Impact of Human Body Temperature on Stress Tolerance and Transcriptome of Cronobacter sakazakii
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. Construction of Gene Deletion Mutants
2.3. Growth Curves
2.4. Analysis of Acid Resistance
2.5. Analysis of Osmotic Stress Tolerance
2.6. Autoaggregation, Cell Surface Hydrophobicity
2.7. Sample Preparation for RNA-Seq
2.8. Mapping of RNA-Seq Libraries and Differential Gene Expression Analysis
2.9. Quantitative Real-Time PCR Verification
2.10. Statistical Analysis
3. Results
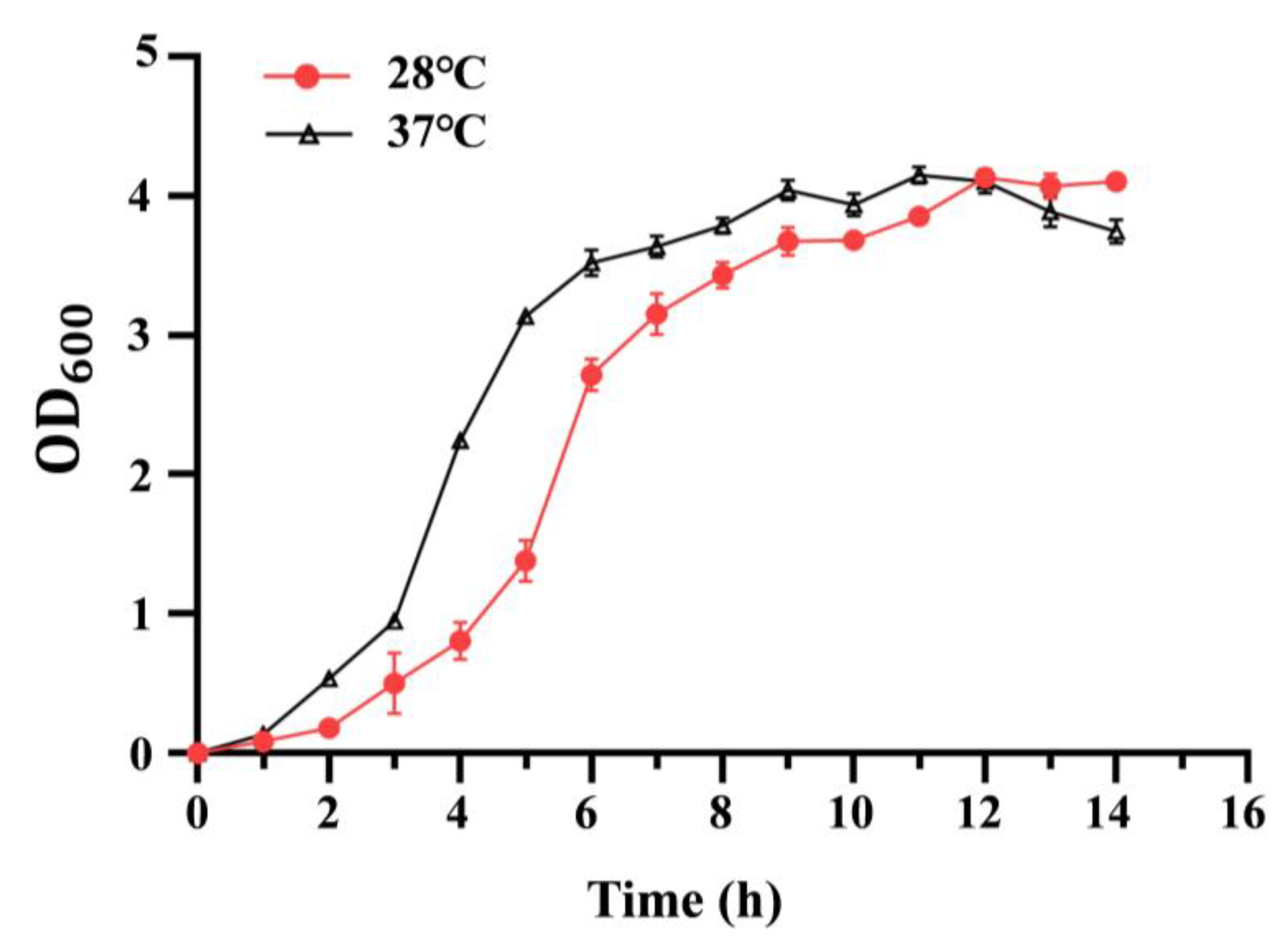
3.1. Effect of Temperature on Growth
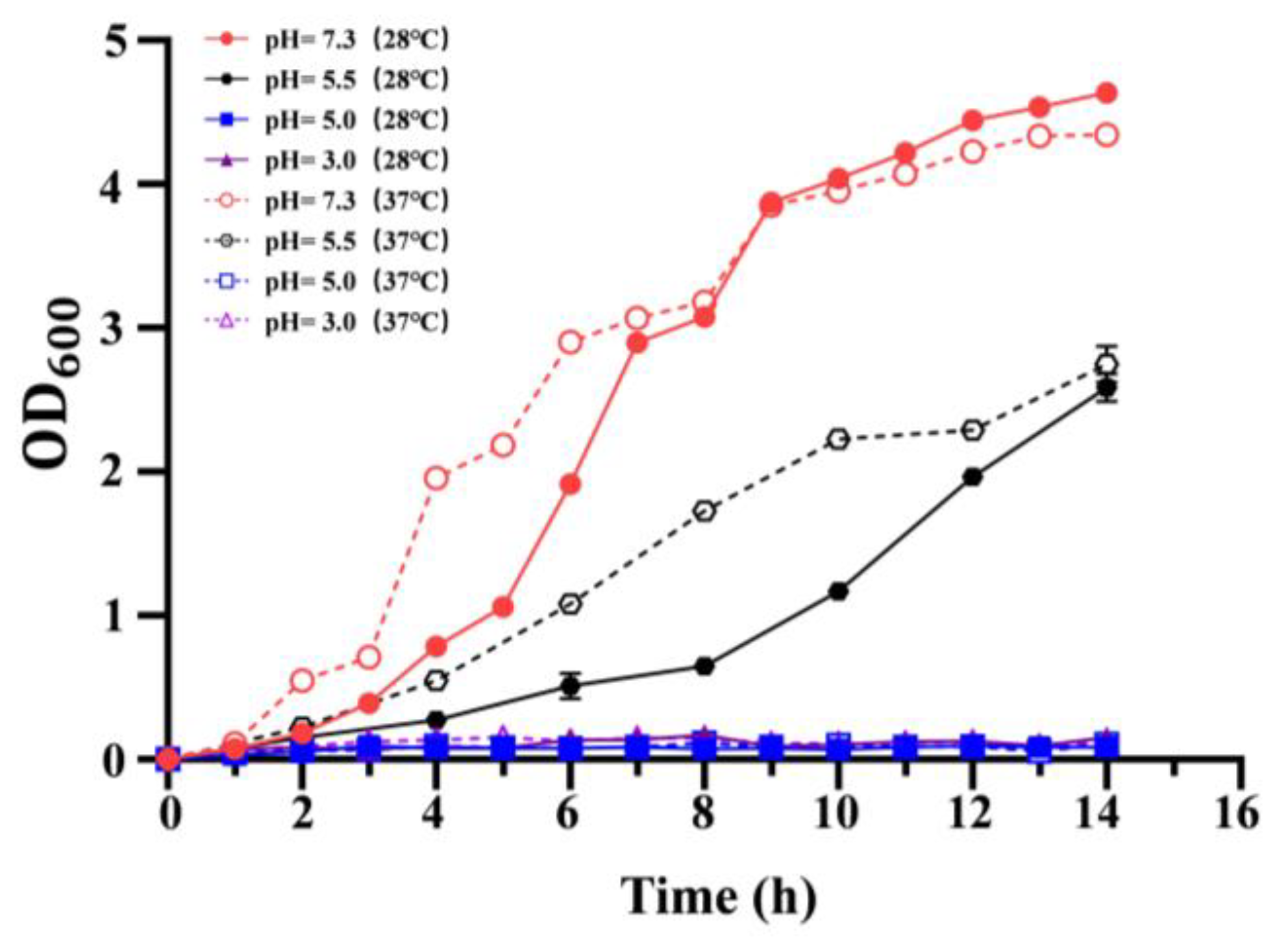
3.2. Effect of Temperature on the Acid Resistance
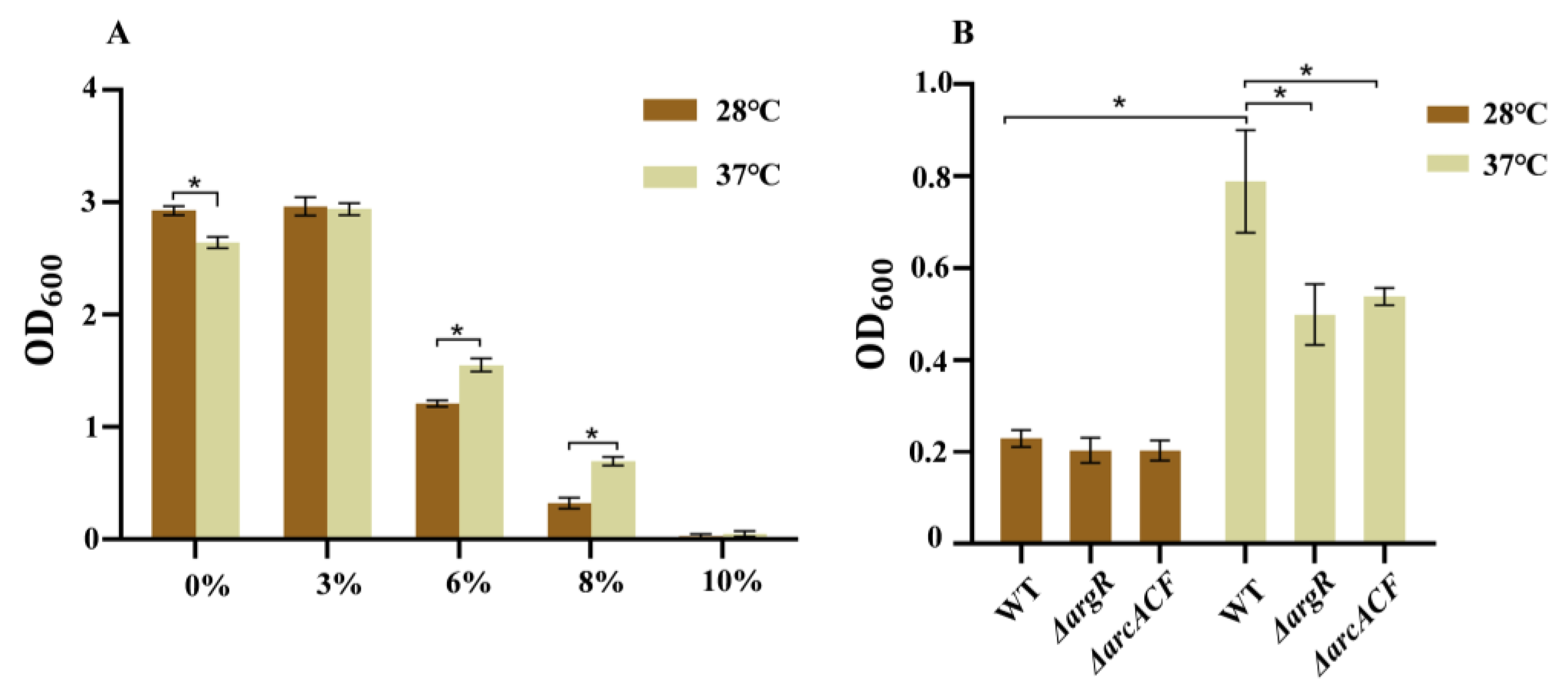
3.3. Effects of Temperature on Growth Under Osmotic Stress
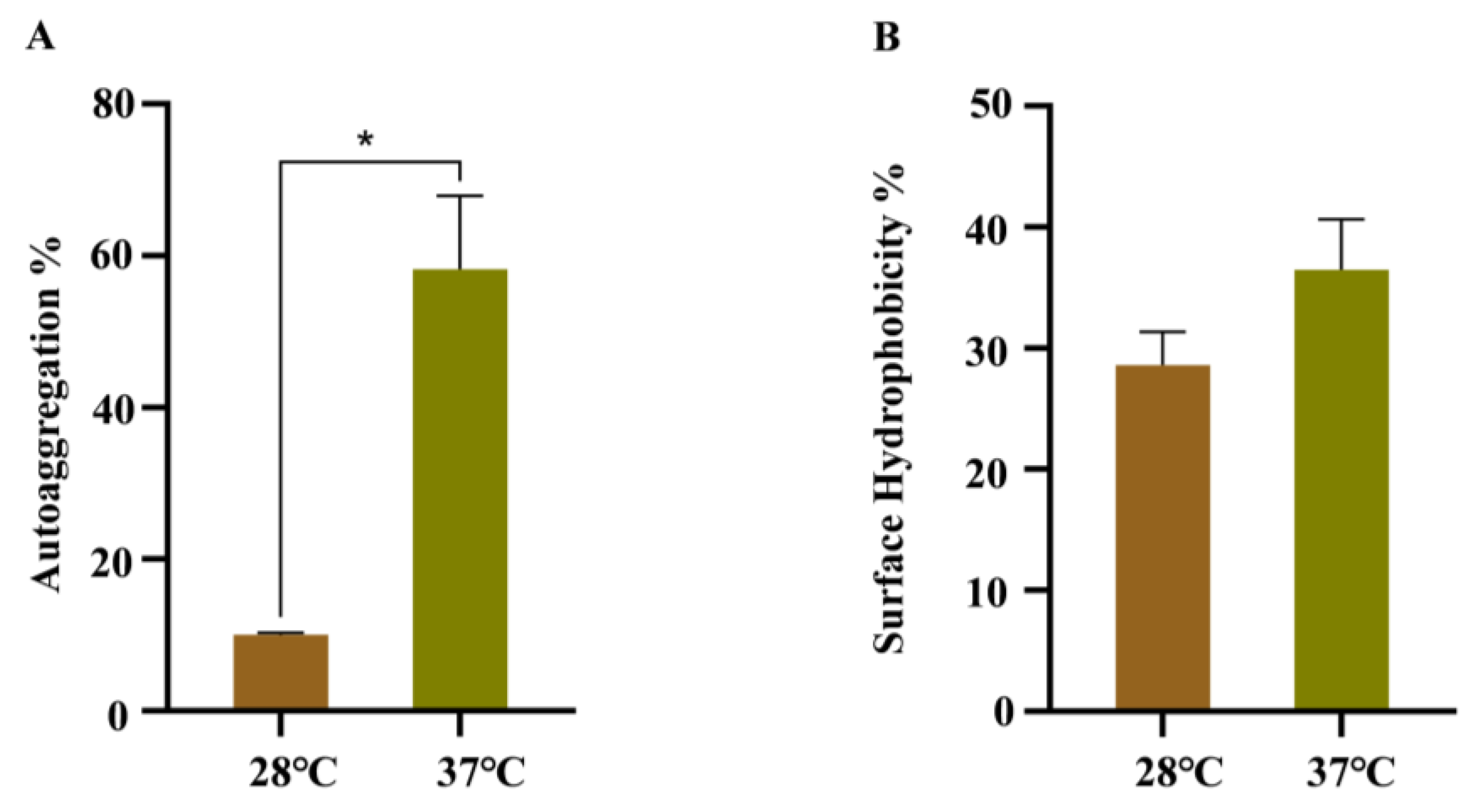
3.4. Effects of Temperature on the Autoaggregation, Cell Surface Hydrophobicity
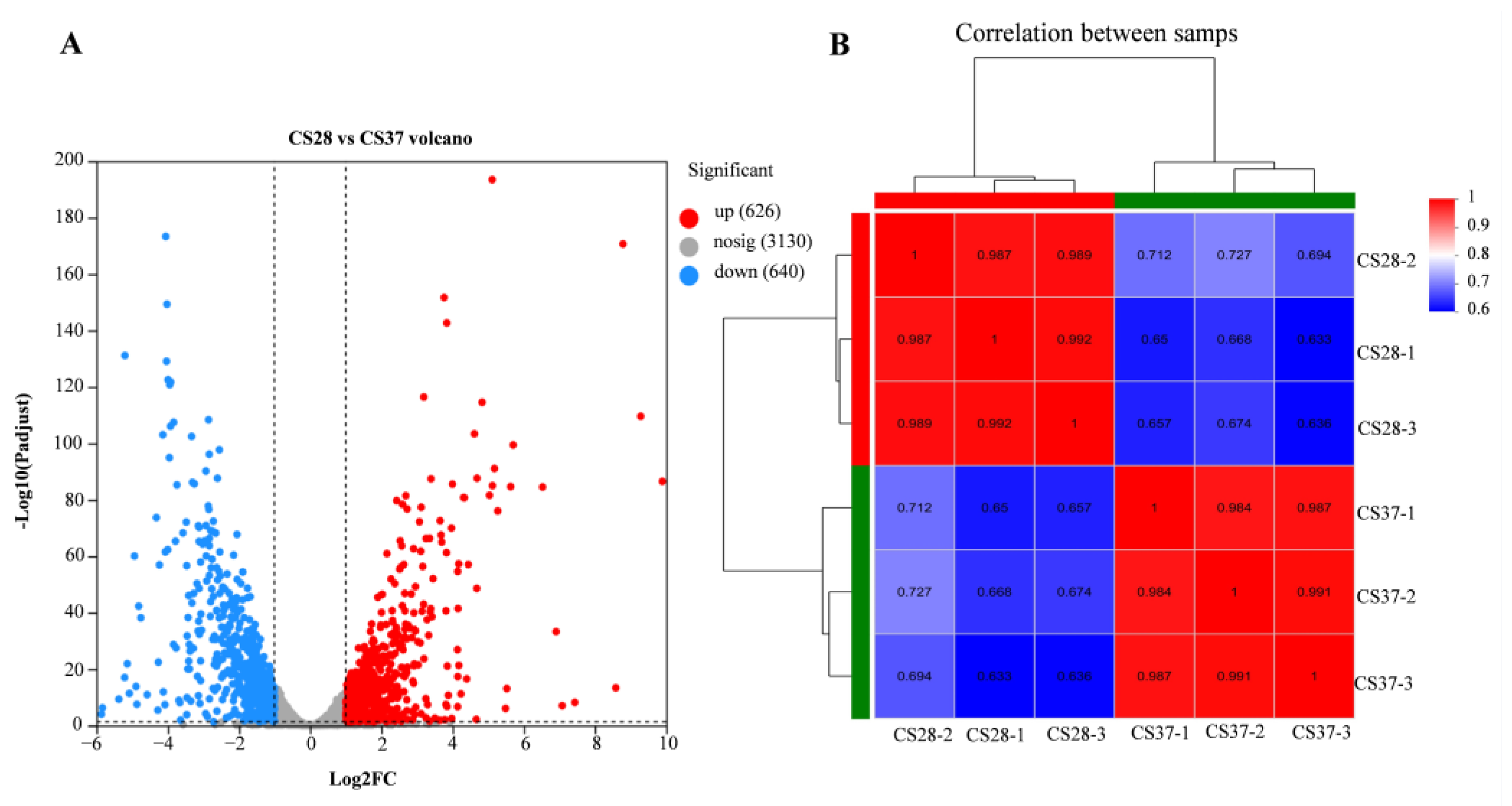
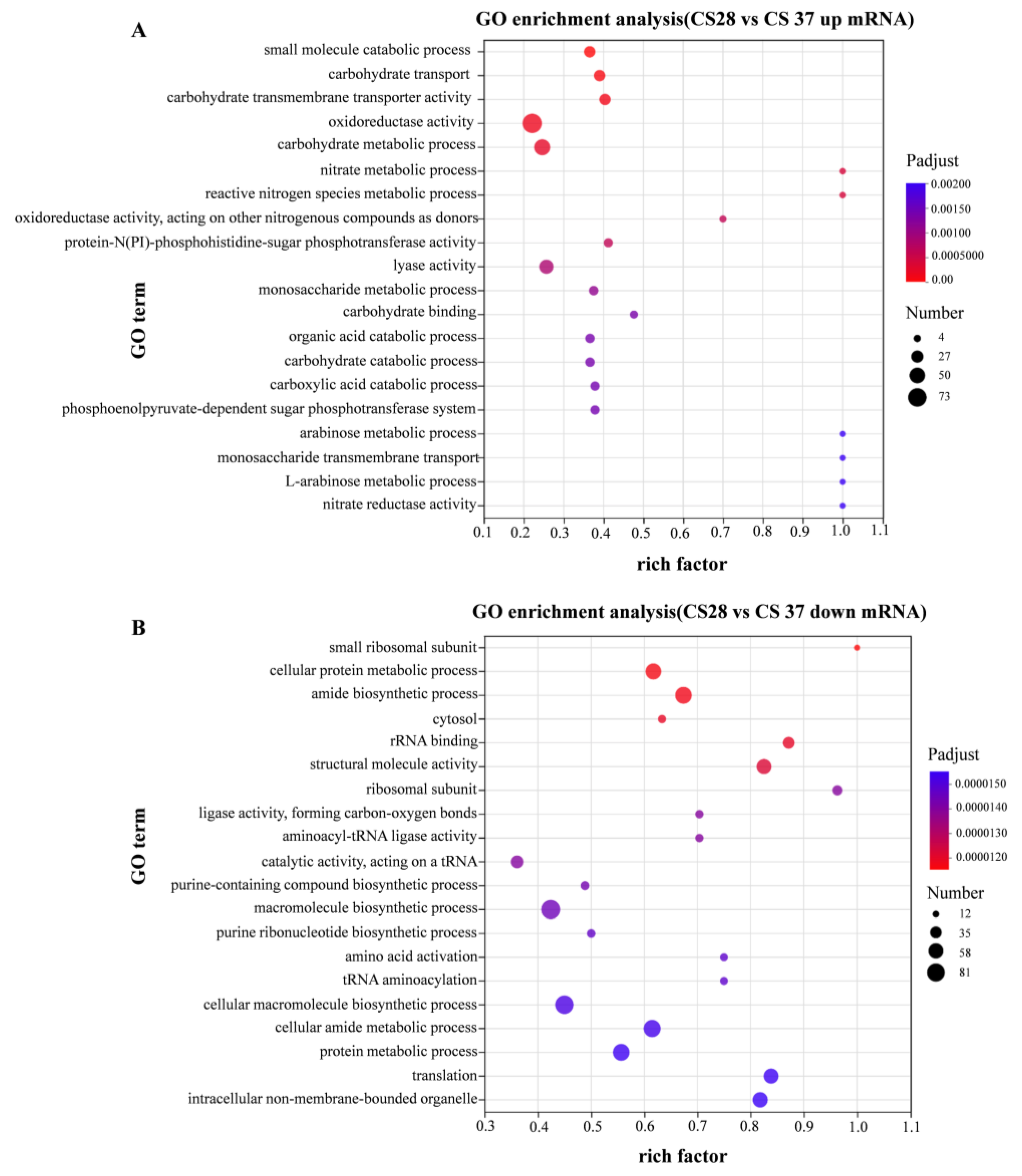
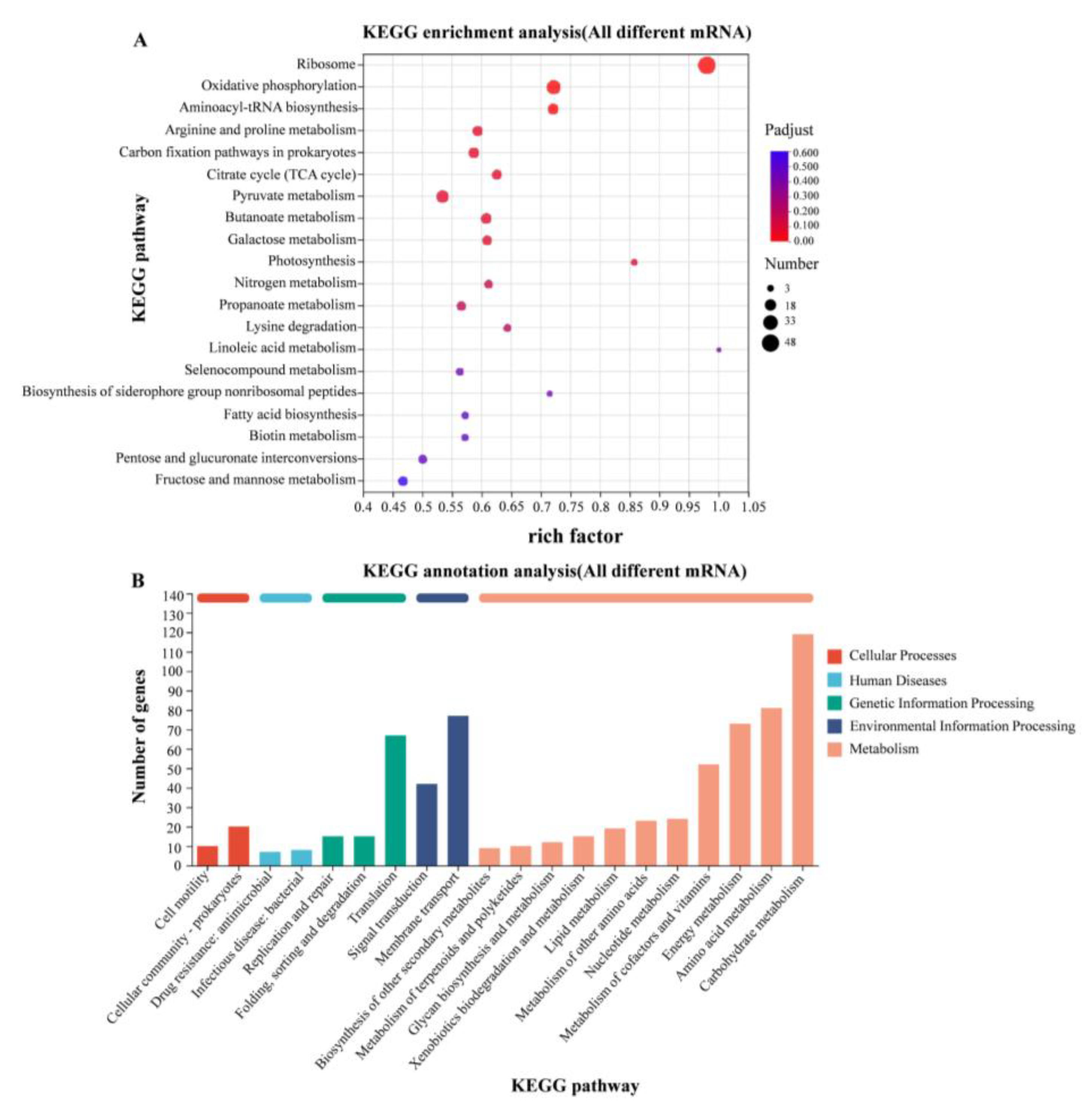
3.5. RNA-Seq Analysis of WT at 37 °C and 28 °C
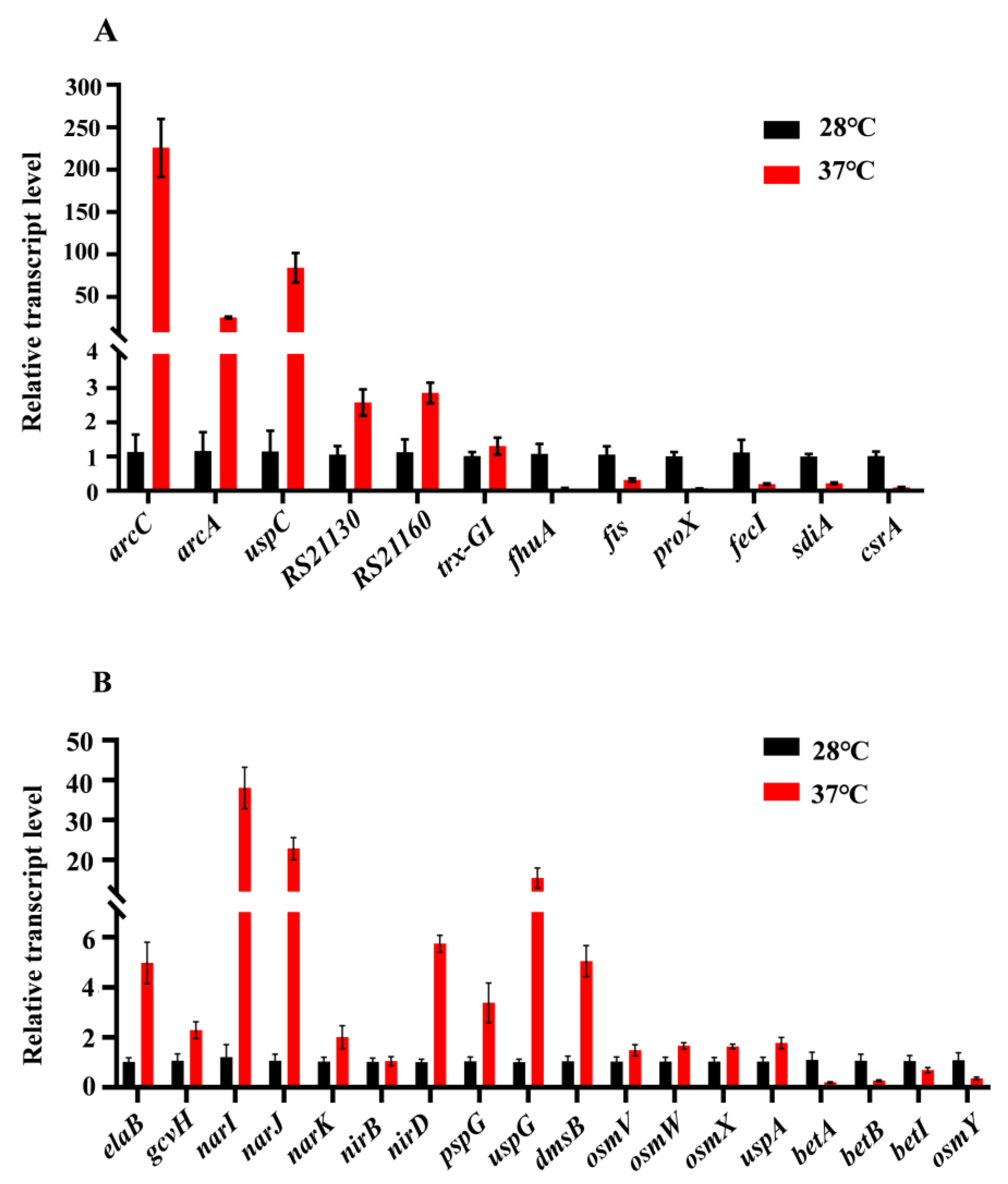
3.6. Verification of RNA-Seq Results by qRT-PCR
3.7. Definition of Transcriptional Units and the Analysis of 5′ Untranslated Regions (5′ UTRs)
3.8. Detection of Expressed Small RNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Du, X.; Wang, X.; Dong, X.; Li, P.; Wang, S. Characterization of the Desiccation Tolerance of Cronobacter Sakazakii Strains. Front. Microbiol. 2018, 9, 2867. [Google Scholar] [CrossRef] [PubMed]
- Strysko, J.; Cope, J.R.; Martin, H.; Tarr, C.; Hise, K.; Collier, S.; Bowen, A. Food Safety and Invasive Cronobacter Infections during Early Infancy, 1961–2018. Emerg. Infect. Dis. 2020, 26, 857–865. [Google Scholar] [CrossRef]
- Phair, K.; Pereira, S.G.; Kealey, C.; Fanning, S.; Brady, D.B. Insights into the Mechanisms of Cronobacter Sakazakii Virulence. Microb. Pathog. 2022, 169, 105643. [Google Scholar] [CrossRef] [PubMed]
- Almajed, F.S.; Forsythe, S.J. Cronobacter Sakazakii Clinical Isolates Overcome Host Barriers and Evade the Immune Response. Microb. Pathog. 2016, 90, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Henry, M.; Fouladkhah, A. Outbreak History, Biofilm Formation, and Preventive Measures for Control of Cronobacter Sakazakii in Infant Formula and Infant Care Settings. Microorganisms 2019, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, Y.-T.; Yoon, H.; Lee, J.-H.; Ryu, S. The Complete Genome Sequence of Cronobacter Sakazakii ATCC 29544T, a Food-Borne Pathogen, Isolated from a Child’s Throat. Gut Pathog. 2017, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, K.-P.; Choi, J.; Lim, J.-A.; Lee, J.; Hwang, S.; Ryu, S. Outer Membrane Proteins A (OmpA) and X (OmpX) Are Essential for Basolateral Invasion of Cronobacter Sakazakii. Appl. Environ. Microbiol. 2010, 76, 5188–5198. [Google Scholar] [CrossRef]
