Perspective Overview of Changing Population Immunity to COVID-19 in the Context of Infection, Vaccination, and Emerging SARS-CoV-2 Variants
Abstract
1. Background
1.1. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in the Context of Other Human Coronaviruses
1.2. Functions of Key SARS-CoV-2 and Host Cell Proteins in the Initial Infection of Human Respiratory Tract Epithelial Cells
1.3. Acquisition of SARS-CoV-2 Infections
1.4. Pathology and Prevalence of COVID-19
1.5. The Impact of Vaccination on Mortality and Morbidity During the COVID-19 Pandemic
1.6. Origin, Evolution, and Spread of Early SARS-CoV-2 Variants
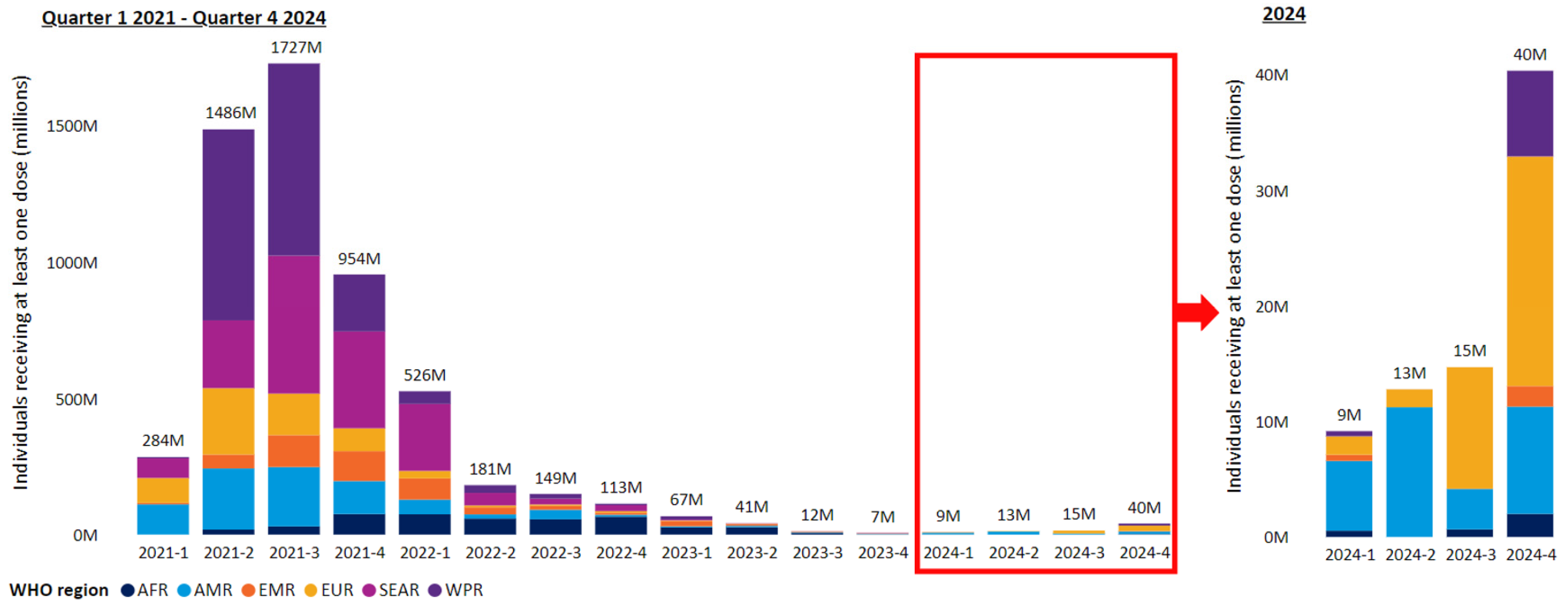
2. COVID-19 Prevalence and Vaccination Status in 2025
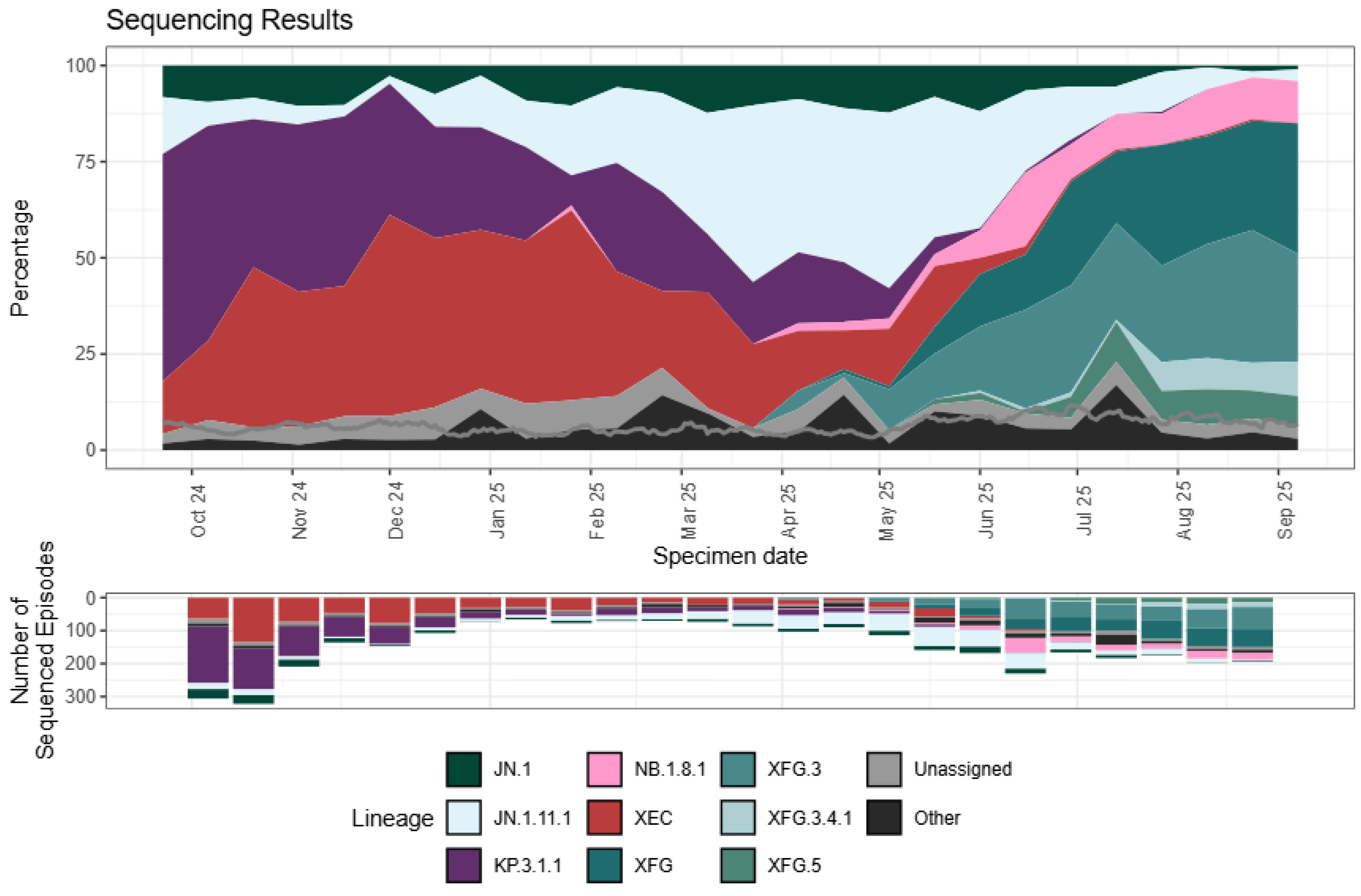
3. SARS-CoV-2 VOI and VUMs in 2025
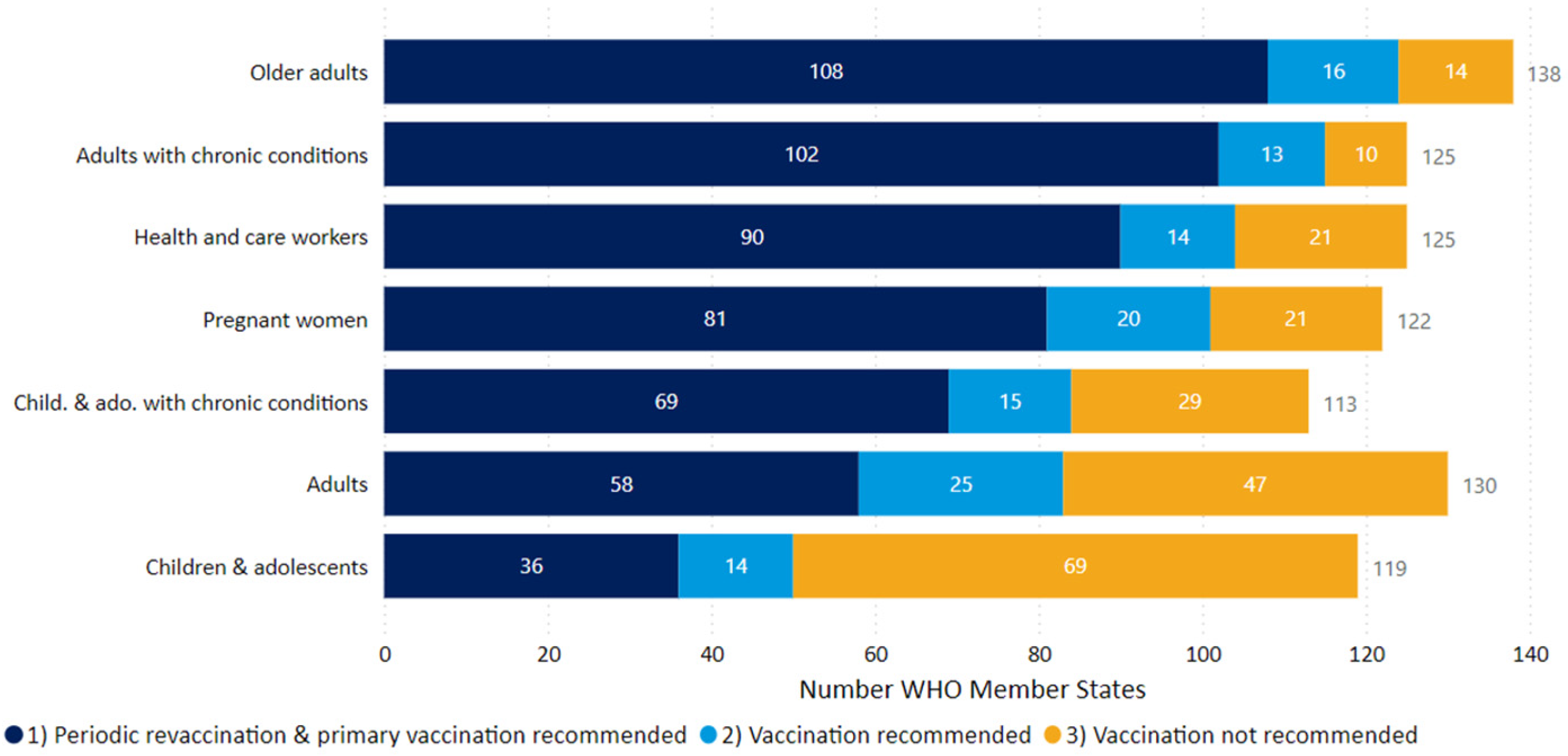
4. Assessment of Population Immunity to COVID-19 in 2025
5. Potential of Presently Circulating SARS-CoV-2 Variants to Generate VOCs
6. Outlook and Strategies for COVID-19 Control in the Future
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Papa, G.; Mallery, D.L.; Albecka, A.; Welch, L.G.; Cattin-Ortolá, J.; Luptak, J.; Paul, D.; McMahon, H.T.; Goodfellow, I.G.; Carter, A.; et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 2021, 17, e1009246. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, R. Nasal conditioning of inspired air, innate immunity in the respiratory tract and SARS-CoV-2 infectivity. Open Sci. Forum 2020. [Google Scholar] [CrossRef]
- Ramasamy, R. Perspective of the relationship between the susceptibility to initial SARS-CoV-2 infectivity and optimal nasal conditioning of inhaled air. Int. J. Mol. Sci. 2021, 22, 7919. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, R. Innate and adaptive immune responses in the upper respiratory tract and the infectivity of SARS-CoV-2. Viruses 2022, 14, 933. [Google Scholar] [CrossRef] [PubMed]
- UK Government Green Book. COVID 19-SARS-CoV-2. 2025. Available online: https://assets.publishing.service.gov.uk/media/68b5be03536d629f9c82a97d/Green-book-chapter-COVID-19_1_9_25.pdf (accessed on 18 November 2025).
