Diagnostic Approaches for Measles Virus: Methods, Advances, and Ongoing Challenges
Abstract
1. Introduction
2. Clinical Symptoms, Vaccination and Management
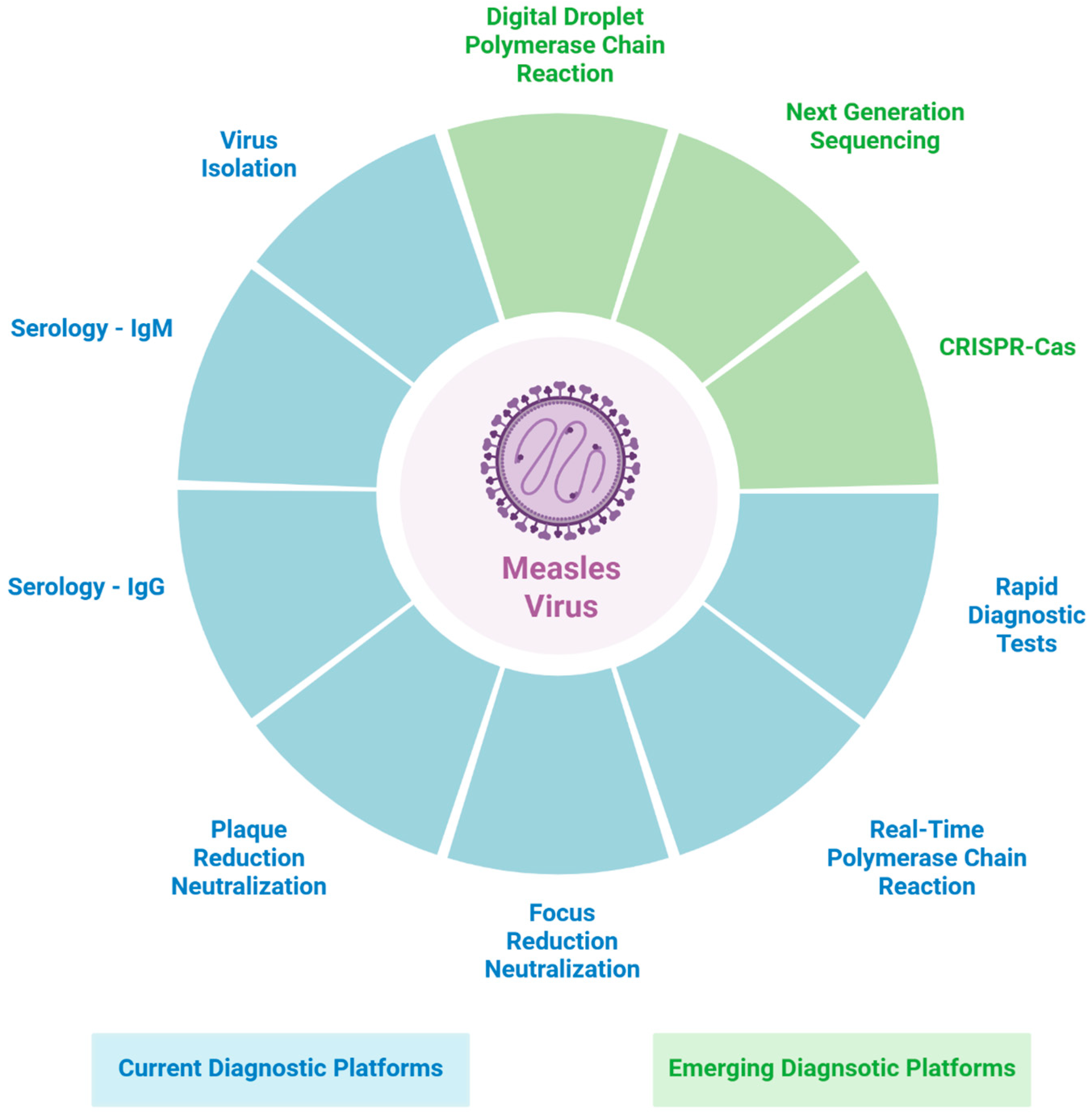
3. Current Diagnostic Methods
3.1. Virus Isolation
3.2. Serology
3.3. Plaque Reduction Neutralization
3.4. Real-Time Reverse Transcriptase-Polymerase Chain Reaction
3.5. Rapid Diagnostic Tests
4. Emerging Diagnostic Platforms
4.1. Digital Droplet Polymerase Chain Reactions
4.2. Next-Generation Sequencing
4.3. CRISPR-Based Diagnostics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BSL-2 | Biosafety level 2 |
| CLIA | Chemiluminescent immunoassay |
| CSF | Cerebrospinal fluid |
| ddPCR | Digital droplet polymerase chain reaction |
| EIA | Enzyme immunoassay |
| FRNT | Focus reduction neutralization test |
| LOD | Limit of detection |
| MeV | Measles virus |
| N | Nucleocapsid protein |
| NGS | Next-generation sequencing |
| PRNT | Plaque reduction neutralization test |
| RDT | Rapid diagnostic test |
| rRT-PCR | Real-time reverse transcriptase polymerase chain reaction |
| SSPE | Subacute sclerosing panencephalitis |
| US CDC | U.S. Centers for Disease Control and Prevention |
| WHO | World Health Organization |
References