- Wang, B. Sialic Acid Is an Essential Nutrient for Brain Development and Cognition. Annu. Rev. Nutr. 2009, 29, 177–222. [Google Scholar] [CrossRef]
- Ji, X.; Lu, P.; Xue, J.; Zhao, N.; Zhang, Y.; Dong, L.; Zhang, X.; Li, P.; Hu, Y.; Wang, J.; et al. The Lipoprotein NlpD in Cronobacter Sakazakii Responds to Acid Stress and Regulates Macrophage Resistance and Virulence by Maintaining Membrane Integrity. Virulence 2021, 12, 415–429. [Google Scholar] [CrossRef]
- Ariadnna, C.; Juan, X.-C.; Bertha, G.-P.; Miriam, B.; Carlos, E.; Irma, R. Virulence Traits in Cronobacter Species Isolated from Different Sources. Can. J. Microbiol. 2011, 57, 9. [Google Scholar] [CrossRef]
- Pagotto, F.J.; Nazarowec-White, M.; Bidawid, S.; Farber, J.M. Enterobacter Sakazakii: Infectivity and Enterotoxin Production In Vitro and In Vivo. J. Food Prot. 2003, 66, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Zhang, X.; Zhang, M.; Ling, N.; Zeng, H.; Gao, J.; Jiao, R.; Wu, Q.; Zhang, J. Potential Factors Involved in Virulence of Cronobacter Sakazakii Isolates by Comparative Transcriptome Analysis. J. Dairy Sci. 2017, 100, 8826–8837. [Google Scholar] [CrossRef] [PubMed]
- Nechooshtan, G.; Elgrably-Weiss, M.; Sheaffer, A.; Westhof, E.; Altuvia, S. A pH-Responsive Riboregulator. Genes. Dev. 2009, 23, 2650–2662. [Google Scholar] [CrossRef] [PubMed]
- Wassarman, K.M. Small RNAs in Bacteria: Diverse Regulators of Gene Expression in Response to Environmental Changes. Cell 2002, 109, 141–144. [Google Scholar] [CrossRef]
- Konkel, M.E.; Tilly, K. Temperature-Regulated Expression of Bacterial Virulence Genes. Microbes Infect. 2000, 2, 157–166. [Google Scholar] [CrossRef]
- Pienkoß, S.; Javadi, S.; Chaoprasid, P.; Nolte, T.; Twittenhoff, C.; Dersch, P.; Narberhaus, F. The Gatekeeper of Yersinia Type III Secretion Is under RNA Thermometer Control. PLoS Pathog. 2021, 17, e1009650. [Google Scholar] [CrossRef]
- Wurtzel, O.; Yoder-Himes, D.R.; Han, K.; Dandekar, A.A.; Edelheit, S.; Greenberg, E.P.; Sorek, R.; Lory, S. The Single-Nucleotide Resolution Transcriptome of Pseudomonas Aeruginosa Grown in Body Temperature. PLoS Pathog. 2012, 8, e1002945. [Google Scholar] [CrossRef]