- Moriyama, M.; Hugentobler, W.J.; Iwasaki, A. Seasonality of respiratory viral infections. Annu. Rev. Virol. 2020, 7, 83–101. [Google Scholar] [CrossRef]
- Baker, R.E.; Yang, W.; Vecchi, G.A.; Metcalf, C.J.E.; Grenfell, B.T. Assessing the influence of climate on wintertime SARS-CoV-2 outbreaks. Nat. Commun. 2021, 12, 846. [Google Scholar] [CrossRef]
- UK Health and Security Agency. 2025. Epidemiology of COVID-19 in England: January 2020 to December 2024. Available online: https://www.gov.uk/government/publications/epidemiology-of-covid-19-in-england/epidemiology-of-covid-19-in-england-january-2020-to-december-2024 (accessed on 29 September 2025).
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef]
- Cao, X. COVID-19: Immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020, 20, 269–270. [Google Scholar] [CrossRef]
- Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022, 20, 270–284. [Google Scholar] [CrossRef]
- De Neck, S.; Penrice-Randal, R.; Clark, J.J.; Sharma, P.; Bentley, E.G.; Kirby, A.; Mega, D.F.; Han, X.; Owen, A.; Hiscox, J.A.; et al. The stereotypic response of the pulmonary vasculature to respiratory viral infections: Findings in mouse models of SARS-CoV-2, influenza A and gammaherpesvirus infections. Viruses 2023, 15, 1637. [Google Scholar] [CrossRef]
- World Health Organization. Coronavirus (COVID-19) Dashboard. 2023. Available online: https://covid19.who.int (accessed on 4 October 2023).
- World Health Organization Emergency Committee. Statement on the Second Meeting of the International Health Regulations Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV). 2020. Available online: https://www.who.int/news/item/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov) (accessed on 8 October 2025).
- World Health Organization. Coronavirus Disease 2019 (COVID-19) Situation Report—51. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf?sfvrsn=1ba62e57) (accessed on 8 October 2025).
- World Health Organization Emergency Committee. Statement on the Fifteenth Meeting of the International Health Regulations (2005) Emergency Committee on the COVID-19 Pandemic. 2023. Available online: https://www.who.int/news-room/speeches/item/who-director-general-s-opening-remarks-at-the-media-briefing---5-may-2023 (accessed on 4 October 2025).
- World Health Organization. WHO COVID-19 Dashboard. 2025. Available online: https://data.who.int/dashboards/covid19/data (accessed on 4 October 2025).
- Ramasamy, R. COVID-19 vaccines for optimizing immunity in the upper respiratory tract. Viruses 2023, 15, 2203. [Google Scholar] [CrossRef]
- Sadarangani, M.; Marchant, A.; Kollmann, T.R. Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nat. Rev. Immunol. 2021, 21, 475–484. [Google Scholar] [CrossRef]
- Zasada, A.A.; Darlinska, A.; Wiatrzyk, A.; Woznica, K.; Forminska, K.; Czajka, U.; Główka, M.; Lis, K.; Górska, P. COVID-19 Vaccines over three years after the outbreak of the COVID-19 Epidemic. Viruses 2023, 15, 1786. [Google Scholar] [CrossRef] [PubMed]
- Plans-Rubió, P. Percentages of vaccination coverage required to establish herd immunity against SARS-CoV-2. Vaccines 2022, 10, 736. [Google Scholar] [CrossRef]
- Knisely, J.M.; Buyon, L.E.; Mandt, R.; Farkas, R.; Balasingam, S.; Bok, K.; Buchholz, U.J.; D’souza, M.P.; Gordon, J.L.; King, D.F.L.; et al. Mucosal vaccines for SARS-CoV-2: Scientific gaps and opportunities—Workshop report. NPJ Vaccines 2023, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Tscherne, A.; Sun, W.; Liu, S.T.H.; Krammer, F. Mucosal COVID-19 vaccines in clinical development. Vaccine 2025, 63, 127602. [Google Scholar] [CrossRef] [PubMed]
- Kiyono, H.; Ernst, P.B. Nasal vaccines for respiratory infections. Nature 2025, 641, 321–330. [Google Scholar] [CrossRef]
- Watson, O.J.; Barnsley, G.; Toor, J.; Hogan, A.B.; Winskill, P.; Ghani, A.C. Global impact of the first year of COVID-19 vaccination: A mathematical modelling study. Lancet Infect. Dis. 2022, 22, 1293–1302. [Google Scholar] [CrossRef]
- Yamana, T.K.; Galanti, M.; Pei, S.; Di Fusco, M.; Angulo, F.J.; Moran, M.M.; Khan, F.; Swerdlow, D.L.; Shaman, J. The impact of COVID-19 vaccination in the US: Averted burden of SARS-CoV-2-related cases, hospitalizations and deaths. PLoS ONE 2023, 18, e0275699. [Google Scholar] [CrossRef]
- Haas, E.J.; McLaughlin, J.M.; Khan, F.; Angulo, F.J.; Anis, E.; Lipsitch, M.; Singer, S.R.; Mircus, G.; Brooks, N.; Smaja, M.; et al. Infections, hospitalisations, and deaths averted via a nationwide vaccination campaign using the Pfizer–BioNTech BNT162b2 mRNA COVID-19 vaccine in Israel: A retrospective surveillance study. Lancet Infect. Dis. 2022, 22, 357–366. [Google Scholar] [CrossRef]
- Ramasamy, R. Overview of immunological and virological factors driving the evolution and global spread of SARS-CoV-2 variants. Indian J. Med. Res. 2023, 158, 257–268. [Google Scholar] [CrossRef]
- Institute for Health Metrics and Evaluation. COVID-19 Model Update: Omicron and Waning Immunity. Available online: https://www.healthdata.org/research-analysis/diseases-injuries/covid/covid-19-model-update-omicron-and-waning-immunity (accessed on 2 October 2025).