- Dunn, J.J.; Baldanti, F.; Puchhammer, E.; Panning, M.; Perez, O.; Harvala, H.; Practice Pan American Society for Clinical Virology Clinical, Committee Public Policy; Committee the European Society for Clinical Virology Executive. Measles is Back—Considerations for Laboratory Diagnosis. J. Clin. Virol. 2020, 128, 104430. [Google Scholar] [CrossRef] [PubMed]
- Do, L.A.H.; Mulholland, K. Measles 2025. N. Engl. J. Med. 2025. [Google Scholar] [CrossRef]
- Ferren, M.; Horvat, B.; Mathieu, C. Measles Encephalitis: Towards New Therapeutics. Viruses 2019, 11, 1017. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Pokharel, Y.R. Measles and rubella: From Global Health Challenges to Advancements in Molecular Diagnostics in the Elimination Era. Mol. Ther. Nucleic Acids 2025, 36, 102698. [Google Scholar] [CrossRef]
- Zubach, V.; Beirnes, J.; Hayes, S.; Severini, A.; Hiebert, J. Diagnostic Accuracy of Commercially Available Serological Tests for the Detection of Measles and Rubella Viruses: A Systematic Review and Meta-Analysis. J. Clin. Microbiol. 2024, 62, e0133923. [Google Scholar] [CrossRef]
- US Centers for Disease Control and Prevention: Laborartory Testing for Measles. Available online: https://www.cdc.gov/measles/php/laboratories/index.html (accessed on 10 December 2025).