- Matanza, X.M.; Osorio, C.R. Exposure of the Opportunistic Marine Pathogen Photobacterium Damselae Subsp. Damselae to Human Body Temperature Is a Stressful Condition That Shapes the Transcriptome, Viability, Cell Morphology, and Virulence. Front. Microbiol. 2020, 11, 1771. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-Step Inactivation of Chromosomal Genes in Escherichia Coli K-12 Using PCR Products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Chen, W.; Liu, Z.; Sun, W.; Li, S.; Liu, J.; Huo, W.; Jia, J.; Shen, W.; Wang, Y.; Chen, G. Electrotransformation of Foodborne Pathogen Cronobacter Sakazakii by a Simple Method. Foodborne Pathog. Dis. 2024, 21, 61–67. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Genom. Biol. 2010, 10, R106. [Google Scholar] [CrossRef]
- Sorek, R.; Cossart, P. Prokaryotic Transcriptomics: A New View on Regulation, Physiology and Pathogenicity. Nat. Rev. Genet. 2010, 11, 9–16. [Google Scholar] [CrossRef]
- Caubilla-Barron, J.; Hurrell, E.; Townsend, S.; Cheetham, P.; Loc-Carrillo, C.; Fayet, O.; Prère, M.-F.; Forsythe, S.J. Genotypic and Phenotypic Analysis of Enterobacter Sakazakii Strains from an Outbreak Resulting in Fatalities in a Neonatal Intensive Care Unit in France. J. Clin. Microbiol. 2007, 45, 3979–3985. [Google Scholar] [CrossRef] [PubMed]
- Lang, E.; Guyot, S.; Peltier, C.; Alvarez-Martin, P.; Perrier-Cornet, J.-M.; Gervais, P. Cellular Injuries in Cronobacter Sakazakii CIP 103183T and Salmonella Enterica Exposed to Drying and Subsequent Heat Treatment in Milk Powder. Front. Microbiol. 2018, 9, 475. [Google Scholar] [CrossRef]
- Osaili, T.; Forsythe, S. Desiccation Resistance and Persistence of Cronobacter Species in Infant Formula. Int. J. Food Microbiol. 2009, 136, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Patil, P.P.; Kumar, S.; Kaur, A.; Midha, S.; Bansal, K.; Patil, P.B. Global Transcriptome Analysis of Stenotrophomonas Maltophilia in Response to Growth at Human Body Temperature. Microb. Genom. 2021, 7, 000600. [Google Scholar] [CrossRef]
- Dhungel, L.; Bonner, R.; Cook, M.; Henson, D.; Moulder, T.; Benbow, M.E.; Jordan, H. Impact of Temperature and Oxygen Availability on Gene Expression Patterns of Mycobacterium ulcerans. Microbiol. Spectr. 2023, 11, e04968-22. [Google Scholar] [CrossRef]
- Yan, T.; Li, M.; Wang, Q.; Wang, M.; Liu, L.; Ma, C.; Xiang, X.; Zhou, Q.; Liu, Z.; Gong, Z. Structures, Functions, and Regulatory Networks of Universal Stress Proteins in Clinically Relevant Pathogenic Bacteria. Cell. Signal. 2024, 116, 111032. [Google Scholar] [CrossRef]
- Nyström, T.; Neidhardt, F.C. Effects of Overproducing the Universal Stress Protein, UspA, in Escherichia Coli K-12. J. Bacteriol. 1996, 178, 927–930. [Google Scholar] [CrossRef]
