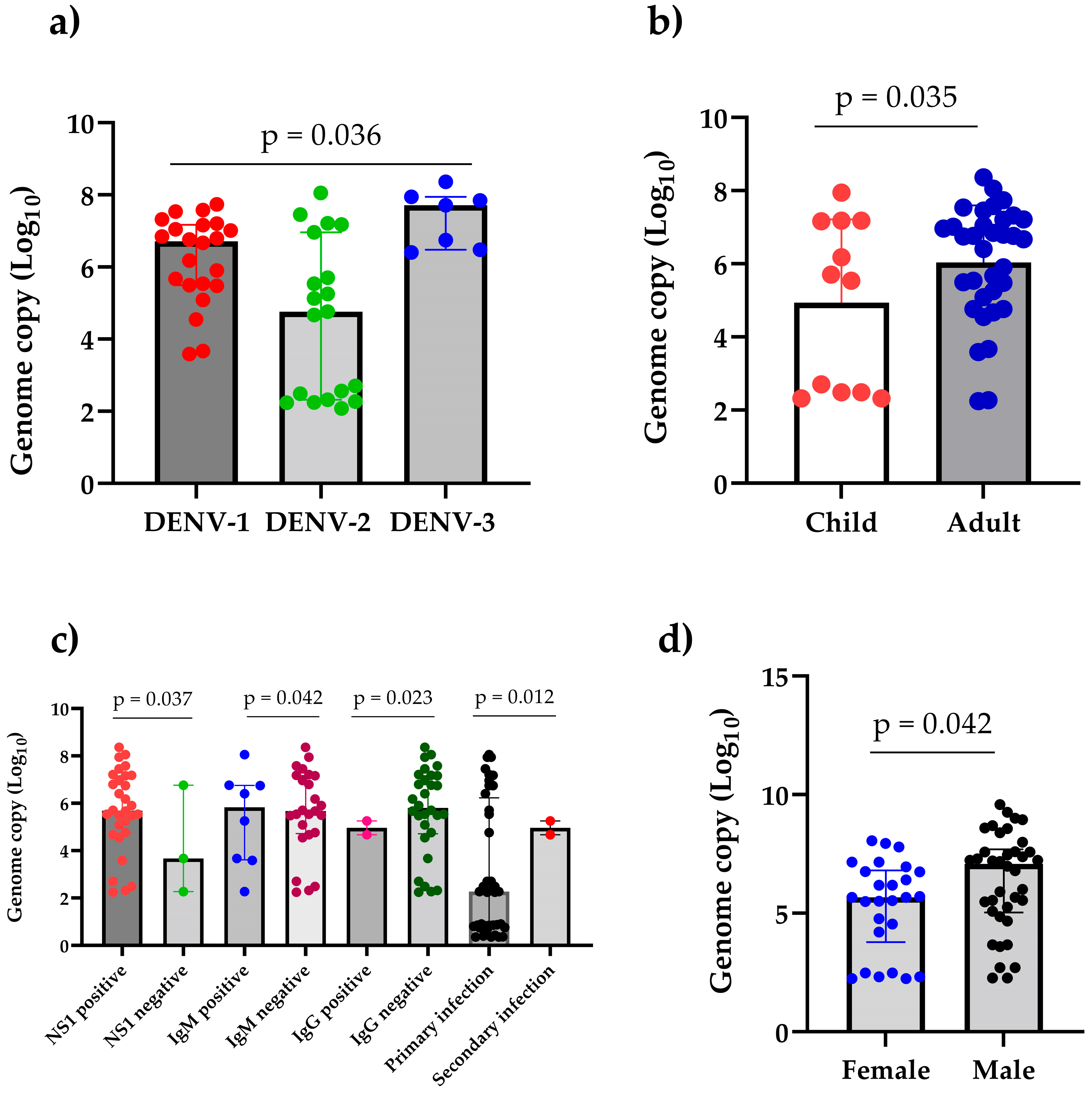
3.7. High Viral Load Detected in 2023 Dengue Outbreak in Mahottari and Surrounding Areas of Nepal
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khetarpal, N.; Khanna, I. Dengue Fever: Causes, Complications, and Vaccine Strategies. J. Immunol. Res. 2016, 2016, 6803098. [Google Scholar] [CrossRef] [PubMed]
- Kularatne, S.A.; Dalugama, C. Dengue Infection: Global Importance, Immunopathology and Management. Clin. Med. 2022, 22, 9–13. [Google Scholar] [CrossRef]
- WHO. Dengue—Global Situation. Disease Outbreak News. 2023. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON498 (accessed on 22 February 2025).