- World Health Organization. Manual for the Laboratory-Based Surveillance of Measles, Rubella, and Congenital Rubella Syndrome; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Fappani, C.; Gori, M.; Canuti, M.; Terraneo, M.; Colzani, D.; Tanzi, E.; Amendola, A.; Bianchi, S. Breakthrough Infections: A Challenge Towards Measles Elimination? Microorganisms 2022, 10, 1567. [Google Scholar] [CrossRef]
- Albrecht, P.; Herrmann, K.; Burns, G.R. Role of Virus Strain in Conventional and Enhanced Measles Plaque Neutralization Test. J. Virol. Methods 1981, 3, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, S.R.; Kumbhar, N.S.; Bhide, V.S. Detection of Measles, Mumps and Rubella Viruses by Immuno-Colorimetric Assay and Its Application in Focus Reduction Neutralization Tests. Microbiol. Immunol. 2014, 58, 666–674. [Google Scholar] [CrossRef]
- Terletskaia-Ladwig, E.; Enders, G.; Meier, S.; Dietz, K.; Enders, M. Development and Evaluation of an Automatable Focus Reduction Neutralisation Test for the Detection of Measles Virus Antibodies Using Imaging Analysis. J. Virol. Methods 2011, 178, 124–128. [Google Scholar] [CrossRef]
- Bankamp, B.; Takeda, M.; Zhang, Y.; Xu, W.; Rota, P.A. Genetic Characterization of Measles Vaccine Strains. J. Infect. Dis. 2011, 204, S533–S548. [Google Scholar] [CrossRef]
- Perez-Rodriguez, F.J.; Cherpillod, P.; Thomasson, V.; Vetter, P.; Schibler, M. Identification of a Measles Variant Displaying Mutations Impacting Molecular Diagnostics, Geneva, Switzerland, 2023. Eurosurveillance 2024, 29, 2400034. [Google Scholar] [CrossRef] [PubMed]
- Roy, F.; Mendoza, L.; Hiebert, J.; McNall, R.J.; Bankamp, B.; Connolly, S.; Ludde, A.; Friedrich, N.; Mankertz, A.; Rota, P.A.; et al. Rapid Identification of Measles Virus Vaccine Genotype by Real-Time PCR. J. Clin. Microbiol. 2017, 55, 735–743. [Google Scholar] [CrossRef]
- Alves, A.D.R.; Raposo, J.V.; de Sousa, R.M.P.; Cardoso, C.A.A.; Costa, P.; Araujo, J.M.; Barreiro, S.T.A.; Bressan, C.D.S.; Calvet, G.A.; de Souza, R.V.; et al. Beyond Arboviruses: A Multicenter Study to Evaluate Differential Diagnosis of Rash Diseases and Acute Febrile Illness Cases in Rio de Janeiro, Brazil. PLoS ONE 2022, 17, e0271758. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.E.; Zubach, V.; Lew, B.; Hasso, M.; Olsha, R.; Salvadori, M.I.; Manoj, N.; Hiebert, J. Measles Vaccine Virus Mutation Following Vaccination in a Healthy Child Resulting in a False Negative Vaccine Specific PCR Test: Ontario, Canada, 2025. Eurosurveillance 2025, 30, 2500774. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.S.; Lopareva, E.N.; Hwang, H.; Hart, D.; de Almeida, M.; Anderson, R.; Rota, P.A.; Bankamp, B. Evaluation of the Sensitivity of a Measles Diagnostic Real-Time RT-PCR Assay Incorporating Recently Observed Priming Mismatch Variants, 2024. Eurosurveillance 2024, 29, 2400410. [Google Scholar] [CrossRef]
- Rachlin, A.; Hampton, L.M.; Rota, P.A.; Mulders, M.N.; Papania, M.; Goodson, J.L.; Krause, L.K.; Hanson, M.; Osborn, J.; Kelly-Cirino, C.; et al. Use of Measles and Rubella Rapid Diagnostic Tests to Improve Case Detection and Targeting of Vaccinations. Vaccines 2024, 12, 823. [Google Scholar] [CrossRef]
- Koczula, K.M.; Gallotta, A. Lateral Flow Assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar] [CrossRef]