- Kim, H.; Goo, E.; Kang, Y.; Kim, J.; Hwang, I. Regulation of Universal Stress Protein Genes by Quorum Sensing and RpoS in Burkholderia Glumae. J. Bacteriol. 2012, 194, 982–992. [Google Scholar] [CrossRef]
- Liu, W.-T.; Karavolos, M.H.; Bulmer, D.M.; Allaoui, A.; Hormaeche, R.D.C.E.; Lee, J.J.; Anjam Khan, C.M. Role of the Universal Stress Protein UspA of Salmonella in Growth Arrest, Stress and Virulence. Microb. Pathog. 2007, 42, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Boll, E.J.; Marti, R.; Hasman, H.; Overballe-Petersen, S.; Stegger, M.; Ng, K.; Knøchel, S.; Krogfelt, K.A.; Hummerjohann, J.; Struve, C. Turn Up the Heat—Food and Clinical Escherichia Coli Isolates Feature Two Transferrable Loci of Heat Resistance. Front. Microbiol. 2017, 8, 579. [Google Scholar] [CrossRef] [PubMed]
- Gajdosova, J.; Benedikovicova, K.; Kamodyova, N.; Tothova, L.; Kaclikova, E.; Stuchlik, S.; Turna, J.; Drahovska, H. Analysis of the DNA Region Mediating Increased Thermotolerance at 58 °C in Cronobacter Sp. and Other Enterobacterial Strains. Antonie Van Leeuwenhoek 2011, 100, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Orieskova, M.; Gajdosova, J.; Oslanecova, L.; Ondreickova, K.; Kaclikova, E.; Stuchlik, S.; Turna, J.; Drahovska, H. Function of Thermotolerance Genomic Island in Increased Stress Resistance of Cronobacter Sakazakii. J. Food Nutr. Res. 2013, 52, 37–44. [Google Scholar]
- Zhu, T.; Wang, Z.; McMullen, L.M.; Raivio, T.; Simpson, D.J.; Gänzle, M.G. Contribution of the Locus of Heat Resistance to Growth and Survival of Escherichia Coli at Alkaline pH and at Alkaline pH in the Presence of Chlorine. Microorganisms 2021, 9, 701. [Google Scholar] [CrossRef]
- Zhou, A.; Cao, Y.; Zhou, D.; Hu, S.; Tan, W.; Xiao, X.; Yu, Y.; Li, X. Global Transcriptomic Analysis of Cronobacter Sakazakii CICC 21544 by RNA-Seq under Inorganic Acid and Organic Acid Stresses. Food Res. Int. 2020, 130, 108963. [Google Scholar] [CrossRef]
- Luiz de Freitas, L.; Aparecida dos Santos, C.I.; Carneiro, D.G.; Dantas Vanetti, M.C. Nisin and Acid Resistance in Salmonella Is Enhanced by N-Dodecanoyl-Homoserine Lactone. Microb. Pathog. 2020, 147, 104320. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; Chen, K.; Zhang, L.; Zhang, Y.; Wang, X.; Xia, X. The Role of PhoP/PhoQ Two Component System in Regulating Stress Adaptation in Cronobacter Sakazakii. Food Microbiol. 2021, 100, 103851. [Google Scholar] [CrossRef]
