Enterococcus faecium as an Emerging Pathogen: Molecular Epidemiology and Antimicrobial Resistance in Clinical Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Selection and Antimicrobial Susceptibility Testing
2.2. Molecular Typing
2.3. Detection of Resistance-Associated and Virulence Genes
3. Results
3.1. Characteristics of Strains and Patients and Antimicrobial Resistance Profiles
3.2. PFGE Analysis
3.3. Typing by MLST
3.4. Prevalence of Resistance and Virulence-Associated Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| VRE | Vancomycin-resistant enterococci |
| VREfm | Vancomycin-resistant Enterococcus faecium |
| WHO | World Health Organization |
| HAIs | Healthcare-associated infections |
| ECDC | European Centre for Disease Prevention and Control |
| ICU | Intensive care unit |
| TSA | Tryptic Soy Agar |
| PFGE | Pulsed-field gel electrophoresis |
| CDC | Centers for Disease Control and Prevention |
| UPGMA | Unweighted pair group method with arithmetic mean |
| MLST | Multilocus sequence typing |
| PCR | Polymerase chain reaction |
| PT | Pulsotype |
| MDR | Multidrug resistant |
| NICU | Neonatal intensive care unit |
| CC | Clonal complex |
References
- Hashimoto, Y.; Suzuki, M.; Kobayashi, S.; Hirahara, Y.; Kurushima, J.; Hirakawa, H.; Nomura, T.; Tanimoto, K.; Tomita, H. Enterococcal Linear Plasmids Adapt to Enterococcus faecium and Spread within Multidrug-Resistant Clades. Antimicrob. Agents Chemother. 2023, 67, e0161922. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.R.; Murray, B.E.; Rice, L.B.; Arias, C.A. Resistance in Vancomycin-Resistant Enterococci. Infect. Dis. Clin. N. Am. 2020, 34, 751–771. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, Y.; He, Z.; Wang, Y.; Wang, J.; Jin, D. A Molecular Study Regarding the Spread of vanA Vancomycin-Resistant Enterococcus faecium in a Tertiary Hospital in China. J. Glob. Antimicrob. Resist. 2022, 31, 270–278. [Google Scholar] [CrossRef]
- World Health Organization (WHO). WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance. Available online: https://www.who.int/publications/i/item/9789240093461 (accessed on 31 March 2025).
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use in European Acute Care Hospitals; ECDC: Stockholm, Sweden, 2024.
- Khan, K.; Jalal, K.; Uddin, R. Pangenome Profiling of Novel Drug Target Against Vancomycin-Resistant Enterococcus faecium. J. Biomol. Struct. Dyn. 2023, 41, 15647–15660. [Google Scholar] [CrossRef]
- Alotaibi, G.; Khan, K.; Al Mouslem, A.K.; Ahmad Khan, S.; Naseer Abbas, M.; Abbas, M.; Ali Shah, S.; Jalal, K. Pangenome-Based Reverse Vaccinology Approach to Explore Enterococcus faecium (VRE) Strains for Identification of Novel Multi-Epitope Vaccine Candidate. Immunobiology 2022, 227, 152221. [Google Scholar] [CrossRef] [PubMed]
- Iacchini, S.; Boros, S.; Pezzotti, P.; Errico, G.; Del Grosso, M.; Camilli, R.; Giufrè, M.; Pantosti, A.; Maraglino, F.; Palamara, A.T.; et al. AR-ISS: Sorveglianza Nazionale dell’Antibiotico-Resistenza. Dati 2023; Rapporti ISS Sorveglianza RIS 5/2024; Istituto Superiore di Sanità: Roma, Italy, 2024. [Google Scholar]
- Wardal, E.; Żabicka, D.; Skalski, T.; Kubiak-Pulkowska, J.; Hryniewicz, W.; Sadowy, E. Characterization of a Tigecycline-, Linezolid- and Vancomycin-Resistant Clinical Enterococcus faecium Isolate, Carrying vanA and vanB Genes. Infect. Dis. Ther. 2023, 12, 2545–2565. [Google Scholar] [CrossRef]
