Genomic Evidence for the Rise of Salmonella Typhimurium ST34 with Increased Plasmid-Mediated Resistance in the Thailand Pork Chain
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Collection and Study Design
2.2. DNA Extraction and Whole-Genome Sequencing
2.3. Functional Annotation and Comparative Genomics
2.4. Genomic Characterization
2.5. Detection of ARGs, Plasmids
3. Results
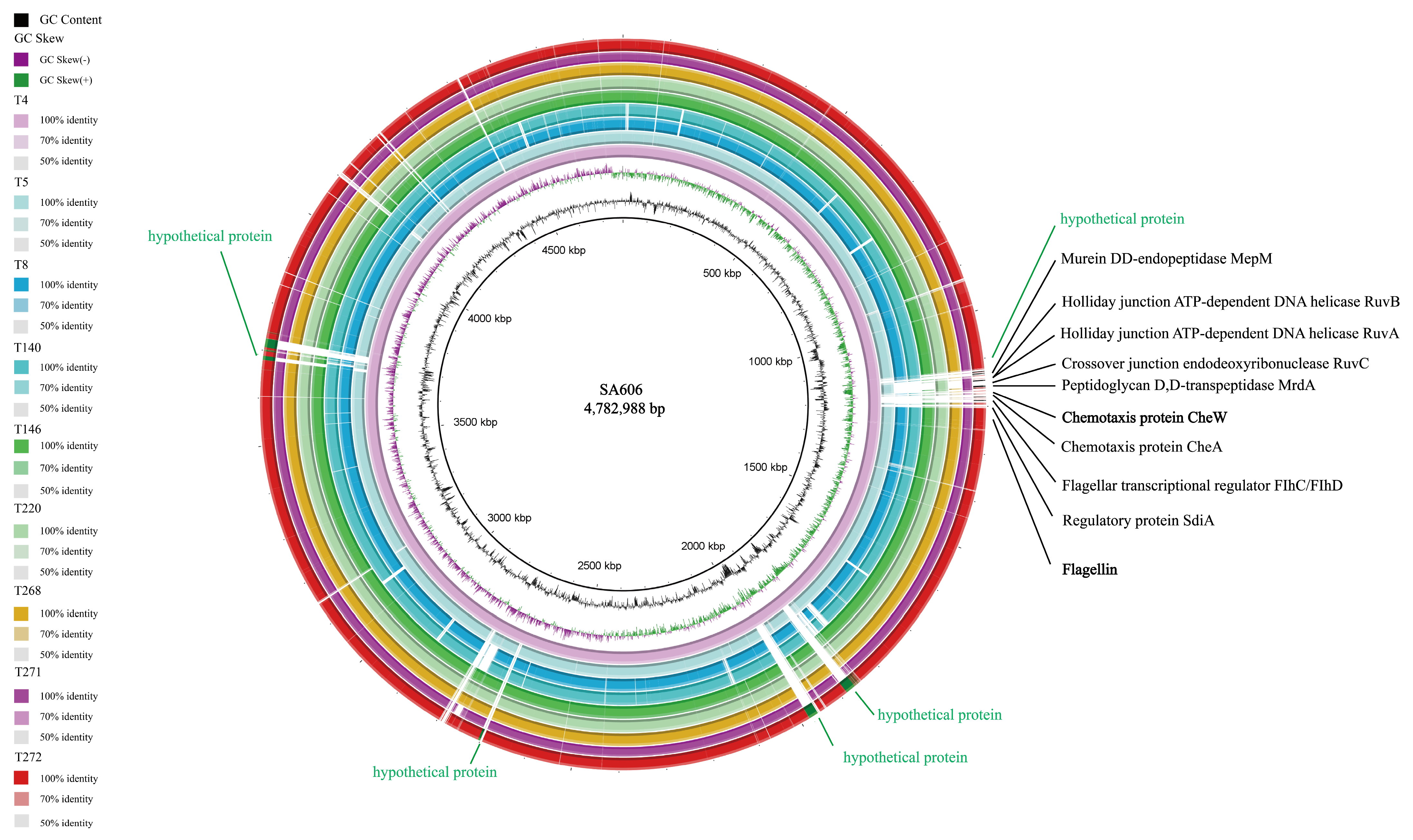
3.1. Comparative Whole-Genome Analysis
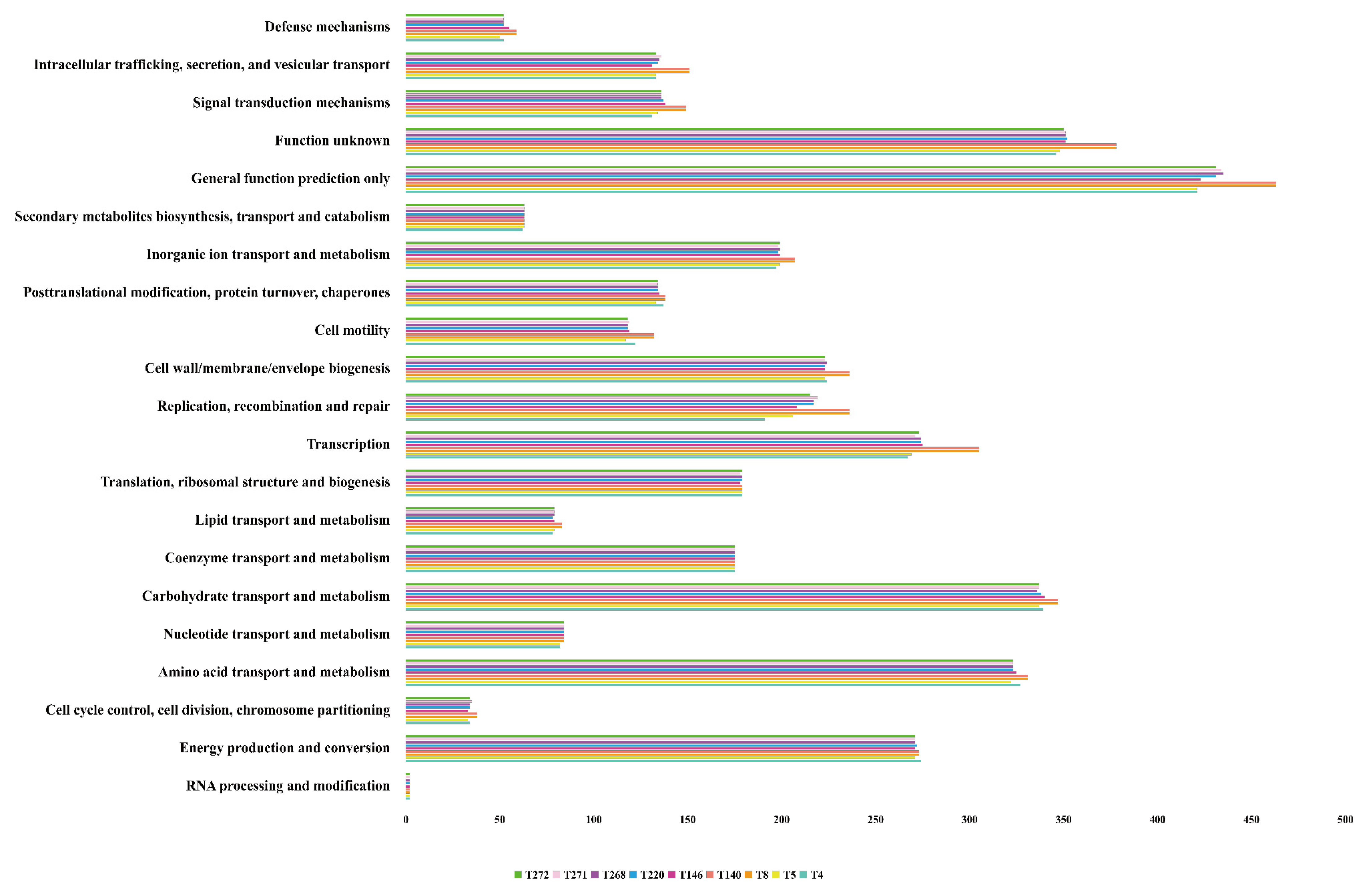
3.2. Functional Annotation of Coding Sequences
3.3. Genome Sequencing and Assembly Quality