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Yang, H.; Guo, H.; Wang, A.; Cao, L.; Fan, Q.; Jiang, J.; Wang, M.; Lin, L.; Ge, X.; Wang, H.; et al. Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants. Nat. Commun. 2024, 15, 7715. [Google Scholar] [CrossRef]
- Abbad, A.; Lerman, B.; Ehrenhaus, J.; Monahan, B.; Singh, G.; Wilson, A.; Slamanig, S.; Aracena, A.; Lyttle, N.; Nardulli, J.; et al. Antibody responses to SARS-CoV-2 variants LP.8.1, LF.7.1, NB.1.8.1, XFG and BA.3.2 following KP.2 monovalent mRNA vaccination. Medrxiv 2025. [Google Scholar] [CrossRef]
- Tang, H.; Zhuo, Y.; Chen, J.; Zhang, R.; Zheng, M.; Huang, X.; Chen, Y.; Huang, M.; Zeng, Z.; Huang, X.; et al. Immune evasion, infectivity, and membrane fusion of the SARS-CoV-2 JN.1 variant. Virol. J. 2025, 22, 162. [Google Scholar] [CrossRef]
- Tian, J.; Shang, B.; Zhang, J.; Guo, Y.; Li, M.; Hu, Y.; Bai, D.; She, J.; Han, Y.; Guo, P.; et al. T cell immune evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots. Nat. Immunol. 2025, 26, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Amicone, M.; Borges, V.; Alves, M.J.; Isidro, J.; Zé-Zé, L.; Duarte, S.; Vieira, L.; Guiomar, R.; Gomes, J.P.; Gordo, I. Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution. Evol. Med. Public Health 2022, 10, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Global Initiative on Sharing All Influenza Data (GISAID). Available online: https://gisaid.org/phylodynamics/global/ (accessed on 8 October 2025).
- World Health Organization. Updated Working Definitions and Primary Actions for SARS-CoV-2 Variants. 2025. Available online: https://www.who.int/publications/m/item/updated-working-definitions-and-primary-actions-for--sars-cov-2-variants (accessed on 4 October 2025).
- Centers for Disease Control and Prevention. COVID. 2025. Available online: https://www.cdc.gov/covid/php/variants/variants-and-genomic-surveillance.html (accessed on 4 October 2025).
- Guo, C.; Yu, Y.; Liu, J.; Jian, F.; Yang, S.; Song, W.; Yu, L.; Shao, F.; Cao, Y. Antigenic and virological characteristics of SARS-CoV-2 variants BA.3.2, XFG, and NB.1.8.1. Lancet Infect. Dis. 2025, 25, e374–e377. [Google Scholar] [CrossRef] [PubMed]
- UK Health Security Agency. National Flu and COVID-19 Surveillance Report: 25 September 2025 (Week 39). 2025. Available online: https://www.gov.uk/government/statistics/national-flu-and-covid-19-surveillance-reports-2025-to-2026-season/national-flu-and-covid-19-surveillance-report-25-september-2025-week-39 (accessed on 6 October 2025).