- O’Toole, R.F.; Leong, K.W.C.; Cumming, V.; Van Hal, S.J. Vancomycin-Resistant Enterococcus faecium and the Emergence of New Sequence Types Associated with Hospital Infection. Res. Microbiol. 2023, 174, 104046. [Google Scholar] [CrossRef]
- Wan, T.W.; Yeo, H.H.; Lee, T.F.; Huang, Y.T.; Hsueh, P.R.; Chiu, H.C. Molecular Epidemiology of Bacteraemic Vancomycin-Resistant Enterococcus faecium Isolates and In Vitro Activities of SC5005 and Other Comparators Against These Isolates Collected from a Medical Centre in Northern Taiwan, 2019–2020. J. Antimicrob. Chemother. 2023, 78, 457–465. [Google Scholar] [CrossRef]
- Gagetti, P.; Bonofiglio, L.; García Gabarrot, G.; Kaufman, S.; Mollerach, M.; Vigliarolo, L.; von Specht, M.; Toresani, I.; Lopardo, H.A. Resistance to β-Lactams in Enterococci. Rev. Argent. Microbiol. 2019, 51, 179–183. [Google Scholar] [CrossRef]
- Darehkordi, H.; Saffari, F.; Mollaei, H.R.; Ahmadrajabi, R. Amino Acid Substitution Mutations and mRNA Expression Levels of the pbp5 Gene in Clinical Enterococcus faecium Isolates Conferring High Level Ampicillin Resistance. APMIS 2019, 127, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Harada, S.; Shibue, Y.; Aoki, K.; Ishii, Y.; Tateda, K. Prevalence of High-Level Aminoglycoside Resistance and Genes Encoding Aminoglycoside-Modifying Enzymes in Enterococcus faecalis and Enterococcus faecium Isolated in a University Hospital in Tokyo. Jpn. J. Infect. Dis. 2020, 73, 476–480. [Google Scholar] [CrossRef] [PubMed]
- Das, A.K.; Dudeja, M.; Kohli, S.; Ray, P.; Nandy, S. High-Level Gentamicin Resistance Mediated by Aac(6′)-Ie-aph(2″)-Ia Gene in Enterococcus Species Isolated from Clinical Samples in Northern India. Indian J. Pharmacol. 2022, 54, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Abu Lila, A.S.; Alharby, T.N.; Alanazi, J.; Alanazi, M.; Abdallah, M.H.; Rizvi, S.M.D.; Moin, A.; Khafagy, E.S.; Tabrez, S.; Al Balushi, A.A.; et al. Clinical Resistant Strains of Enterococci and Their Correlation to Reduced Susceptibility to Biocides: Phenotypic and Genotypic Analysis of Macrolides, Lincosamides, and Streptogramins. Antibiotics 2023, 12, 461. [Google Scholar] [CrossRef]
- Lin, P.Y.; Chan, S.Y.; Stern, A.; Chen, P.H.; Yang, H.C. Epidemiological Profiles and Pathogenicity of Vancomycin-Resistant Enterococcus faecium Clinical Isolates in Taiwan. PeerJ 2023, 11, e14859. [Google Scholar] [CrossRef]
- Tang, Y.F.; Lin, Y.S.; Su, L.H.; Liu, J.W. Increasing Trend of Healthcare-Associated Infections Due to Vancomycin-Resistant Enterococcus faecium (VRE-fm) Paralleling Escalating Community-Acquired VRE-fm Infections in a Medical Center Implementing Strict Contact Precautions: An Epidemiologic and Pathogenic Genotype Analysis and Its Implications. J. Microbiol. Immunol. Infect. 2023, 56, 1045–1053. [Google Scholar] [CrossRef]
- Tokano, M.; Tarumoto, N.; Sakai, J.; Imai, K.; Kodana, M.; Kawamura, T.; Maeda, T.; Maesaki, S. Vancomycin-Resistant Enterococcus faecium in Japan, 2007–2015: A Molecular Epidemiology Analysis Focused on Examining Strain Characteristics Over Time. Microbiol. Spectr. 2024, 12, e0244423. [Google Scholar] [CrossRef]
- Rogers, L.A.; Strong, K.; Cork, S.C.; McAllister, T.A.; Liljebjelke, K.; Zaheer, R.; Checkley, S.L. The Role of Whole Genome Sequencing in the Surveillance of Antimicrobial Resistant Enterococcus spp.: A Scoping Review. Front. Public Health 2021, 9, 599285. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Clinical Breakpoints—Breakpoints and Guidance. 2025. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 9 May 2024).