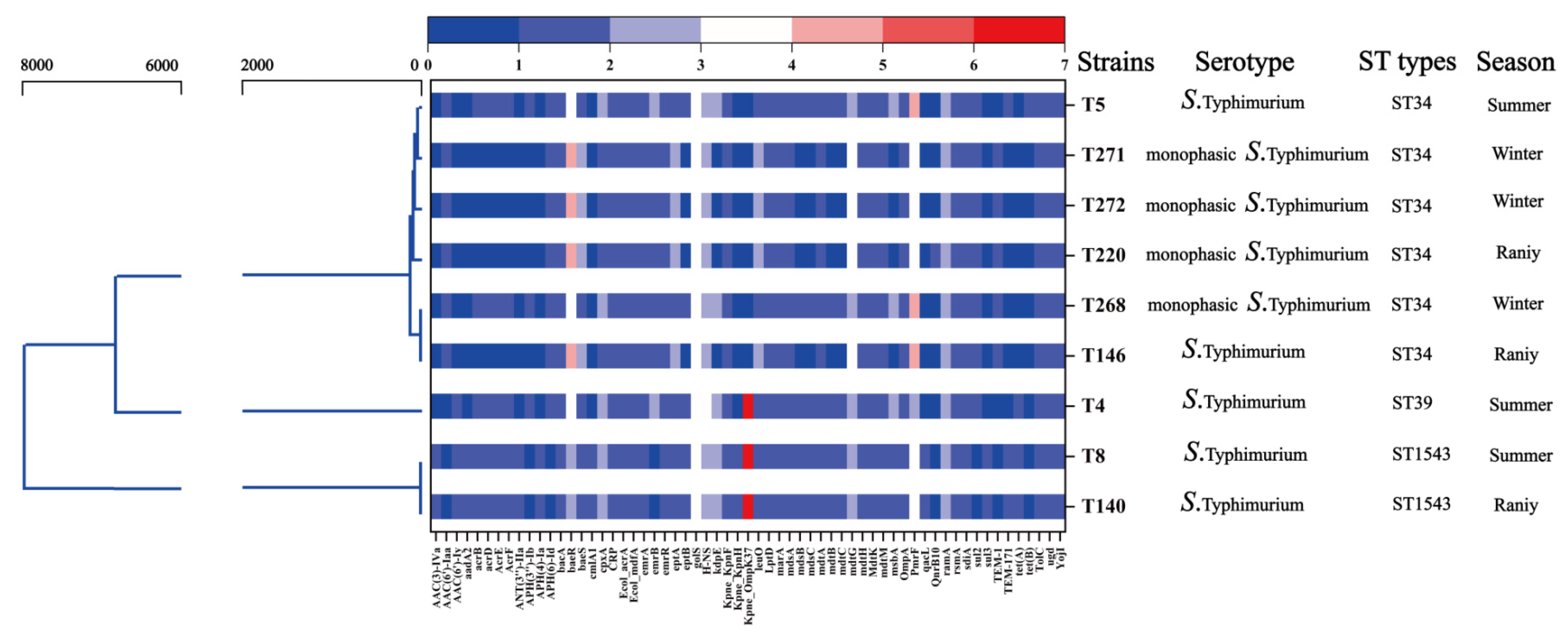
3.4. Phylogenetic Relationships and Sequence Types
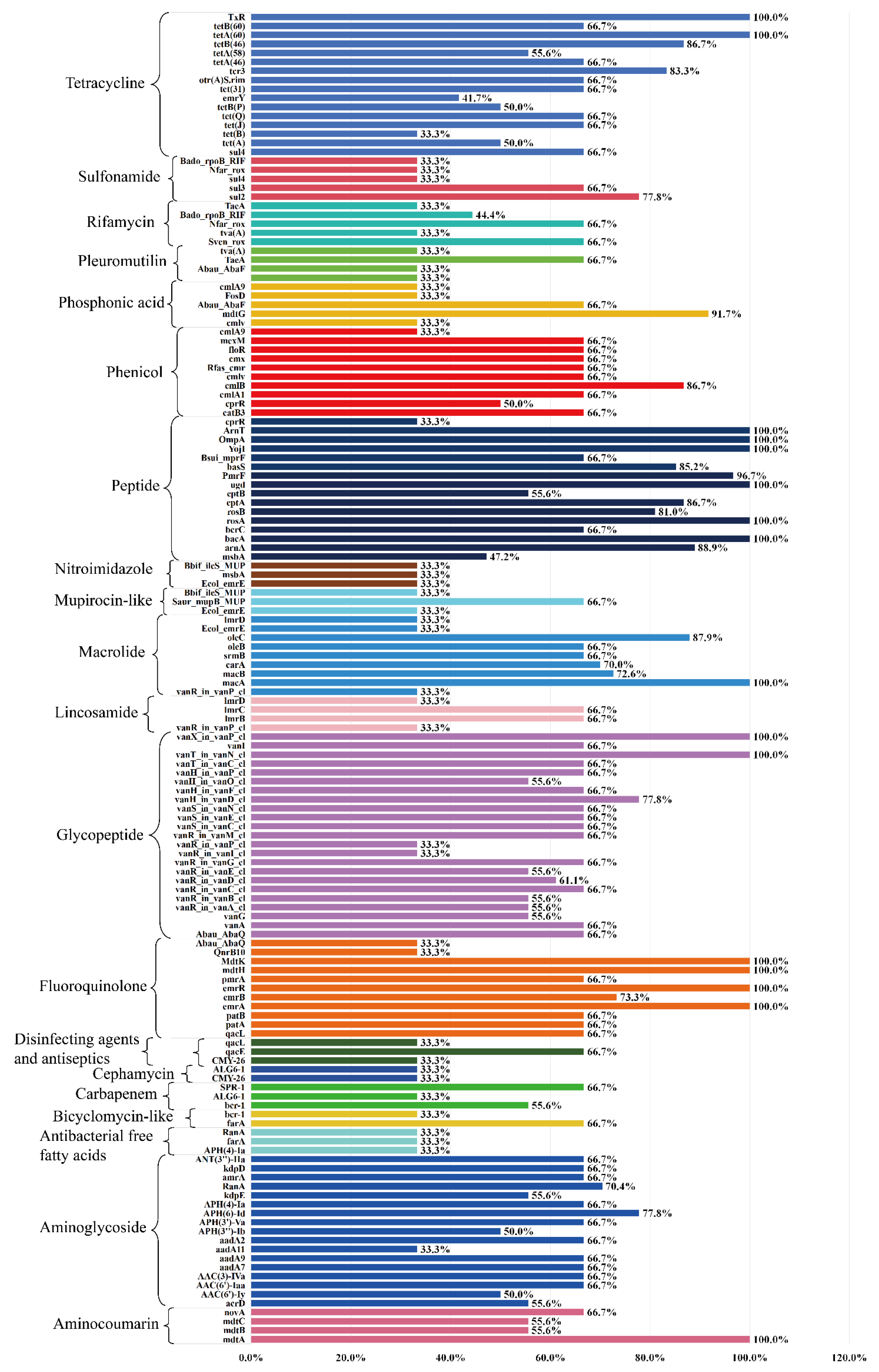
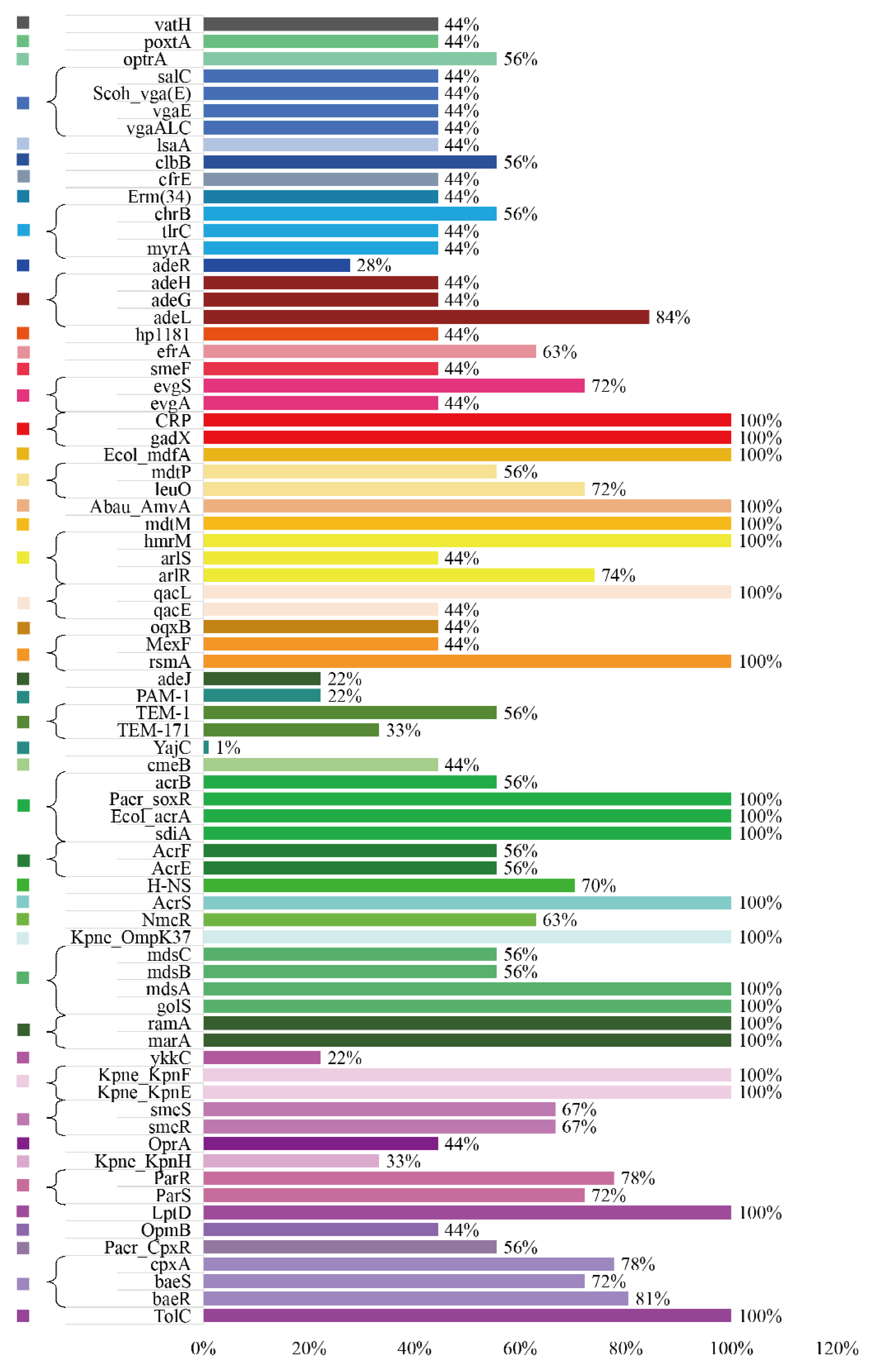
3.5. ARGs Profiles
3.6. Plasmid Replicons and Mobility Types
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, G.; Jian, S.; Li, W.; Yan, L.; Chen, T.; Cheng, T.; Liu, Z.; Ye, G.; Tang, H.; Zhang, L. Epigallocatechin gallate protects mice from Salmonella enterica ser. Typhimurium infection by modulating bacterial virulence through quorum sensing inhibition. Front. Cell. Infect. Microbiol. 2024, 14, 1432111. [Google Scholar] [CrossRef]