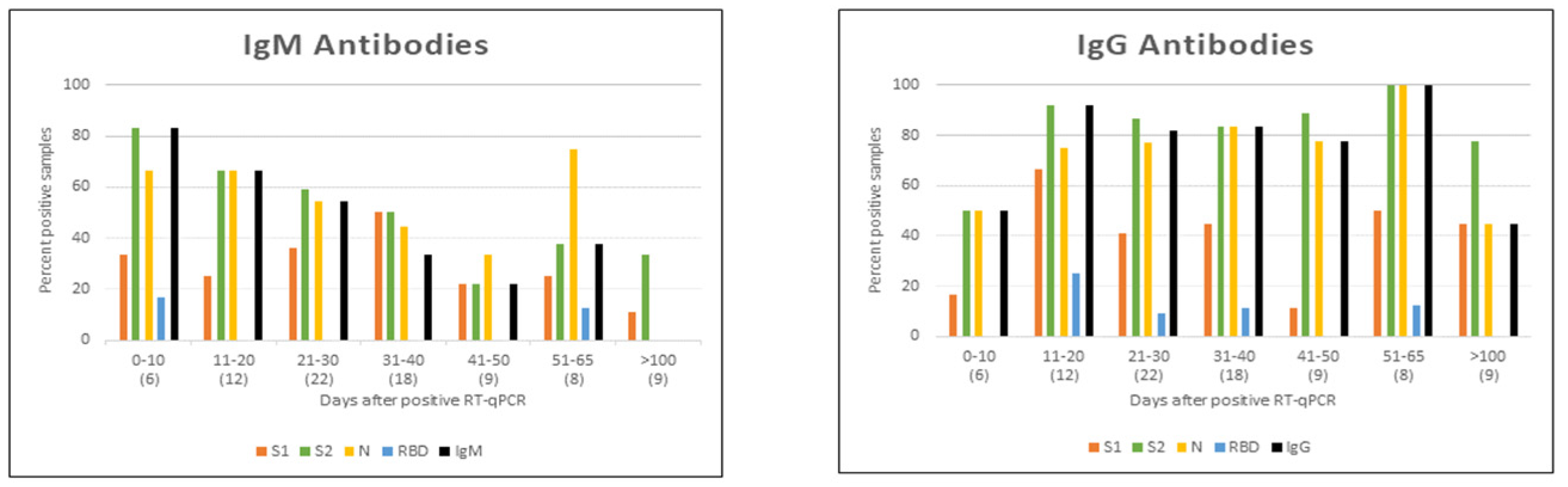
- Shah, J.; Liu, S.; Potula, H.H.; Bhargava, P.; Cruz, I.; Force, D.; Bazerbashi, A.; Ramasamy, R. IgG and IgM antibody formation to spike and nucleocapsid proteins in COVID-19 characterized by multiplex immunoblot assays. BMC Infect. Dis. 2021, 21, 325. [Google Scholar] [CrossRef]
- Notarte, K.I.; Guerrero-Arguero, I.; Velasco, J.V.; Ver, A.T.; Santos de Oliveira, M.H.; Catahay, J.A.; Khan, M.S.R.; Pastrana, A.; Juszczyk, G.; Torrelles, J.B.; et al. Characterization of the significant decline in humoral immune response six months post-SARS-CoV-2 mRNA vaccination: A systematic review. J. Med. Virol. 2022, 94, 2939–2961. [Google Scholar] [CrossRef]
- Moore, M.; Anderson, L.; Schiffer, J.T.; Matrajt, L.; Dimitrov, D. Durability of COVID-19 vaccine and infection induced immunity: A systematic review and meta-regression analysis. Vaccine 2025, 54, 126966. [Google Scholar] [CrossRef]
- Feldstein, L.R.; Ruffin, J.; Wiegand, R.E.; Borkowf, C.B.; James-Gist, J.; Babu, T.M.; Briggs-Hagen, M.; Chappell, J.; Chu, H.Y.; Englund, J.A.; et al. Effectiveness of mRNA COVID-19 vaccines and hybrid immunity in preventing SARS-CoV-2 infection and symptomatic COVID-19 among adults in the United States. J. Infect. Dis. 2025, 231, e743–e753. [Google Scholar] [CrossRef]
- Klaassen, F.; Swartwood, N.A.; Chitwood, M.H.; Lopes, R.; Haraguchi, M.; Salomon, J.A.; Cohen, T.; Menzies, N.A. National- and state-level SARS-CoV-2 immunity trends from January 2020 to December 2023: A mathematical modeling analysis. J. Infect. Dis. 2025. [Google Scholar] [CrossRef]
- National Health Service. The COVID-19 Vaccine. 2025. Available online: https://www.nhs.uk/vaccinations/covid-19-vaccine/ (accessed on 8 October 2025).
- European Medicines Agency. 2025. Available online: https://www.ema.europa.eu/en/documents/other/ema-recommendation-update-antigenic-composition-authorised-covid-19-vaccines-2025-2026_en.pdf (accessed on 8 October 2025).
- Swadling, L.; Diniz, M.O.; Schmidt, N.M.; Amin, O.E.; Chandran, A.; Shaw, E.; Pade, C.; Gibbons, J.M.; Le Bert, N.; Tan, A.T.; et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 2022, 601, 110–117. [Google Scholar] [CrossRef]
- Wang, Y.; Xia, B.; Gao, Z. A comprehensive review of current insights into the virulence factors of SARS-CoV-2. J. Virol. 2025, 99, e0204924. [Google Scholar] [CrossRef] [PubMed]
- Bilev, E.; Wild, N.; Momayyezi, P.; Sala, B.M.; Sun, R.; Sandalova, T.; Marquardt, N.; Ljunggren, H.G.; Achour, A.; Hammer, Q. Emerging mutation in SARS-CoV-2 facilitates escape from NK cell recognition and associates with enhanced viral fitness. PLoS Pathog. 2024, 20, e1012755. [Google Scholar] [CrossRef] [PubMed]
- Abay, Z.; Sadikaliyeva, S.; Nurpeisova, A.; Jekebekov, K.; Shorayeva, K.; Yespembetov, B.; Nurabayev, S.; Kerimbayev, A.; Khairullin, B.; Yoo, H.; et al. Breaking the barrier: SARS-CoV-2 infections in wild and companion animals show implications for public health. Viruses 2024, 16, 956. [Google Scholar] [CrossRef]
- Garten, R.J.; Davis, C.T.; Russell, C.A.; Shu, B.; Lindstrom, S.; Balish, A.; Sessions, W.M.; Xu, X.; Skepner, E.; Deyde, V.; et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009, 325, 197–201. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Khatri, R.; Siddqui, G.; Sadhu, S.; Maithil, V.; Vishwakarma, P.; Lohiya, B.; Goswami, A.; Ahmed, S.; Awasthi, A.; Samal, S. Intrinsic D614G and P681R/H mutations in SARS-CoV-2 VoCs alpha, delta, omicron and viruses with D614G plus key signature mutations in spike protein alters fusogenicity and infectivity. Med. Microbiol. Immunol. 2023, 212, 103–122. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H., 3rd; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020, 370, 1464–1468. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.; Shum, M.H.; Leung, G.M.; Lam, T.T.; Wu, J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro. Surveill. 2021, 26, 2002106. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C.; et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 2022, 602, 294–299. [Google Scholar] [CrossRef]
- Meng, B.; Kemp, S.A.; Papa, G.; Datir, R.; Ferreira, I.A.T.M.; Marelli, S.; Harvey, W.T.; Lytras, S.; Mohamed, A.; Gallo, G.; et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021, 35, 109292. [Google Scholar] [CrossRef]
- Xie, X.; Liu, Y.; Liu, J.; Zhang, X.; Zou, J.; Fontes-Garfias, C.R.; Xia, H.; Swanson, K.A.; Cutler, M.; Cooper, D.; et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021, 27, 620–621. [Google Scholar] [CrossRef]
- Sarkar, P.; Banerjee, S.; Saha, S.A.; Mitra, P.; Sarkar, S. Genome surveillance of SARS-CoV-2 variants and their role in pathogenesis focusing on second wave of COVID-19 in India. Sci. Rep. 2023, 13, 4692. [Google Scholar] [CrossRef]