- Centers for Disease Control and Prevention. Unified Pulsed-Field Gel Electrophoresis (PFGE) Protocol for Gram-Positive Bacteria. Available online: https://www.cdc.gov/healthcare-associated-infections/media/pdfs/Unified_PFGE_Protocol.pdf (accessed on 31 March 2025).
- Homan, W.L.; Tribe, D.; Poznanski, S.; Li, M.; Hogg, G.; Spalburg, E.; Van Embden, J.D.; Willems, R.J. Multilocus Sequence Typing Scheme for Enterococcus faecium. J. Clin. Microbiol. 2002, 40, 1963–1971. [Google Scholar] [CrossRef]
- Shirvani, F.; Behzad, A.; Abdollahi, N.; Mohkam, M.; Sharifian, M.; Esfandiar, N.; Fallah, F. Frequency and Co-Colonization of Vancomycin-Resistant Enterococci and Candida in ICU-Hospitalized Children. New Microbes New Infect. 2021, 41, 100881. [Google Scholar] [CrossRef]
- Haghi, F.; Lohrasbi, V.; Zeighami, H. High Incidence of Virulence Determinants, Aminoglycoside and Vancomycin Resistance in Enterococci Isolated from Hospitalized Patients in Northwest Iran. BMC Infect. Dis. 2019, 19, 744. [Google Scholar] [CrossRef] [PubMed]
- Portillo, A.; Ruiz-Larrea, F.; Zarazaga, M.; Alonso, A.; Martinez, J.L.; Torres, C. Macrolide Resistance Genes in Enterococcus spp. Antimicrob. Agents Chemother. 2000, 44, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Poeta, P.; Costa, D.; Igrejas, G.; Sáenz, Y.; Zarazaga, M.; Rodrigues, J.; Torres, C. Polymorphisms of the pbp5 Gene and Correlation with Ampicillin Resistance in Enterococcus faecium Isolates of Animal Origin. J. Med. Microbiol. 2007, 56, 236–240. [Google Scholar] [CrossRef]
- Weng, P.L.; Ramli, R.; Hamat, R.A. Antibiotic Susceptibility Patterns, Biofilm Formation and esp Gene Among Clinical Enterococci: Is There Any Association? Int. J. Environ. Res. Public Health 2019, 16, 3439. [Google Scholar] [CrossRef] [PubMed]
- Jovanović, M.; Velebit, B.; Tošić, T.; Maki, G.; Pavić, S.; Jovanović, S.; Stošović, R.; Zervos, M.J. Comparative Study of Virulence Factor Genes, β-Hemolysis and Biofilm Production in Invasive and Colonizing Enterococci. Eur. J. Inflamm. 2023, 21, 1721727X231156333. [Google Scholar] [CrossRef]
- Trautmannsberger, I.; Kolberg, L.; Meyer-Buehn, M.; Huebner, J.; Werner, G.; Weber, R.; Heselich, V.; Schroepf, S.; Muench, H.G.; von Both, U. Epidemiological and genetic characteristics of vancomycin-resistant Enterococcus faecium isolates in a university children’s hospital in Germany: 2019 to 2020. Antimicrob. Resist. Infect. Control 2022, 11, 48. [Google Scholar] [CrossRef]
- Esme, M.; Topeli, A.; Yavuz, B.B.; Akova, M. Infections in elderly critically ill patients. Front. Med. 2019, 6, 118. [Google Scholar] [CrossRef]
- De Waele, J.J.; Akova, M.; Antonelli, M.; Canton, R.; Carlet, J.; De Backer, D.; Dimopoulos, G.; Garnacho-Montero, J.; Kesecioglu, J.; Lipman, J.; et al. Antimicrobial Resistance and Antibiotic Stewardship Programs in the ICU: Insistence and Persistence in the Fight Against Resistance. A Position Statement from ESICM/ESCMID/WAAAR Round Table on Multi-Drug Resistance. Intensive Care Med. 2018, 44, 189–196. [Google Scholar] [CrossRef]