- Pierson, T.C.; Diamond, M.S. The Continued Threat of Emerging Flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef]
- Postler, T.S.; Beer, M.; Blitvich, B.J.; Bukh, J.; De Lamballerie, X.; Drexler, J.F.; Imrie, A.; Kapoor, A.; Karganova, G.G.; Lemey, P.; et al. Renaming of the Genus Flavivirus to Orthoflavivirus and Extension of Binomial Species Names within the Family Flaviviridae. Arch. Virol. 2023, 168, 224. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV) Approved Proposals. 2023. Available online: https://ictv.global/files/proposals/approved (accessed on 22 June 2025).
- WHO. Dengue. Guidelines for Diagnosis, Treatment, Prevention and Control; WHO: Geneva, Switzerland, 2009; pp. 10–12. [Google Scholar]
- ECDC. Dengue Cases January–December 2023; European Center for Disease Prevention and Control: Solna, Sweden, 2024; Available online: https://www.ecdc.europa.eu/en/publications-data/dengue-cases-january-december-2023 (accessed on 13 March 2025).
- ECDC. Twelve-Month Dengue Virus Disease Case Notification Rate per 100,000 Population, January–December 2024; European Center for Disease Prevention and Control: Solna, Sweden, 2025; Available online: www.ecdc.europa.eu/en/publications-data/twelve-month-dengue-virus-disease-case-notification-rate-100-000-population (accessed on 13 March 2025).
- Cattarino, L.; Rodriguez-Barraquer, I.; Imai, N.; Cummings, D.A.T.; Ferguson, N.M. Mapping Global Variation in Dengue Transmission Intensity. Sci. Transl. Med. 2020, 12, eaax4144. [Google Scholar] [CrossRef]
- Asish, P.R.; Dasgupta, S.; Rachel, G.; Bagepally, B.S.; Girish Kumar, C.P. Global Prevalence of Asymptomatic Dengue Infections—A Systematic Review and Meta-Analysis. Int. J. Infect. Dis. 2023, 134, 292–298. [Google Scholar] [CrossRef] [PubMed]
- WHO Dengue Haemorrhagic Fever: Diagnosis, Treatment, Prevention and Control; World Health Organization: Geneva, Switzerland, 1997; ISBN 92-4-154500-3.