- Senin, A.; Noordin, N.M.; Sani, J.A.M.; Mahat, D.; Donadel, M.; Scobie, H.M.; Omar, A.; Chem, Y.K.; Zahari, M.I.; Ismail, F.; et al. A Measles IgM Rapid Diagnostic Test to Address Challenges with National Measles Surveillance and Response in Malaysia. PLoS ONE 2024, 19, e0298730. [Google Scholar] [CrossRef]
- Quan, P.L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef]
- Kojabad, A.A.; Farzanehpour, M.; Galeh, H.E.G.; Dorostkar, R.; Jafarpour, A.; Bolandian, M.; Nodooshan, M.M. Droplet Digital PCR of viral DNA/RNA, Current Progress, Challenges, and Future Perspectives. J. Med. Virol. 2021, 93, 4182–4197. [Google Scholar] [CrossRef]
- Javornik Cregeen, S.; Tisza, M.J.; Hanson, B.; Cook, M.; Surathu, A.; Schneider, R.; Wu, J.; Short, K.; Domakonda, K.; Hopkins, L.; et al. Sequencing-Based Detection of Measles in Wastewater: Texas, January 2025. Am. J. Public Health 2025, 115, 1115–1119. [Google Scholar] [CrossRef]
- Joseph, K.M.; Chen, X.; Parikh, D.; Rios, J.; Troisi, C.L.; Tisza, M.J.; Maresso, A.W.; Hanson, B.M.; Gitter, A.; Deegan, J.; et al. Detection of Measles in Texas Wastewater. medRxiv 2025. [Google Scholar] [CrossRef]
- Näyhä, S. Environmental temperature and mortality. Int. J. Circumpolar Health 2005, 64, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, M.X.; Kalvapalle, P.; Nute, M.; Treangen, T.J.; Ensor, K.; Hopkins, L.; Poretsky, R.; Stadler, L.B. Multiplexed Detection, Partitioning, and Persistence of Wild-Type and Vaccine Strains of Measles, Mumps, and Rubella Viruses in Wastewater. Environ. Sci. Technol. 2024, 58, 21930–21941. [Google Scholar] [CrossRef] [PubMed]
- Langan, L.M.; Bain, F.L.; Snow, C.C.; Oldfather, J.; Sagvold, O.; Kaneshiro, K.; Nwagwu, C.; Choi, H.; Wronski, A.R.; Alamin, M.; et al. Spatially Informed Wastewater Differentiation Among Locations During an Ongoing Measles Outbreak in Texas, USA. ACS Environ. Au 2025, 5, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Gwinn, M.; MacCannell, D.; Armstrong, G.L. Next-Generation Sequencing of Infectious Pathogens. JAMA 2019, 321, 893–894. [Google Scholar] [CrossRef]
- NIHR Global Health Research Unit on Genomic Surveillance of AMR. Whole-Genome Sequencing as Part of National and International Surveillance Programmes for Antimicrobial Resistance: A Roadmap. BMJ Glob. Health 2020, 5, e002244. [Google Scholar] [CrossRef]
- Bankamp, B.; Kim, G.; Hart, D.; Beck, A.; Ben Mamou, M.; Penedos, A.; Zhang, Y.; Evans, R.; Rota, P.A. Global Update on Measles Molecular Epidemiology. Vaccines 2024, 12, 810. [Google Scholar] [CrossRef]
- Gruber, C.E.M.; Gioacchini, S.; Fabeni, L.; Rueca, M.; Bordi, L.; Lalle, E.; Baggieri, M.; Bucci, P.; Fioravanti, R.; Giuseppetti, R.; et al. Molecular Investigation on Measles Cases Rise and Variants Co-Circulation in the Lazio Region, Italy. Virol. J. 2025, 22, 288. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Miller, S.A. Clinical Metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, Y. Clinical Applications of Metagenomics Next-Generation Sequencing in Infectious Diseases. J. Zhejiang Univ. Sci. B 2024, 25, 471–484. [Google Scholar] [CrossRef]
- Gaston, D.C.; Chiang, A.D.; Dee, K.; Dulek, D.; Banerjee, R.; Humphries, R.M. Diagnostic Stewardship for Next-Generation Sequencing Assays in Clinical Microbiology: An Appeal for Thoughtful Collaboration. Clin. Lab. Med. 2024, 44, 63–73. [Google Scholar] [CrossRef]
- Gaston, D.C. Clinical Metagenomics for Infectious Diseases: Progress Toward Operational Value. J. Clin. Microbiol. 2023, 61, e0126722. [Google Scholar] [CrossRef] [PubMed]