- Weisser, M.; Oostdijk, E.A.; Willems, R.J.; Bonten, M.J.; Frei, R.; Elzi, L.; Halter, J.; Widmer, A.F.; Top, J. Dynamics of ampicillin-resistant Enterococcus faecium clones colonizing hospitalized patients: Data from a prospective observational study. BMC Infect. Dis. 2012, 12, 68. [Google Scholar] [CrossRef]
- Póvoa, P.; Ramirez, P.; Blot, S. Decolonization Strategies Against Multidrug Resistant Organisms in the ICU. Intensive Care Med. 2024, 50, 577–579. [Google Scholar] [CrossRef]
- Domingo, M.C.; Huletsky, A.; Giroux, R.; Boissinot, K.; Picard, F.J.; Lebel, P.; Ferraro, M.J.; Bergeron, M.G. High prevalence of glycopeptide resistance genes vanB, vanD, and vanG not associated with Enterococci in human fecal flora. Antimicrob. Agents Chemother. 2005, 49, 4784–4786. [Google Scholar] [CrossRef] [PubMed]
- Ghazvinian, M.; Asgharzadeh Marghmalek, S.; Gholami, M.; Amir Gholami, S.; Amiri, E.; Goli, H.R. Antimicrobial Resistance Patterns, Virulence Genes, and Biofilm Formation in Enterococci Strains Collected from Different Sources. BMC Infect. Dis. 2024, 24, 274. [Google Scholar] [CrossRef] [PubMed]
- Mirzaii, M.; Alebouyeh, M.; Sohrabi, M.B.; Eslami, P.; Fazli, M.; Ebrahimi, M.; HajiAsgarli, P.; Rashidan, M. Antibiotic resistance assessment and multi-drug efflux pumps of Enterococcus faecium isolated from clinical specimens. J. Infect. Dev. Ctries. 2023, 17, 649–655. [Google Scholar] [CrossRef]
- Pan, P.; Sun, L.; Shi, X.; Huang, X.; Yin, Y.; Pan, B.; Hu, L.; Shen, Q. Analysis of molecular epidemiological characteristics and antimicrobial susceptibility of vancomycin-resistant and linezolid-resistant Enterococcus in China. BMC Med. Genom. 2024, 17, 174. [Google Scholar] [CrossRef]
- Arredondo-Alonso, S.; Top, J.; McNally, A.; Puranen, S.; Pesonen, M.; Pensar, J.; Marttinen, P.; Braat, J.C.; Rogers, M.R.C.; van Schaik, W.; et al. Plasmid-mediated horizontal gene transfer and the emergence of the vancomycin-resistant Enterococcus faecium ST80 lineage. mBio 2021, 12, e02786-20. [Google Scholar]
- Fioriti, S.; Simoni, S.; Caucci, S.; Morroni, G.; Ponzio, E.; Coccitto, S.N.; Brescini, L.; Cirioni, O.; Menzo, S.; Biavasco, F.; et al. Trend of Clinical Vancomycin-Resistant Enterococci Isolated in a Regional Italian Hospital from 2001 to 2018. Braz. J. Microbiol. 2020, 51, 1607–1613. [Google Scholar] [CrossRef]
- Stampone, L.; Del Grosso, M.; Boccia, D.; Pantosti, A. Clonal spread of a vancomycin-resistant Enterococcus faecium strain among bloodstream-infecting isolates in Italy. J. Clin. Microbiol. 2005, 43, 1575–1580. [Google Scholar] [CrossRef]