- Malla, S.; Thakur, G.D.; Shrestha, S.K.; Banjeree, M.K.; Thapa, L.B.; Gongal, G.; Ghimire, P.; Upadhyay, B.P.; Gautam, P.; Khanal, S.; et al. Identification of All Dengue Serotypes in Nepal. Emerg. Infect. Dis. 2008, 14, 1669–1670. [Google Scholar] [CrossRef] [PubMed]
- Rimal, S.; Shrestha, S.; Pandey, K.; Nguyen, T.V.; Bhandari, P.; Shah, Y.; Acharya, D.; Adhikari, N.; Rijal, K.R.; Ghimire, P.; et al. Co-Circulation of Dengue Virus Serotypes 1, 2, and 3 during the 2022 Dengue Outbreak in Nepal: A Cross-Sectional Study. Viruses 2023, 15, 507. [Google Scholar] [CrossRef]
- Rimal, S.; Shrestha, S.; Paudel, S.W.; Shah, Y.; Bhandari, G.; Pandey, K.; Kharbuja, A.; Kapandji, M.; Gautam, I.; Bhujel, R.; et al. Molecular and Entomological Characterization of 2023 Dengue Outbreak in Dhading District, Central Nepal. Viruses 2024, 16, 594. [Google Scholar] [CrossRef]
- Dumre, S.; Bhandari Dumre, R.; Shakya, G.; Shrestha, S.; Cherif, M.; Ghimire, P.; Klungthong, C.; Yoon, I.-K.; Hirayama, K.; Na-bangchang, K.; et al. Dengue Virus Serotypes 1 and 2 Responsible for Major Dengue Outbreaks in Nepal: Clinical, Laboratory, and Epidemiological Features. Am. J. Trop. Med. Hyg. 2017, 97, 1062–1069. [Google Scholar] [CrossRef]
- Ngwe Tun, M.M.; Pandey, K.; Nabeshima, T.; Kyaw, A.K.; Adhikari, M.; Raini, S.K.; Inoue, S.; Dumre, S.P.; Pandey, B.D.; Morita, K. An Outbreak of Dengue Virus Serotype 2 Cosmopolitan Genotype in Nepal, 2017. Viruses 2021, 13, 1444. [Google Scholar] [CrossRef] [PubMed]
- Khetan, R.P.; Stein, D.A.; Chaudhary, S.K.; Rauniyar, R.; Upadhyay, B.P.; Gupta, U.P.; Gupta, B.P. Profile of the 2016 Dengue Outbreak in Nepal. BMC Res. Notes 2018, 11, 423. [Google Scholar] [CrossRef]
- EDCD. Situation Update of Dengue 2023; Epidemiology and Disease Control Division: Kathmandu, Nepal, 2023. Available online: https://edcd.gov.np/uploads/news/pdf/657e90834a34c.pdf (accessed on 2 February 2025).
- EDCD Dengue Situation Update, Epidemiology and Disease Control Division, Kathmandu, Nepal 2024. Available online: https://www.edcd.gov.np/news/20241203dengue-situation-update (accessed on 15 January 2025).
- Paudel, N.; Naharki, A.R.; Upreti, R.; Larvae Destruction Drive Launched to Fight Dengue. The Rising Nepal. 2023. Available online: https://risingnepaldaily.com/news/33280 (accessed on 3 June 2024).
- EDCD. Situation Update of Dengue 2022; Epidemiology and Disease Control Division: Kathmandu, Nepal, 2022. Available online: https://edcd.gov.np/uploads/news/pdf/6347833a92f00.pdf (accessed on 1 January 2025).
- Toan, N.T.; Rossi, S.; Prisco, G.; Nante, N.; Viviani, S. Dengue Epidemiology in Selected Endemic Countries: Factors Influencing Expansion Factors as Estimates of Underreporting. Trop. Med. Int. Health 2015, 20, 840–863. [Google Scholar] [CrossRef]
- A Brief Introduction: Bardibas Municipality. Available online: https://bardibasmun.gov.np/ne/node/3 (accessed on 5 June 2024).
- Global Historical Weather and Climate Data, Mahottari, Janakpur, Nepal Climate. Available online: https://weatherandclimate.com/nepal/janakpur/mahottari (accessed on 12 July 2024).