- Ling, N.; Zhang, J.; Li, C.; Zeng, H.; He, W.; Ye, Y.; Wu, Q. The Glutaredoxin Gene, grxB, Affects Acid Tolerance, Surface Hydrophobicity, Auto-Aggregation, and Biofilm Formation in Cronobacter Sakazakii. Front. Microbiol. 2018, 9, 133. [Google Scholar] [CrossRef]
- Alvarez-Ordóñez, A.; Cummins, C.; Deasy, T.; Clifford, T.; Begley, M.; Hill, C. Acid Stress Management by Cronobacter Sakazakii. Int. J. Food Microbiol. 2014, 178, 21–28. [Google Scholar] [CrossRef]
- Waterman, S.R.; Small, P.L.C. Acid-Sensitive Enteric Pathogens Are Protected from Killing under Extremely Acidic Conditions of pH 2.5 When They Are Inoculated onto Certain Solid Food Sources. Appl. Environ. Microbiol. 1998, 64, 3882–3886. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, Y.; Zhan, W.; Wood, T.K.; Wang, X. Resistance to Oxidative Stress by Inner Membrane Protein ElaB Is Regulated by OxyR and RpoS. Microb. Biotechnol. 2019, 12, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Flores-Kim, J.; Darwin, A.J. The Phage Shock Protein Response. Annu. Rev. Microbiol. 2016, 70, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Darwin, A.J.; Miller, V.L. The Psp Locus of Yersinia Enterocolitica Is Required for Virulence and for Growth in Vitro When the Ysc Type III Secretion System Is Produced. Mol. Microbiol. 2001, 39, 429–445. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Teng, J.L.L.; Watt, R.M.; Liu, C.; Lau, S.K.P.; Woo, P.C.Y. Molecular Characterization of Arginine Deiminase Pathway in Aribacter Hongkongensis and Unique Regulation of Arginine Catabolism and Anabolism by Multiple Environmental Stresses. Environ. Microbiol. 2015, 17, 4469–4483. [Google Scholar] [CrossRef]
- Xu, B.; Yang, X.; Zhang, P.; Ma, Z.; Lin, H.; Fan, H. The Arginine Deiminase System Facilitates Environmental Adaptability of Streptococcus Equi Ssp. Zooepidemicus through pH Adjustment. Res. Microbiol. 2016, 167, 403–412. [Google Scholar] [CrossRef]
- Burne, R.A.; Parsons, D.T.; Marquis, R.E. Cloning and Expression in Escherichia Coli of the Genes of the Arginine Deiminase System of Streptococcus Sanguis NCTC 10904. Infect. Immun. 1989, 57, 3540–3548. [Google Scholar] [CrossRef]
- Okamura-Ikeda, K.; Ohmura, Y.; Fujiwara, K.; Motokawa, Y. Cloning and Nucleotide Sequence of the Gcv Operon Encoding the Escherichia Coli Glycine-Cleavage System. Eur. J. Biochem. 1993, 216, 539–548. [Google Scholar] [CrossRef]
- Yadav, U.; Sundd, M. Backbone Chemical Shift Assignments of the Glycine Cleavage Complex H Protein of Escherichia Coli. Biomol. NMR Assign. 2018, 12, 163–165. [Google Scholar] [CrossRef]