- Hammerum, A.M.; Karstensen, K.T.; Roer, L.; Kaya, H.; Lindegaard, M.; Porsbo, L.J.; Kjerulf, A.; Pinholt, M.; Holzknecht, B.J.; Worning, P.; et al. Surveillance of Vancomycin-Resistant Enterococci Reveals Shift in Dominating Clusters from vanA to vanB Enterococcus faecium Clusters, Denmark, 2015 to 2022. Euro Surveill. 2024, 29, 2300633. [Google Scholar] [CrossRef]
- Boccella, M.; Santella, B.; Pagliano, P.; De Filippis, A.; Casolaro, V.; Galdiero, M.; Borrelli, A.; Capunzo, M.; Boccia, G.; Franci, G. Prevalence and antimicrobial resistance of Enterococcus species: A retrospective cohort study in Italy. Antibiotics 2021, 10, 1552. [Google Scholar] [CrossRef]
- Gök, Ş.M.; Türk Dağı, H.; Kara, F.; Arslan, U.; Fındık, D. Investigation of antibiotic resistance and virulence factors of Enterococcus faecium and Enterococcus faecalis strains isolated from clinical samples. Mikrobiyol Bul. 2020, 54, 26–39. [Google Scholar] [CrossRef]
- Sattari-Maraji, A.; Jabalameli, F.; Node Farahani, N.; Beigverdi, R.; Emaneini, M. Antimicrobial Resistance Pattern, Virulence Determinants and Molecular Analysis of Enterococcus faecium Isolated from Children Infections in Iran. BMC Microbiol. 2019, 19, 156. [Google Scholar] [CrossRef]
- Singh, K.V.; Galloway-Peña, J.; Montealegre, M.C.; Dong, X.; Murray, B.E. Genomic Context as Well as Sequence of Both psr and Penicillin-Binding Protein 5 Contributes to β-Lactam Resistance in Enterococcus faecium. mBio 2024, 15, e0017024. [Google Scholar] [CrossRef] [PubMed]
- Ligozzi, M.; Pittaluga, F.; Fontana, R. Modification of penicillin-binding protein 5 associated with high-level ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 1996, 40, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Paganelli, F.L.; Bierschenk, D.; Kuipers, A.; Bonten, M.J.; Willems, R.J.; van Schaik, W. Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium. PLoS Genet. 2012, 8, e1002804. [Google Scholar] [CrossRef]
- Herrero, I.A.; Issa, N.C.; Patel, R. Nosocomial Spread of Linezolid-Resistant, Vancomycin-Resistant Enterococcus faecium. N. Engl. J. Med. 2002, 346, 867–869. [Google Scholar] [CrossRef]
- Kiruthiga, A.; Padmavathy, K.; Shabana, P.; Naveenkumar, V.; Gnanadesikan, S.; Malaiyan, J. Improved Detection of esp, hyl, asa1, gelE, cylA Virulence Genes Among Clinical Isolates of Enterococci. BMC Res. Notes 2020, 13, 170. [Google Scholar] [CrossRef]
- Strateva, T.; Mitov, I. Enterococcus faecalis—A Dangerous Opponent in the Nosocomial World. J. Glob. Antimicrob. Resist. 2016, 4, 65–69. [Google Scholar] [CrossRef]
- Heikens, E.; Bonten, M.J.; Willems, R.J. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J. Bacteriol. 2007, 189, 8233–8240. [Google Scholar] [CrossRef]
- Russotto, V.; Cortegiani, A.; Raineri, S.M.; Giarratano, A. Bacterial contamination of inanimate surfaces and equipment in the intensive care unit. J. Intensive Care 2015, 3, 54. [Google Scholar] [CrossRef]
- Dancer, S.J. Controlling hospital-acquired infection: Focus on the role of the environment and new technologies for decontamination. Clin. Microbiol. Rev. 2014, 27, 665–690. [Google Scholar] [CrossRef]