- Ngwe Tun, M.M.; Muthugala, R.; Nabeshima, T.; Soe, A.M.; Dumre, S.P.; Rajamanthri, L.; Jayawardana, D.; Attanayake, S.; Inoue, S.; Morita, K. Complete Genome Analysis and Characterization of Neurotropic Dengue Virus 2 Cosmopolitan Genotype Isolated from the Cerebrospinal Fluid of Encephalitis Patients. PLoS ONE 2020, 15, e0234508. [Google Scholar] [CrossRef]
- Ito, M.; Takasaki, T.; Yamada, K.-I.; Nerome, R.; Tajima, S.; Kurane, I. Development and Evaluation of Fluorogenic TaqMan Reverse Transcriptase PCR Assays for Detection of Dengue Virus Types 1 to 4. J. Clin. Microbiol. 2004, 42, 5935–5937. [Google Scholar] [CrossRef]
- Cao-Lormeau, V.-M.; Roche, C.; Aubry, M.; Teissier, A.; Lastere, S.; Daudens, E.; Mallet, H.-P.; Musso, D.; Aaskov, J. Recent Emergence of Dengue Virus Serotype 4 in French Polynesia Results from Multiple Introductions from Other South Pacific Islands. PLoS ONE 2011, 6, e29555. [Google Scholar] [CrossRef] [PubMed]
- Halstead, S.B. Pathogenesis of Dengue: Challenges to Molecular Biology. Science 1988, 239, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Katzelnick, L.C.; Fonville, J.M.; Gromowski, G.D.; Arriaga, J.B.; Green, A.; James, S.L.; Lau, L.; Montoya, M.; Wang, C.; VanBlargan, L.A.; et al. Dengue Viruses Cluster Antigenically but Not as Discrete Serotypes. Science 2015, 349, 1338–1343. [Google Scholar] [CrossRef]
- Brower, V. Vector-borne Diseases and Global Warming: Are Both on an Upward Swing?: Scientists Are Still Debating Whether Global Warming Will Lead to a Further Spread of Mosquitoes and the Diseases They Transmit. EMBO Rep. 2001, 2, 755–757. [Google Scholar] [CrossRef]
- Dumre, S.P.; Acharya, D.; Lal, B.K.; Brady, O.J. Dengue Virus on the Rise in Nepal. Lancet Infect. Dis. 2020, 20, 889–890. [Google Scholar] [CrossRef]
- Prajapati, S.; Napit, R.; Bastola, A.; Rauniyar, R.; Shrestha, S.; Lamsal, M.; Adhikari, A.; Bhandari, P.; Yadav, S.R.; Manandhar, K.D. Molecular Phylogeny and Distribution of Dengue Virus Serotypes Circulating in Nepal in 2017. PLoS ONE 2020, 15, e0234929. [Google Scholar] [CrossRef] [PubMed]
- Bharati, N.; Dumre, S.P.; Shah, Y.; Nabesima, T.; Dhimal, M.; Pandey, S.; Kapandji, M.; Takamatsu, Y.; Urano, T.; Pandey, B.D.; et al. Circulating Serotypes and Genotypes of Dengue Virus during the 2023 Outbreak in Eastern Nepal. J. Clin. Virol. 2024, 174, 105721. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, N.; Johnson, B.J.; Dixit, S.M.; Devine, G.J. Patterns of Dengue in Nepal from 2010–2019 in Relation to Elevation and Climate. Trans. R. Soc. Trop. Med. Hyg. 2021, 115, 741–749. [Google Scholar] [CrossRef]
- Tuladhar, R.; Singh, A.; Varma, A.; Choudhary, D.K. Climatic Factors Influencing Dengue Incidence in an Epidemic Area of Nepal. BMC Res. Notes 2019, 12, 131. [Google Scholar] [CrossRef]
- Banu, S.; Hu, W.; Hurst, C.; Tong, S. Dengue Transmission in the Asia-Pacific Region: Impact of Climate Change and Socio-environmental Factors. Trop. Med. Int. Health 2011, 16, 598–607. [Google Scholar] [CrossRef]
- Mashal, M.; Sharma, B. An Indian State Banned Alcohol. The Drinking Moved to Nearby Nepal. The Irish Times, 22 May 2022. Available online: https://www.irishtimes.com/news/world/asia-pacific/an-indian-state-banned-alcohol-the-drinking-moved-to-nearby-nepal-1.4885277 (accessed on 23 September 2024).