- Elhosseiny, N.M.; Amin, M.A.; Yassin, A.S.; Attia, A.S. Acinetobacter Baumannii Universal Stress Protein A Plays a Pivotal Role in Stress Response and Is Essential for Pneumonia and Sepsis Pathogenesis. Int. J. Med. Microbiol. 2015, 305, 114–123. [Google Scholar] [CrossRef]
- Seifart Gomes, C.; Izar, B.; Pazan, F.; Mohamed, W.; Mraheil, M.A.; Mukherjee, K.; Billion, A.; Aharonowitz, Y.; Chakraborty, T.; Hain, T. Universal Stress Proteins Are Important for Oxidative and Acid Stress Resistance and Growth of Listeria Monocytogenes EGD-e In Vitro and In Vivo. PLoS ONE 2011, 6, e24965. [Google Scholar] [CrossRef] [PubMed]
- Burgess, C.M.; Gianotti, A.; Gruzdev, N.; Holah, J.; Knøchel, S.; Lehner, A.; Margas, E.; Esser, S.S.; Sela (Saldinger), S.; Tresse, O. The Response of Foodborne Pathogens to Osmotic and Desiccation Stresses in the Food Chain. Int. J. Food Microbiol. 2016, 221, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Frossard, S.M.; Khan, A.A.; Warrick, E.C.; Gately, J.M.; Hanson, A.D.; Oldham, M.L.; Sanders, D.A.; Csonka, L.N. Identification of a Third Osmoprotectant Transport System, the OsmU System, in Salmonella Enterica. J. Bacteriol. 2012, 194, 3861–3871. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Ling, N.; Gao, J.; Zhang, X.; Zhang, M.; Tong, L.; Zeng, H.; Zhang, J.; Wu, Q. Roles of Outer Membrane Protein W (OmpW) on Survival, Morphology, and Biofilm Formation under NaCl Stresses in Cronobacter Sakazakii. J. Dairy Sci. 2018, 101, 3844–3850. [Google Scholar] [CrossRef]
- Heermann, R.; Weber, A.; Mayer, B.; Ott, M.; Hauser, E.; Gabriel, G.; Pirch, T.; Jung, K. The Universal Stress Protein UspC Scaffolds the KdpD/KdpE Signaling Cascade of Escherichia Coli under Salt Stress. J. Mol. Biol. 2009, 386, 134–148. [Google Scholar] [CrossRef]
- Martínez, L.; Vadyvaloo, V. Mechanisms of Post-Transcriptional Gene Regulation in Bacterial Biofilms. Front. Cell. Infect. Microbiol. 2014, 4, 38. [Google Scholar] [CrossRef]
- Schulmeyer, K.H.; Yahr, T.L. Post-Transcriptional Regulation of Type III Secretion in Plant and Animal Pathogens. Curr. Opin. Microbiol. 2017, 36, 30–36. [Google Scholar] [CrossRef]
- Han, Y.; Li, C.; Yan, Y.; Lin, M.; Ke, X.; Zhang, Y.; Zhan, Y. Post-Transcriptional Control of Bacterial Nitrogen Metabolism by Regulatory Noncoding RNAs. World J. Microbiol. Biotechnol. 2022, 38, 126. [Google Scholar] [CrossRef]
- Mellin, J.R.; Koutero, M.; Dar, D.; Nahori, M.-A.; Sorek, R.; Cossart, P. Sequestration of a Two-Component Response Regulator by a Riboswitch-Regulated Noncoding RNA. Science 2014, 345, 940–943. [Google Scholar] [CrossRef]