- Marin, C.; Chinillac, M.C.; Cerdà-Cuéllar, M.; Montoro-Dasi, L.; Sevilla-Navarro, S.; Ayats, T.; Marco-Jimenez, F.; Vega, S. Contamination of pig carcass with Salmonella enterica serovar Typhimurium monophasic variant 1,4[5],12:i:- originates mainly in live animals. Sci. Total Environ. 2020, 703, 134609. [Google Scholar] [CrossRef]
- Harrison, O.L.; Rensing, S.; Jones, C.K.; Trinetta, V. Salmonella enterica 4,[5],12:i:-, an Emerging Threat for the Swine Feed and Pork Production Industry. J. Food Prot. 2022, 85, 660–663. [Google Scholar] [CrossRef]
- Wachino, J.I. Horizontal Gene Transfer Systems for Spread of Antibiotic Resistance in Gram-Negative Bacteria. Microbiol. Immunol. 2025, 69, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, R.A.; Msefula, C.L.; Thomson, N.R.; Kariuki, S.; Holt, K.E.; Gordon, M.A.; Harris, D.; Clarke, L.; Whitehead, S.; Sangal, V.; et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009, 19, 2279–2287. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Z.X.; Feng, Y.L.; Zhang, X.W.; Liu, H.; Zeng, F.Y.; Li, X.Y. Whole-Genome Sequencing Reveals the Population Structure and Genetic Diversity of Salmonella Typhimurium ST34 and ST19 Lineages. J. Microbiol. 