- Tricou, V.; Minh, N.N.; Farrar, J.; Tran, H.T.; Simmons, C.P. Kinetics of Viremia and NS1 Antigenemia Are Shaped by Immune Status and Virus Serotype in Adults with Dengue. PLoS Negl. Trop. Dis. 2011, 5, e1309. [Google Scholar] [CrossRef] [PubMed]
- Yung, C.-F.; Lee, K.-S.; Thein, T.-L.; Tan, L.-K.; Gan, V.C.; Wong, J.G.X.; Lye, D.C.; Ng, L.-C.; Leo, Y.-S. Dengue Serotype-Specific Differences in Clinical Manifestation, Laboratory Parameters and Risk of Severe Disease in Adults, Singapore. Am. Soc. Trop. Med. Hyg. 2015, 92, 999–1005. [Google Scholar] [CrossRef]
- Rodriguez-Roche, R.; Gould, E.A. Understanding the Dengue Viruses and Progress towards Their Control. BioMed Res. Int. 2013, 2013, 1–20. [Google Scholar] [CrossRef]
- Perera, D.R.; Ranadeva, N.D.; Sirisena, K.; Wijesinghe, K.J. Roles of NS1 Protein in Flavivirus Pathogenesis. ACS Infect. Dis. 2024, 10, 20–56. [Google Scholar] [CrossRef]
- Erra, E.O.; Korhonen, E.M.; Voutilainen, L.; Huhtamo, E.; Vapalahti, O.; Kantele, A. Dengue in Travelers: Kinetics of Viremia and NS1 Antigenemia and Their Associations with Clinical Parameters. PLoS ONE 2013, 8, e65900. [Google Scholar] [CrossRef]
- Endy, T.P.; Nisalak, A.; Chunsuttitwat, S.; Vaughn, D.W.; Green, S.; Ennis, F.A.; Rothman, A.L.; Libraty, D.H. Relationship of Preexisting Dengue Virus (DV) Neutralizing Antibody Levels to Viremia and Severity of Disease in a Prospective Cohort Study of DV Infection in Thailand. J. Infect. Dis. 2004, 189, 990–1000. [Google Scholar] [CrossRef]
- Vaughn, D.W.; Green, S.; Kalayanarooj, S.; Innis, B.L.; Nimmannitya, S.; Suntayakorn, S.; Endy, T.P.; Raengsakulrach, B.; Rothman, A.L.; Ennis, F.A.; et al. Dengue Viremia Titer, Antibody Response Pattern, and Virus Serotype Correlate with Disease Severity. J. Infect. Dis. 2000, 181, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Dayarathna, S.; Kuruppu, H.; Silva, T.; Gomes, L.; Shyamali, N.L.A.; Jeewandara, C.; Ariyaratne, D.; Ramu, S.T.; Wijewickrama, A.; Ogg, G.S.; et al. Are Viral Loads in the Febrile Phase a Predictive Factor of Dengue Disease Severity? BMC Infect. Dis. 2024, 24, 1248. [Google Scholar] [CrossRef] [PubMed]
- De Souza, L.J.; Bastos Pessanha, L.; Carvalho Mansur, L.; Assed De Souza, L.; Barbosa Tâmega Ribeiro, M.; Do Vale Da Silveira, M.; Damian Souto Filho, J.T. Comparison of Clinical and Laboratory Characteristics between Children and Adults with Dengue. Braz. J. Infect. Dis. 2013, 17, 27–31. [Google Scholar] [CrossRef]