- Earnest, R.; Uddin, R.; Matluk, N.; Renzette, N.; Turbett, S.E.; Siddle, K.J.; Loreth, C.; Adams, G.; Tomkins-Tinch, C.H.; Petrone, M.E.; et al. Comparative transmissibility of SARS-CoV-2 variants Delta and Alpha in New England, USA. Cell Rep. Med. 2022, 3, 100583. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, T.; Cai, Y.; Lavine, C.L.; Peng, H.; Zhu, H.; Anand, K.; Tong, P.; Gautam, A.; Mayer, M.L.; et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science 2021, 374, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021, 29, 1124–1136.e11. [Google Scholar] [CrossRef]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: A cohort study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.B.; Rophina, M.; Chaudhry, R.; Senthivel, V.; Bala, K.; Bhoyar, R.C.; Jolly, B.; Jamshed, N.; Imran, M.; Gupta, R.K.; et al. Variants of concern responsible for SARS-CoV-2 vaccine breakthrough infections from India. J. Med. Virol. 2022, 94, 1696–1700. [Google Scholar] [CrossRef]
- He, P.; Liu, B.; Gao, X.; Yan, Q.; Pei, R.; Sun, J.; Chen, Q.; Hou, R.; Li, Z.; Zhang, Y.; et al. SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope. Nat. Microbiol. 2022, 7, 1635–1649. [Google Scholar] [CrossRef]
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Makatini, Z.; Moyo, S.; Amoako, D.G.; et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022, 28, 1785–1790. [Google Scholar] [CrossRef]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef]
- Chen, D.Y.; Chin, C.V.; Kenney, D.; Tavares, A.H.; Khan, N.; Conway, H.L.; Liu, G.; Choudhary, M.C.; Gertje, H.P.; O’Connell, A.K.; et al. Spike and nsp6 are key determinants of SARS-CoV-2 Omicron BA.1 attenuation. Nature 2023, 615, 143–150. [Google Scholar] [CrossRef]
- Sheward, D.J.; Kim, C.; Fischbach, J.; Sato, K.; Muschiol, S.; Ehling, R.A.; Björkström, N.K.; Karlsson Hedestam, G.B.; Reddy, S.T.; Albert, J.; et al. Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Lancet Infect. Dis. 2022, 22, 1538–1540. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.; Peacock, T.P.; Harvey, W.T.; Hughes, J.; Wright, D.W.; COVID-19 Genomics UK (COG-UK) Consortium; Willett, B.J.; Thomson, E.; Gupta, R.K.; Peacock, S.J.; et al. SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nat. Rev. Microbiol. 2023, 21, 112–124. [Google Scholar] [CrossRef]
- Cao, Y.; Jian, F.; Wang, J.; Yu, Y.; Song, W.; Yisimayi, A.; Wang, J.; An, R.; Chen, X.; Zhang, N.; et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 2023, 614, 521–529. [Google Scholar] [CrossRef]
- Tuekprakhon, A.; Nutalai, R.; Dijokaite-Guraliuc, A.; Zhou, D.; Ginn, H.M.; Selvaraj, M.; Liu, C.; Mentzer, A.J.; Supasa, P.; Duyvesteyn, H.M.E.; et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 2022, 185, 2422–2433.e13. [Google Scholar] [CrossRef] [PubMed]
- Uraki, R.; Ito, M.; Furusawa, Y.; Yamayoshi, S.; Iwatsuki-Horimoto, K.; Adachi, E.; Saito, M.; Koga, M.; Tsutsumi, T.; Yamamoto, S.; et al. Humoral immune evasion of the omicron subvariants BQ.1.1 and XBB. Lancet Infect. Dis. 2023, 23, 30–32. [Google Scholar] [CrossRef]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; COVID-19 Genomics UK Consortium; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Uraki, R.; Halfann, P.J.; Iida, S.; Yamayoshi, S.; Furusawa, Y.; Kiso, M.; Ito, M.; Iwatsuki-Horimoto, K.; Mine, S.; Kuroda, M.; et al. Characterization of SARS-CoV-2 Omicron BA.4 and BA.5 isolates in rodents. Nature 2022, 612, 540–545. [Google Scholar] [CrossRef]
- Lewnard, J.A.; Hong, V.X.; Patel, M.M.; Kahn, R.; Lipsitch, M.; Tartof, S.Y. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in Southern California. Nat. Med. 2022, 28, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Lewnard, J.A.; Hong, V.; Kim, J.S.; Shaw, S.F.; Lewin, B.; Takhar, H.; Tartof, S.Y. Association of SARS-CoV-2 BA.4/BA.5 Omicron lineages with immune escape and clinical outcome. Nat. Commun. 2023, 14, 1407. [Google Scholar] [CrossRef]
- Ricciardi, S.; Guarino, A.M.; Giaquinto, L.; Polishchuk, E.V.; Santoro, M.; Di Tullio, G.; Wilson, C.; Panariello, F.; Soares, V.C.; Dias, S.S.G.; et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature 2022, 606, 761–768. [Google Scholar] [CrossRef]
- Sun, X.; Liu, Y.; Huang, Z.; Xu, W.; Hu, W.; Yi, L.; Liu, Z.; Chan, H.; Zeng, J.; Liu, X.; et al. SARS-CoV-2 non-structural protein 6 triggers NLRP3-dependent pyroptosis by targeting ATP6AP1. Cell Death Differ. 2022, 29, 1240–1254. [Google Scholar] [CrossRef]
- Alfi, O.; Hamdan, M.; Wald, O.; Yakirevitch, A.; Wandel, O.; Oiknine-Djian, E.; Gvili, B.; Knoller, H.; Rozendorn, N.; Golan Berman, H.; et al. SARS-CoV-2 Omicron induces enhanced mucosal interferon response compared to other variants of concern, associated with restricted replication in human lung tissues. Viruses 2022, 14, 1583. [Google Scholar] [CrossRef]
- Siddiqui, A.N.; Musharaf, I.; Gulumbe, B.H. The JN.1 variant of COVID-19: Immune evasion, transmissibility, and implications for global health. Ther. Adv. Infect. Dis. 2025, 12, 20499361251314763. [Google Scholar] [CrossRef]
- Wickenhagen, A.; Flagg, M.; Port, J.R.; Yinda, C.K.; Goldin, K.; Gallogly, S.; Schulz, J.E.; Lutterman, T.; Williamson, B.N.; Kaiser, F.; et al. Evolution of Omicron lineage towards increased fitness in the upper respiratory tract in the absence of severe lung pathology. Nat. Commun. 2025, 16, 594. [Google Scholar] [CrossRef]
- World Health Organization. Risk Evaluation for SARS-CoV-2 Variant Under Monitoring: XFG. Available online: https://www.who.int/publications/m/item/risk-evaluation-for-sars-cov-2-variant-under-monitoring-xfg (accessed on 14 November 2025).