- Mellin, J.R.; Tiensuu, T.; Bécavin, C.; Gouin, E.; Johansson, J.; Cossart, P. A Riboswitch-Regulated Antisense RNA in Listeria Monocytogenes. Proc. Natl. Acad. Sci. USA 2013, 110, 13132–13137. [Google Scholar] [CrossRef]
- Grosso-Becerra, M.V.; Croda-García, G.; Merino, E.; Servín-González, L.; Mojica-Espinosa, R.; Soberón-Chávez, G. Regulation of Pseudomonas Aeruginosa Virulence Factors by Two Novel RNA Thermometers. Proc. Natl. Acad. Sci. USA 2014, 111, 15562–15567. [Google Scholar] [CrossRef]
- Pienkoß, S.; Javadi, S.; Chaoprasid, P.; Holler, M.; Roßmanith, J.; Dersch, P.; Narberhaus, F. RNA Thermometer-Coordinated Assembly of the Yersinia Injectisome. J. Mol. Biol. 2022, 434, 167667. [Google Scholar] [CrossRef] [PubMed]
| Strain or Plasmids | Genotype or Characteristics | Sources |
|---|---|---|
| Cronobacter sakazakii | ||
| WT | Wild-type strain C. sakazakii ATCC29544 | ATCC |
| ΔargR | argR deletion mutant | this study |
| ΔargACF | argACF deletion mutant | this study |
| Pasmids | ||
| pKD46 | λ red recombinase, AmpR | [19] |
| pKD3 | template plasmid for CmR | [19] |
| Primer | Primer Sequences (5′-3′) | Amplification Size (bp) |
|---|---|---|
| pKD3-argR-H1U | CGCAGGAGTGACGGTCCATGAAGGATTACGGTGATTATTCCGTGTAGGCTGGAGCTGCT | 1115 |
| pKD3-argR-H1D | GAAATGCACACTTTAGCCGCAGGAATCGATTGCTGTGAATAATGGGAATTAGCCATGGT | |
| pKD3-argR-H2U | CGGGCCGGGAAAACAATATCGTTTTTCTTCAACTTTCATCAACGCAGGAGTGACGGTCC | 1197 |
| pKD3-argR-H2D | TGTCCGGCATTATACGCATGTGCGGTTAGCTGACAAGCAGGAAATGCACACTTTAGCCG | |
| pKD3-arcACF-H1U | ATGCCGCCGTTTTTATTCTGAAAATCAGCCAAGGAATAAATGTGTAGGCTGGAGCTGCT | 1115 |
| pKD3-arcACF-H1D | TCTCCGAAGCGGCGGGCGCACCGGCGGCGCGCCCGCAAAGGATGGGAATTAGCCATGGT | |
| pKD3-arcACF-H2U | ATCTGCACGGTAACTCGTTATGCGAACACGCGTCGCATAATGCCGCCGTTTTTATTCTG | 1196 |
| pKD3-arcACF-H2D | ATGGTGTAAGCGGAAGGAAATTTGAACCTGTGCATGAGGGTATCTCCGAAGCGGCGGGC | |
| pKD3-F | GTGTAGGCTGGAGCTGCT | 1033 |
| pKD3-R | ATGGGAATTAGCCATGGT | |
| arcC-F | CGATATTCAGCGTCATAAC | 161 |
| arcC-R | GAATGTCGAGCGGATAAG | |
| argA-F | GATATTGACACCTTCTCC | 160 |
| argA-R | TGATGAGTCGTATCTGAT | |
| uspC-F | GGAACTCTTTAATCAAATGT | 151 |
| uspC-R | TATATTCTCCAGGCTCTC | |
| RS21130-F | GACAAGCAGTACAAGATTA | 124 |
| RS21130-R | CTTCCTTCTTCTCCTGTT | |
| RS21160-F | TATCAAACAGCCACAATAC | 158 |
| RS21160-R | CCTTGTCATCTACCTGTG | |
| trx-GI-F | AAGAATGAGCTTCACCTC | 81 |
| trx-GI-R | GTTGGGATGTTCCGATAG | |
| fhuA-F | GTGAACTTCCTCTATGAC | 165 |
| fhuA-R | ATCGTTAATCTCAGCAAG | |
| proX-F | CTTTCAAACGGCAATAAC | 122 |
| proX-R | GTAATCGGCAACTTCATT | |
| fecI-F | TTGATGCAGATGACATTG | 160 |
| fecI-R | ATCTCCAGATACGCTTTT | |
| sdiA-F | CTGGAAATGAAACTGAGTAAAC | 179 |