- Ben-Shachar, R.; Schmidler, S.; Koelle, K. Drivers of Inter-Individual Variation in Dengue Viral Load Dynamics. PLOS Comput. Biol. 2016, 12, e1005194. [Google Scholar] [CrossRef] [PubMed]
- Handel, A.; Rohani, P. Crossing the Scale from Within-Host Infection Dynamics to between-Host Transmission Fitness: A Discussion of Current Assumptions and Knowledge. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140302. [Google Scholar] [CrossRef]
- Martina, B.E.E.; Koraka, P.; Osterhaus, A.D.M.E. Dengue Virus Pathogenesis: An Integrated View. Clin. Microbiol. Rev. 2009, 22, 564–581. [Google Scholar] [CrossRef]
- Gupta, B.P.; Haselbeck, A.; Kim, J.H.; Marks, F.; Saluja, T. The Dengue Virus in Nepal: Gaps in Diagnosis and Surveillance. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 32. [Google Scholar] [CrossRef]


| Blood Parameters | Total Number | Dengue Patient, Median (IQR) | Non-Dengue Patient, Median (IQR) | p-Value |
|---|---|---|---|---|
| Hemoglobin (g/dL) | 1878 | 12.8 (11.6–14.2) | 12.4 (11.3–13.7) | 0.001 |
| WBC (cells/µL) | 1882 | 4000 (3000–5820) | 7200 (5300–9502.5) | <0.001 |
| RBC (million/µL) | 1880 | 4.4 (4–4.8) | 4.4 (4–4.8) | 0.468 |
| Platelet (cells/µL) | 1880 | 131,000 (102,500–165,500) | 175,000 (139,000–223,000) | <0.001 |
| Neutrophil (%) | 1872 | 63 (54.5–71) | 70 (62–78) | <0.001 |
| Lymphocyte (%) | 1872 | 30 (23–39) | 25 (17–31) | <0.001 |
| Eosinophil (%) | 1872 | 3 (2–3) | 2 (2–3) | <0.001 |
| Monocyte (%) | 1872 | 3 (2–4) | 3 (2–4) | <0.001 |
| PCV (%) | 1593 | 42.7 (37.5–48.4) | 41.2 (36.1–46.55) | 0.038 |
| MCV (femtoliters) | 1593 | 97 (87.75–106.5) | 93.9 (86.1–102.8) | <0.001 |
| MCH (pg) | 1592 | 28.9 (26.6–30.6) | 28 (26.1–30) | 0.003 |
| MCHC (g/dL) | 1591 | 29.9 (27–32.6) | 30.1 (27.3–32.6) | 0.627 |
| Bilirubin, total (mg/dL) | 176 | 0.8 (0.7–0.9) | 0.8 (0.7–0.9) | 0.52 |
| Bilirubin, direct (mg/dL) | 176 | 0.2 (0.2–0.3) | 0.2 (0.2–0.3) | 0.54 |
| ALP (U/L) | 160 | 168.9 (130.5–205.1) | 185.32 (118.2–234.8) | 0.388 |
| SGOT (U/L) | 397 | 51 (34–88.1) | 33.125 (25–50.6) | <0.001 |
| SGPT (U/L) | 401 | 40.3 (29.2–73.3) | 28.6 (21.6–42.8) | <0.001 |
| Total protein (g/dL) | 168 | 6.5 (6.2–7) | 6.7 (6.1–7) | 0.652 |
| Albumin (g/dL) | 165 | 3.9 (3.8–4) | 3.8 (3.7–4) | 0.353 |
| Creatinine (mg/dL) | 584 | 0.895 (0.8–1) | 0.86 (0.7–1) | 0.187 |
| Urea (mg/dL) | 494 | 24.9 (20.5–30.4) | 25.07 (20–30.8) | 0.85 |