- Plans-Rubió, P. Effectiveness of adapted COVID-19 vaccines and ability to establish herd immunity against omicron BA.1 and BA.4-5 variants of SARS-CoV-2. Vaccines 2023, 11, 1836. [Google Scholar] [CrossRef] [PubMed]
- Plans-Rubió, P. What COVID-19 vaccination strategy should be implemented and which vaccines should be used in the post-pandemic era? Vaccines 2024, 12, 1180. [Google Scholar] [CrossRef]
- UK Government Green Book. Chapter 19. Influenza. 2025. Available online: https://assets.publishing.service.gov.uk/media/6838317b5150d70c85aafab9/Green-book-chapter-19-influenza-28May2025.pdf (accessed on 18 November 2025).
- Sette, A.; Sidney, J.; Crotty, S. T Cell Responses to SARS-CoV-2. Annu. Rev. Immunol. 2023, 41, 343–373. [Google Scholar] [CrossRef] [PubMed]
- Britzke, T.; Halwe, N.J.; Ulrich, L.; Breithaupt, A.; Barut, G.T.; Wylezich, C.; Ebert, N.; Trüeb, B.S.; Thiel, V.; Hoffmann, D.; et al. Live attenuated SARS-CoV-2 vaccine OTS-228 demonstrates efficacy, safety, and stability in preclinical model. NPJ Vaccines 2025, 10, 104. [Google Scholar] [CrossRef]
- Suzuki Okutani, M.; Okamura, S.; Gis, T.; Sasaki, H.; Lee, S.; Kashiwabara, A.; Goto, S.; Matsumoto, M.; Yamawaki, M.; Miyazaki, T.; et al. Immunogenicity and safety of a live-attenuated SARS-CoV-2 vaccine candidate based on multiple attenuation mechanisms. eLife 2025, 13, RP97532. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.W.; Ong, C.P.; Cao, H.; Tang, K.; Gray, V.S.; Hinson Cheung, P.H.; Wang, J.; Li, W.; Zhang, H.; Luo, P.; et al. A live attenuated SARS-CoV-2 vaccine constructed by dual inactivation of NSP16 and ORF3a. eBioMedicine 2025, 114, 105662. [Google Scholar] [CrossRef]
- Spearman, P.; Jin, H.; Xiao, P.; Knopp, K.; Radziewicz, H.; Tellier, M.; Larsen, S.E.; Berube, B.J.; Song, X.; Kidd, J.; et al. Safety and immunogenicity of intranasal parainfluenza virus type 5 (PIV5)-vectored COVID-19 vaccine in adults and teens in an open-label phase 1 trial. Sci. Adv. 2025, 11, eadw0896. [Google Scholar] [CrossRef]
- Jeyanathan, M.; Afkhami, S.; D’Agostino, M.R.; Satia, I.; Fritz, D.K.; Miyasaki, K.; Ang, J.C.; Zganiacz, A.; Howie, K.J.; Swinton, M.; et al. Induction of lung mucosal immunity by a next-generation inhaled aerosol COVID-19 vaccine: An open-label, multi-arm phase 1 clinical trial. Nat. Commun. 2025, 16, 6000. [Google Scholar] [CrossRef] [PubMed]
- UK Health Security Agency. National Flu and COVID-19 Surveillance Report: 6 November 2025 (Week 45). Available online: https://www.gov.uk/government/statistics/national-flu-and-covid-19-surveillance-reports-2025-to-2026-season/national-flu-and-covid-19-surveillance-report-6-november-2025-week-45 (accessed on 19 November 2025).