- Joshi, S.; Shallal, A.; Zervos, M. Vancomycin-Resistant Enterococci: Epidemiology, Infection Prevention, and Control. Infect. Dis. Clin. N. Am. 2021, 35, 953–968. [Google Scholar] [CrossRef] [PubMed]
- Cinthi, M.; Coccitto, S.N.; Simoni, S.; D’Achille, G.; Zeni, G.; Mazzariol, A.; Pocognoli, A.; Di Lodovico, S.; Di Giulio, M.; Morroni, G.; et al. Molecular characterization of Enterococcus faecium clinical isolates harbouring erm(T) from an Italian hospital. Curr. Microbiol. 2024, 81, 431. [Google Scholar] [CrossRef] [PubMed]
- Cinthi, M.; Coccitto, S.N.; Simoni, S.; Gherardi, G.; Palamara, A.T.; Di Lodovico, S.; Di Giulio, M.; Du, X.D.; Vignaroli, C.; Brenciani, A.; et al. The optrA, cfr(D) and vanA genes are co-located on linear plasmids in linezolid- and vancomycin-resistant enterococcal clinical isolates in Italy. J. Antimicrob. Chemother. 2025, 80, 1362–1370. [Google Scholar] [CrossRef]
- Coccitto, S.N.; Cinthi, M.; Simoni, S.; Pocognoli, A.; Zeni, G.; Mazzariol, A.; Morroni, G.; Mingoia, M.; Giovanetti, E.; Brenciani, A.; et al. Genetic analysis of vancomycin-variable Enterococcus faecium clinical isolates in Italy. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 673–682. [Google Scholar] [CrossRef]
- Tamburro, M.; Lombardi, A.; Sammarco, M.L.; Ripabelli, G. Molecular Epidemiology Revealed Distinct Patterns Among Multidrug Resistant Clinical Acinetobacter baumannii Strains in Different Periods in the Main Hospital in Molise Region, Central Italy. Appl. Microbiol. 2025, 5, 9. [Google Scholar] [CrossRef]
- Karino, M.; Yanagihara, M.; Harada, T.; Sugo, M.; Karino, M.; Ohtaki, H.; Hanada, H.; Takano, T.; Yamato, M.; Okamoto, S. New Multilocus Sequence Typing Scheme for Enterococcus faecium Reveals Sequential Outbreaks of Vancomycin-Resistant E. faecium ST1162 and ST610 in a Japanese Tertiary Medical Center. Microbiol. Spectr. 2025, 13, e0213124. [Google Scholar] [CrossRef]
- Salzo, A.; Ripabelli, G.; Sammarco, M.L.; Mariano, A.; Niro, C.; Tamburro, M. Healthcare-Associated Infections and Antibiotics Consumption: A Comparison of Point Prevalence Studies and Intervention Strategies. Hosp. Top. 2021, 99, 140–150. [Google Scholar] [CrossRef]
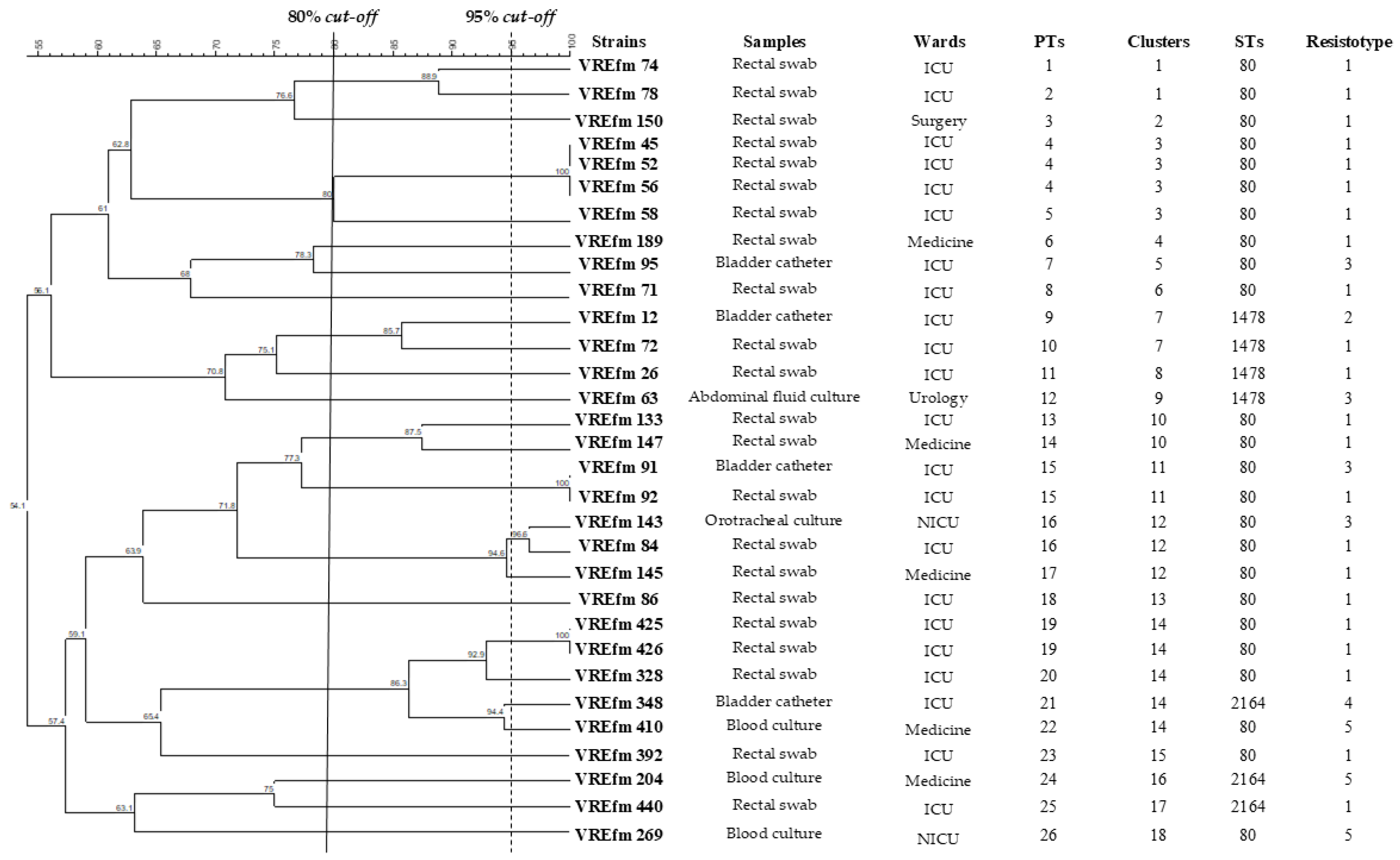
| Strains | Ward | Hospital | Age | Sex | Sample | Collection Date |
|---|---|---|---|---|---|---|
| VREfm 12 | ICU | Hub | 70 | M | Bladder catheter | November 2022 |
| VREfm 26 | ICU | Hub | 53 | M | Rectal swab | November 2022 |
| VREfm 45 | ICU | Hub | 82 | M | Rectal swab | February 2023 |
| VREfm 52 | ICU | Spoke | 78 | F | Rectal swab | February 2023 |
| VREfm 56 | ICU | Hub | 49 | M | Rectal swab | February 2023 |
| VREfm 58 | ICU | Spoke | 69 | F | Rectal swab | February 2023 |
| VREfm 63 | Urology | Hub | 59 | M | Abdominal fluid culture | February 2023 |
| VREfm 71 | ICU | Spoke | 90 | M | Rectal swab | February 2023 |