| Sodium (mmol/L) | 466 | 135.6 (134–138.3) | 135.2 (134.2–136.9) | 0.356 |
| Potassium (mmol/L) | 463 | 3.8 (3.7–3.9) | 3.8 (3.7–4) | 0.324 |
| Blood Parameters | Total Number | Inpatients, Median (IQR) | Outpatients, Median (IQR) | p-Value |
|---|---|---|---|---|
| Hemoglobin (g/dL) | 405 | 11.7 (9–12.3) | 12.8 (11.7–14.2) | 0.005 |
| WBC (cells/µL) | 405 | 3320 (2645–4360) | 4000 (3010–5835) | 0.196 |
| RBC (million/µL) | 405 | 4.0 (3.5–4.4) | 4.4 (4–4.8) | 0.081 |
| Platelet (cells/µL) | 403 | 110,000 (71,500–156,000) | 131,000 (103,000–166,750) | 0.604 |
| Neutrophil (%) | 403 | 54.5 (44.8–72.5) | 64 (55–71) | 0.420 |
| Lymphocyte (%) | 403 | 37.5 (21.5–50.0) | 30 (23–39) | 0.328 |
| Eosinophil (%) | 403 | 3 (2–3) | 3 (2–3) | 0.907 |
| Monocyte (%) | 403 | 3 (2.8–4.3) | 3 (2–4) | 0.665 |
| PCV (%) | 313 | 33.2 (28.6–43.4) | 42.9 (37.7–48.4) | 0.110 |
| MCV (femtoliters) | 313 | 90.5 (77.8–104.4) | 97 (87.8–106.6) | 0.788 |
| MCH (pg) | 313 | 25.7 (22.1–28.1) | 28.9 (26.8–30.6) | 0.130 |
| MCHC (g/dL) | 312 | 30.1 (22.5–34.4) | 29.9 (27.1–32.6) | 0.764 |
| Bilirubin, total (mg/dL) | 26 | 0.73 (0.6–0.8) | 0.8 (0.7–1.0) | 1.000 |
| Bilirubin, direct (mg/dL) | 26 | 0.205 (0.18–0.23) | 0.2 (0.2–0.3) | 0.483 |
| ALP (U/L) | 25 | 161.3 (148.4–174.2) | 168.9 (125.8–208.5) | 1.000 |
| SGOT (U/L) | 65 | 67.7 (28.5–216.0) | 49.6 (34.0–88.0) | 0.584 |
| SGPT (U/L) | 65 | 34.9 (13.5–63.1) | 41.9 (29.3–78.2) | 0.628 |
| Total protein (g/dL) | 25 | 6.6 (6.2–6.9) | 6.5 (6.2–7.0) | 1.000 |
| Albumin (g/dL) | 25 | 3.7 (3.6–3.9) | 3.9 (3.8–4.0) | 0.520 |
| Creatinine (mg/dL) | 107 | 0.9 (0.9–1.0) | 0.9 (0.8–1.0) | 1.000 |
| Urea (mg/dL) | 84 | 28.4 (23.6–31.4) | 24.6 (20.4–30.4) | 0.409 |
| Sodium (mmol/L) | 81 | 134.6 (133.0–147.9) | 135.7 (134.0–138.3) | 0.766 |
| Potassium (mmol/L) | 81 | 3.5 (3.2–3.8) | 3.8 (3.7–3.9) | 0.453 |
| Blood Parameters | Total Number | Primary, Median (IQR) | Secondary, Median (IQR) | p-Value |
|---|---|---|---|---|
| Hemoglobin (g/dL) | 405 | 12.8 (11.7–14.2) | 11.65 (10.55–13.3) | 0.164 |
| WBC (cells/µL) | 405 | 4000 (3000–5800) | 3880 (2567.5–6327.5) | 0.881 |
| RBC (million/µL) | 405 | 4.4 (4–4.8) | 4.2 (3.7–4.8) | 0.34 |
| Platelet (cells/µL) | 403 | 132,000 (103,000–166,000) | 120,000 (89,750–160,000) | 0.276 |
| Neutrophil (%) | 403 | 63 (55–71) | 65.5 (54–72) | 0.81 |
| Lymphocyte (%) | 403 | 30 (23–39) | 29.5 (23.3–37.5) | 0.621 |
| Eosinophil (%) | 403 | 3 (2–3) | 3 (2–3.8) | 0.476 |