- Grassly, N.C.; Shaw, A.G.; Owusu, M. Global wastewater surveillance for pathogens with pandemic potential: Opportunities and challenges. Lancet Microbe 2025, 6, 100939. [Google Scholar] [CrossRef] [PubMed]
- Farkas, K.; Williams, R.C.; Hillary, L.S.; Garcia-Delgado, A.; Jameson, E.; Kevill, J.L.; Wade, M.J.; Grimsley, J.M.S.; Jones, D.L. Harnessing the power of next-generation sequencing in wastewater-based epidemiology and global disease surveillance. Food Environ. Virol. 2024, 17, 5. [Google Scholar] [CrossRef]
- Morfino, R.; Gawlik, B.M.; Tavazzi, S.; Tessarolo, A.; Gutierrez, A.B.; Madhav, N.K.; Grimsley, J.; Schierhorn, A.; Franklin, A.; Vargha, M.; et al. Establishing a European wastewater pathogen monitoring network employing aviation samples: A proof of concept. Hum. Genom. 2025, 19, 24. [Google Scholar] [CrossRef]
- Nooruzzaman, M.; Johnson, K.E.E.; Rani, R.; Finkelsztein, E.J.; Caserta, L.C.; Kodiyanplakkal, R.P.; Wang, W.; Hsu, J.; Salpietro, M.T.; Banakis, S.; et al. Emergence of transmissible SARS-CoV-2 variants with decreased sensitivity to antivirals in immunocompromised patients with persistent infections. Nat. Commun. 2024, 15, 7999. [Google Scholar] [CrossRef]
- Gan, M.; Cao, J.; Ouyang, Q.; Xu, X.; Wang, X.; Dan, P.; Yao, Y.; Fu, H.; Yao, X.; Lin, X.; et al. Extensive cross-reactive T cell epitopes across SARS-CoV-2 Omicron variant spikes with finite immune evasion mutations. J. Transl. Med. 2025, 23, 1027. [Google Scholar] [CrossRef] [PubMed]
| Location | Cumulative Cases Until 14 September 2025 | Cases in the 28 d Preceding 14 September 2025 | Cases in the 28 d Preceding 16 August 2025 | Cumulative Deaths Until 14 September 2025 | Deaths in the 28 d Preceding 14 September 2025 | Deaths in the 28 d Preceding 16 August 2025 |
|---|---|---|---|---|---|---|
| Worldwide | 778.7 million | 132,801 | 79,253 | 7.1 million | 1671 | 1187 |
| COVID-19 Phase | Variant and Its Origin and Prevalence | Mutations | Immune Evasion | Transmissibility | Disease Severity |
|---|---|---|---|---|---|
| Before vaccination | Variant with D614G mutation in S. Multiple global origins around March 2020 [38,55]. Evolved into later VOCs [56]. | D614G in S and linked mutation in RNA polymerase [55,56,57,58,59]. | Not seen with convalescent serum antibodies [57,58,59]. | Greater than the ancestral Wuhan strain [57,58,59]. | Marginally greater than the ancestral strain [57,58]. |
| Alpha VOC, B.1.1.7 Originated in UK mid-2020 and then spread worldwide, but is no longer in circulation [38]. | In total, 17 mutations in S including D614G, N501Y, ΔH69-ΔV70, and P681H [56,60,61,62]. | Small neutralization reduction with ancestral S mRNA vaccine-elicited sera compared with ancestral strain [63]. | Increased compared to earlier strains [60,61,62]. | More severe than ancestral strain [60,61,62]. | |
| Early post-vaccination | Delta VOC, B.1.617.2 Originated in India in April 2021 and then spread widely but is no longer in circulation [38,64]. | Additional mutations in S including P681R, L452R, and others in RNA polymerase and additional viral proteins [65,66,67,68,69]. | Markedly reduced sensitivity to immune antibodies [70,71]. | Increased compared to Alpha [65]. | More severe than Alpha [68,69]. |
| After large-scale vaccination and COVID-19 infection | Omicron B.1.1.529 VOC first identified in Africa in November 2021 [72], and the precursor for other Omicron variants | Increasing number of mutations compared to earlier VOCs in S, notably in RBD, and also in other viral proteins [73,74,75,76,77]. | Evasion of antibodies in convalescent and vaccinee sera, and monoclonal antibodies used in therapy [75,76,77,78,79]. | Changes in intrinsic transmissibility unclear [73]. | Less than Delta and earlier VOCs [73,74,80,81,82,83,84,85,86]. |
| Omicron JN.1 first characterized in August 2023 and designated VOI in December 2023 [87]. Largely replaced in 2025 by more fit offspring variants, e.g., XFG (Figure 3) [41,42]. | More mutations in S compared to earlier Omicrons, notably L555S [35,41,87]. | Greater evasion of invasion inhibiting antibodies [35,36,87], and possibly cytotoxic T cells recognizing S and N [36]. | More transmissible than earlier Omicrons to other persons and also animals [35]. | Greater transmissibility and infectibility causing more infections [35,36,47,87]. Evidence for preferential URT replication and lower LRT pathology than earlier VOCs [88]. | |
| Omicron XFG first identified in January 2025. Designated a VUM. XFG and its sublineages dominant globally in late 2025 [89]. | XFG has 11 additional mutations in S compared to JN.1 [89]. | Available data suggest greater evasion of neutralizing antibodies compared to LP.8.1.1 [41]. | High relative growth advantage compared to co-circulating variants [41]. | No evidence for greater disease severity than other co-circulating Omicron variants [89]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramasamy, R. Perspective Overview of Changing Population Immunity to COVID-19 in the Context of Infection, Vaccination, and Emerging SARS-CoV-2 Variants. Pathogens 2025, 14, 1197. https://doi.org/10.3390/pathogens14121197
Ramasamy R. Perspective Overview of Changing Population Immunity to COVID-19 in the Context of Infection, Vaccination, and Emerging SARS-CoV-2 Variants. Pathogens. 2025; 14(12):1197. https://doi.org/10.3390/pathogens14121197
Chicago/Turabian StyleRamasamy, Ranjan. 2025. "Perspective Overview of Changing Population Immunity to COVID-19 in the Context of Infection, Vaccination, and Emerging SARS-CoV-2 Variants" Pathogens 14, no. 12: 1197. https://doi.org/10.3390/pathogens14121197
APA StyleRamasamy, R. (2025). Perspective Overview of Changing Population Immunity to COVID-19 in the Context of Infection, Vaccination, and Emerging SARS-CoV-2 Variants. Pathogens, 14(12), 1197. https://doi.org/10.3390/pathogens14121197