| VREfm 72 | ICU | Hub | 67 | M | Rectal swab | February 2023 |
| VREfm 74 | ICU | Spoke | 74 | M | Rectal swab | February 2023 |
| VREfm 78 | ICU | Hub | 41 | F | Rectal swab | March 2023 |
| VREfm 84 | ICU | Hub | 75 | F | Rectal swab | March 2023 |
| VREfm 86 | ICU | Hub | 53 | M | Rectal swab | March 2023 |
| VREfm 91 | ICU | Hub | 41 | F | Bladder catheter | March 2023 |
| VREfm 92 | ICU | Spoke | 64 | M | Rectal swab | March 2023 |
| VREfm 95 | ICU | Hub | 41 | F | Bladder catheter | March 2023 |
| VREfm 133 | ICU | Hub | 80 | M | Rectal swab | April 2023 |
| VREfm 143 | NICU | Hub | 1 | F | Orotracheal culture | May 2023 |
| VREfm 145 | Medicine | Hub | 82 | M | Rectal swab | May 2023 |
| VREfm 147 | Medicine | Hub | 72 | M | Rectal swab | May 2023 |
| VREfm 150 | Surgery | Hub | 74 | M | Rectal swab | May 2023 |
| VREfm 189 | Medicine | Hub | 49 | M | Rectal swab | June 2023 |
| VREfm 204 | Medicine | Hub | 79 | F | Blood culture | July 2023 |
| VREfm 269 | NICU | Hub | 0 | F | Blood culture | August 2023 |
| VREfm 328 | ICU | Hub | 67 | M | Rectal swab | November 2023 |
| VREfm 348 | ICU | Hub | 69 | M | Bladder catheter | November 2023 |
| VREfm 392 | ICU | Hub | 69 | M | Rectal swab | December 2023 |
| VREfm 410 | Medicine | Hub | 89 | F | Blood culture | January 2024 |
| VREfm 425 | ICU | Hub | 83 | M | Rectal swab | February 2024 |
| VREfm 426 | ICU | Hub | 83 | F | Rectal swab | February 2024 |
| VREfm 440 | ICU | Hub | 83 | M | Rectal swab | February 2024 |
| Strains | Sample | Ward | atpA Allele | ddl Allele | gdh Allele | Purk Allele | gyd Allele | pstS Allele | adk Allele | ST |
|---|---|---|---|---|---|---|---|---|---|---|
| VREfm 12 | Bladder catheter | ICU | 9 | 1 | 1 | 1 | 1 | 0 | 1 | 1478 |
| VREfm 26 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 1 | 0 | 1 | 1478 |
| VREfm 45 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 52 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 56 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 58 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 63 | Abdominal fluid culture | Urology | 9 | 1 | 1 | 1 | 1 | 0 | 1 | 1478 |
| VREfm 71 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 72 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 1 | 0 | 1 | 1478 |
| VREfm 74 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 78 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 84 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 86 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 91 | Bladder catheter | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 92 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 95 | Bladder catheter | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 133 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 143 | Orotracheal culture | NICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 145 | Rectal swab | Medicine | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 147 | Rectal swab | Medicine | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 150 | Rectal swab | Surgery | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 189 | Rectal swab | Medicine | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 204 | Blood culture | Medicine | 9 | 141 | 1 | 1 | 12 | 1 | 1 | 2164 |