- Getchell, M.; Wulandari, S.; de Alwis, R.; Agoramurthy, S.; Khoo, Y.K.; Mak, T.M.; Moe, L.; Stona, A.C.; Pang, J.; Momin, M.; et al. Pathogen Genomic Surveillance Status Among Lower Resource Settings in Asia. Nat. Microbiol. 2024, 9, 2738–2747. [Google Scholar] [CrossRef] [PubMed]
- Pronyk, P.M.; de Alwis, R.; Rockett, R.; Basile, K.; Boucher, Y.F.; Pang, V.; Sessions, O.; Getchell, M.; Golubchik, T.; Lam, C.; et al. Advancing Pathogen Genomics in Resource-Limited Settings. Cell Genom. 2023, 3, 100443. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-Based Diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, Y.; Teng, X.; Hou, H.; Deng, R.; Li, J. CRISPR-Based Nucleic Acid Diagnostics for Pathogens. Trends Anal. Chem. 2023, 160, 116980. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-Based Detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Huang, T.; Zhang, R.; Li, J. CRISPR-Cas-Based Techniques for Pathogen Detection: Retrospect, Recent Advances, and Future Perspectives. J. Adv. Res. 2023, 50, 69–82. [Google Scholar] [CrossRef]
- Pinchon, E.; Henry, S.; Leon, F.; Fournier-Wirth, C.; Foulongne, V.; Cantaloube, J.F. Rapid Detection of Measles Virus Using Reverse Transcriptase/Recombinase Polymerase Amplification Coupled with CRISPR/Cas12a and a Lateral Flow Detection: A Proof-of-Concept Study. Diagnostics 2024, 14, 517. [Google Scholar] [CrossRef]
- Rodriguez-Manzano, J.; Subramaniam, S.; Uchea, C.; Szostak-Lipowicz, K.M.; Freeman, J.; Rauch, M.; Tinto, H.; Zar, H.J.; D’Alessandro, U.; Holmes, A.H.; et al. Innovative Diagnostic Technologies: Navigating Regulatory Frameworks Through Advances, Challenges, and Future Prospects. Lancet Digit. Health 2024, 6, e934–e943. [Google Scholar] [CrossRef] [PubMed]
| Method | Target | Sensitivity (%) | Specificity (%) | Technical Requirements | Time To Result |
|---|---|---|---|---|---|
| Serology–EIA | IgM/IgG antibodies | 75–98 | 87–99 | Specialized virology expertise | 3–5 days |
| Serology-CLIA | IgM/IgG antibodies | 94–97 | 95–98 | Automated platform training | 0.5–1 h |
| PRNT | Neutralizing antibodies | 98–100 | 99–100 | Specialized virology expertise | 3–5 days |
| FRNT | Neutralizing antibodies | 95–100 | 98–100 | Specialized virology expertise | 2–3 days |
| rRT-PCR | RNA | 94–99 | 99–100 | Molecular biology expertise or automated platform training | 2–6 h |
| RDT | IgM antibodies | 90–95 | 94–96 | Minimal training | 0.5 h |
| ddPCR | RNA | 95–100 | 98–100 | Molecular biology and dPCR expertise | 4–6 h |
| NGS | RNA | 70–100 | 90–100 | NGS and bioinformatics expertise | 6 h–5 days |
| CRISPR | RNA | 96 | 100 | Molecular biology expertise | 0.5–1 h |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, Y.-C.; Ren, P. Diagnostic Approaches for Measles Virus: Methods, Advances, and Ongoing Challenges. Pathogens 2025, 14, 1295. https://doi.org/10.3390/pathogens14121295
Xue Y-C, Ren P. Diagnostic Approaches for Measles Virus: Methods, Advances, and Ongoing Challenges. Pathogens. 2025; 14(12):1295. https://doi.org/10.3390/pathogens14121295
Chicago/Turabian StyleXue, Yuan-Chao, and Ping Ren. 2025. "Diagnostic Approaches for Measles Virus: Methods, Advances, and Ongoing Challenges" Pathogens 14, no. 12: 1295. https://doi.org/10.3390/pathogens14121295
APA StyleXue, Y.-C., & Ren, P. (2025). Diagnostic Approaches for Measles Virus: Methods, Advances, and Ongoing Challenges. Pathogens, 14(12), 1295. https://doi.org/10.3390/pathogens14121295