| Monocyte (%) | 403 | 3 (2–4) | 3.5 (2.3–4) | 0.351 |
| PCV (%) | 313 | 42.8 (37.6–48.6) | 41 (35–47.8) | 0.99 |
| MCV (femtoliters) | 313 | 96.9 (87.7–105.8) | 104 (89.1–113.1) | 0.952 |
| MCH (pg) | 313 | 28.9 (26.7–30.6) | 28.7 (23.6–30.3) | 0.646 |
| MCHC (g/dL) | 312 | 30 (27.1–32.6) | 28.4 (22.7–31.05) | 0.587 |
| Bilirubin, total (mg/dL) | 26 | 0.8 (0.7–0.8) | 1.1 (1–1.9) | 0.032 |
| Bilirubin, direct (mg/dL) | 26 | 0.23 (0.2–0.3) | 0.4 (0.3–0.4) | 0.085 |
| ALP (U/L) | 25 | 168.0 (124.4–203.4) | 188.6 (137.9–202.6) | 0.593 |
| SGOT (U/L) | 65 | 50.3 (33.1–93.9) | 52 (44.4–98.6) | 0.978 |
| SGPT (U/L) | 65 | 41.1 (29.2–77.1) | 32.5 (29–76) | 0.356 |
| Total protein (g/dL) | 25 | 6.66 (6.2–7.0) | 6.2 (6–6.4) | 0.48 |
| Albumin (g/dL) | 25 | 3.9 (3.8–4) | 3.6 (3.6–3.6) | 0.52 |
| Creatinine (mg/dL) | 107 | 0.9 (0.8–1.0) | 0.8 (0.6–1.2) | 0.674 |
| Urea (mg/dL) | 84 | 24.9 (20.52–30.1) | 27.3 (14.4–34.1) | 0.694 |
| Sodium (mmol/L) | 81 | 135.5 (134–137.9) | 137.6 (135.3–138.8) | 0.181 |
| Potassium (mmol/L) | 81 | 3.8 (3.7–3.9) | 3.7 (3.6–4.0) | 0.915 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shrestha, S.; Rimal, S.; Kharbuja, A.; Ray, M.K.; Shrestha, S.; Dulal, A.; Subedi, S.; Khadka, A.; Adhikari, N.; Dhimal, M.; et al. Emergence of Dengue Virus Serotypes 1 and 3 in Mahottari and Adjacent Areas of Southern Nepal. Pathogens 2025, 14, 639. https://doi.org/10.3390/pathogens14070639
Shrestha S, Rimal S, Kharbuja A, Ray MK, Shrestha S, Dulal A, Subedi S, Khadka A, Adhikari N, Dhimal M, et al. Emergence of Dengue Virus Serotypes 1 and 3 in Mahottari and Adjacent Areas of Southern Nepal. Pathogens. 2025; 14(7):639. https://doi.org/10.3390/pathogens14070639
Chicago/Turabian StyleShrestha, Sabin, Sandesh Rimal, Anjana Kharbuja, Manoj Kumar Ray, Susmita Shrestha, Anjali Dulal, Suprabha Subedi, Ashma Khadka, Nabaraj Adhikari, Meghnath Dhimal, and et al. 2025. "Emergence of Dengue Virus Serotypes 1 and 3 in Mahottari and Adjacent Areas of Southern Nepal" Pathogens 14, no. 7: 639. https://doi.org/10.3390/pathogens14070639
APA StyleShrestha, S., Rimal, S., Kharbuja, A., Ray, M. K., Shrestha, S., Dulal, A., Subedi, S., Khadka, A., Adhikari, N., Dhimal, M., Pandey, B. D., Urano, T., Morita, K., Ngwe Tun, M. M., & Dumre, S. P. (2025). Emergence of Dengue Virus Serotypes 1 and 3 in Mahottari and Adjacent Areas of Southern Nepal. Pathogens, 14(7), 639. https://doi.org/10.3390/pathogens14070639