| sdiA-R | GCAGCATAACAGGCAATT | |
| csrA-F | TGACCGTGACAGTTCTTG | 104 |
| csrA-R | CTGGATACGCTGGTAGAT | |
| elaB-F | ATGCCTTTATCTTCACAA | 128 |
| elaB-R | GCTTTCAGTTCAACATAC | |
| gcvH-F | TGAAATACAGCAAAGAACA | 113 |
| gcvH-R | CAGATCGACAAACACCAT | |
| narI-F | GCTGATATTCTGATCCTG | 108 |
| narI-R | CACCAGTTTCATCATCTC | |
| narJ-F | GAAGAGCAGGTGAAATTC | 112 |
| narJ-R | CGGCAGAGATATTCAGATA | |
| narK-F | CGGTCAATCTGAATAAGG | 172 |
| narK-R | CACGGAATAATCAGGATG | |
| nirB-F | GATATGACACGATGACAA | 121 |
| nirB-R | CCAAAGACGATGATATGA | |
| nirD-F | TATGCCATCAGCAATATC | 149 |
| nirD-R | CTTTCGTCTTCCATACAC | |
| pspG-F | GTGGCTTTCTTTCTGATG | 126 |
| pspG-R | CGGCAACACTTTAATCAT | |
| uspG-F | TACAACACCATTCTGATG | 188 |
| uspG-R | GCCTCTAAATGTTCTTCA | |
| dmsB-F | CTCTCAATAGCCTGTAAC | 196 |
| dmsB-R | CGTCGCATTTAGTCATAT | |
| osmV-F | GGTAAAGCGGAAGATAAC | 131 |
| osmV-R | AGATAATCGGCGATATAATC | |
| osmW-F | GATGAATATCGGCGTGAT | 152 |
| osmW-R | GTAACCAGTCCAGAATAATG | |
| osmX-F | GCTGATTATCTTCAACCA | 129 |
| osmX-R | AATGCGTAGGTATTGTTC | |
| uspA-F | TGGTAGATAAAGCGGTAT | 196 |
| uspA-R | GTTTCGGTAATCGGATAG | |
| betA-F | ATCACCTGGAGATGTATC | 200 |
| betA-R | GAAGTGGTACTGAATGTT | |
| betB-F | GCTTGCAGAAATTTACAC | 124 |
| betB-R | GGTGAACGAGACTTTATC | |
| betI-F | GCAGGCAGTTAATTGATG | 120 |
| betI-R | TAAAGTAGTGGCTGATAATG | |
| osmY-F | CTGTTGTATTGGGTTCTG | 200 |
| osmY-R | GGTGCTCTTAATCTGTTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Wang, Y.; Yang, Y.; Yu, X.; Liu, J.; Jiang, M.; Zhang, J.; Yun, G.; Han, Y.; Wang, H.; et al. Impact of Human Body Temperature on Stress Tolerance and Transcriptome of Cronobacter sakazakii. Pathogens 2025, 14, 281. https://doi.org/10.3390/pathogens14030281
Li S, Wang Y, Yang Y, Yu X, Liu J, Jiang M, Zhang J, Yun G, Han Y, Wang H, et al. Impact of Human Body Temperature on Stress Tolerance and Transcriptome of Cronobacter sakazakii. Pathogens. 2025; 14(3):281. https://doi.org/10.3390/pathogens14030281
Chicago/Turabian StyleLi, Siqi, Yuanyuan Wang, Yahao Yang, Xinlu Yu, Jiajia Liu, Meiling Jiang, Jing Zhang, Ge Yun, Yufei Han, Heng Wang, and et al. 2025. "Impact of Human Body Temperature on Stress Tolerance and Transcriptome of Cronobacter sakazakii" Pathogens 14, no. 3: 281. https://doi.org/10.3390/pathogens14030281
APA StyleLi, S., Wang, Y., Yang, Y., Yu, X., Liu, J., Jiang, M., Zhang, J., Yun, G., Han, Y., Wang, H., Xie, Q., & Chen, G. (2025). Impact of Human Body Temperature on Stress Tolerance and Transcriptome of Cronobacter sakazakii. Pathogens, 14(3), 281. https://doi.org/10.3390/pathogens14030281