| VREfm 269 | Blood culture | NICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 328 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 348 | Bladder catheter | ICU | 9 | 141 | 1 | 1 | 12 | 1 | 1 | 2164 |
| 3.3VREfm 392 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 410 | Blood culture | Medicine | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 425 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 426 | Rectal swab | ICU | 9 | 1 | 1 | 1 | 12 | 1 | 1 | 80 |
| VREfm 440 | Rectal swab | ICU | 9 | 141 | 1 | 1 | 12 | 1 | 1 | 2164 |
| STs | PTs | Strains | N. of Strains |
|---|---|---|---|
| ST80 (identified for 24 strains) | 1 | VREfm 74 | 1 |
| 2 | VREfm 78 | 1 | |
| 3 | VREfm 150 | 1 | |
| 4 | VREfm 45, VREfm 52, VREfm 56 | 3 | |
| 5 | VREfm 58 | 1 | |
| 6 | VREfm 189 | 1 | |
| 7 | VREfm 95 | 1 | |
| 8 | VREfm 71 | 1 | |
| 13 | VREfm 133 | 1 | |
| 14 | VREfm 147 | 1 | |
| 15 | VREfm 91, VREfm 92 | 2 | |
| 16 | VREfm 143, VREfm 84 | 2 | |
| 17 | VREfm 145 | 1 | |
| 18 | VREfm 86 | 1 | |
| 19 | VREfm 425, VREfm 426 | 2 | |
| 20 | VREfm 328 | 1 | |
| 22 | VREfm 410 | 1 | |
| 23 | VREfm 392 | 1 | |
| 26 | VREfm 269 | 1 | |
| ST1478 (identified for 4 strains) | 9 | VREfm 12 | 1 |
| 10 | VREfm 72 | 1 | |
| 11 | VREfm 26 | 1 | |
| 12 | VREfm 63 | 1 | |
| ST2164 (identified for 3 strains) | 21 | VREfm 348 | 1 |
| 24 | VREfm 204 | 1 | |
| 25 | VREfm 440 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lombardi, A.; Ripabelli, G.; Sammarco, M.L.; Tamburro, M. Enterococcus faecium as an Emerging Pathogen: Molecular Epidemiology and Antimicrobial Resistance in Clinical Strains. Pathogens 2025, 14, 483. https://doi.org/10.3390/pathogens14050483
Lombardi A, Ripabelli G, Sammarco ML, Tamburro M. Enterococcus faecium as an Emerging Pathogen: Molecular Epidemiology and Antimicrobial Resistance in Clinical Strains. Pathogens. 2025; 14(5):483. https://doi.org/10.3390/pathogens14050483
Chicago/Turabian StyleLombardi, Adele, Giancarlo Ripabelli, Michela Lucia Sammarco, and Manuela Tamburro. 2025. "Enterococcus faecium as an Emerging Pathogen: Molecular Epidemiology and Antimicrobial Resistance in Clinical Strains" Pathogens 14, no. 5: 483. https://doi.org/10.3390/pathogens14050483
APA StyleLombardi, A., Ripabelli, G., Sammarco, M. L., & Tamburro, M. (2025). Enterococcus faecium as an Emerging Pathogen: Molecular Epidemiology and Antimicrobial Resistance in Clinical Strains. Pathogens, 14(5), 483. https://doi.org/10.3390/pathogens14050483