2024, 62, 859–870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hu, M.; Xu, X.; Lv, C.; Shi, C. Dynamic antimicrobial resistance and phylogenomic structure of Salmonella Typhimurium from 2007 to 2019 in Shanghai, China. Microbiol. Spectr. 2024, 12, e0026224. [Google Scholar] [CrossRef]
- Ali, M.S.; Na, S.H.; Moon, B.Y.; Kang, H.Y.; Kang, H.S.; Kim, S.J.; Kim, T.S.; Heo, Y.E.; Hwang, Y.J.; Yoon, S.S.; et al. Antimicrobial Resistance Profiles and Molecular Characteristics of Extended-Spectrum β-Lactamase-Producing Salmonella enterica Serovar Typhimurium Isolates from Food Animals During 2010-2021 in South Korea. Foodborne Pathog. Dis. 2024, 21, 634–642. [Google Scholar] [CrossRef]
- Chung The, H.; Pham, P.; Ha Thanh, T.; Phuong, L.V.K.; Yen, N.P.; Le, S.H.; Vu Thuy, D.; Chau, T.T.H.; Le Phuc, H.; Ngoc, N.M.; et al. Multidrug resistance plasmids underlie clonal expansions and international spread of Salmonella enterica serotype 1,4,[5],12:i:- ST34 in Southeast Asia. Commun. Biol. 2023, 6, 1007. [Google Scholar] [CrossRef]
- Biswas, S.; Li, Y.; Elbediwi, M.; Yue, M. Emergence and Dissemination of mcr-Carrying Clinically Relevant Salmonella Typhimurium Monophasic Clone ST34. Microorganisms 2019, 7, 298. [Google Scholar] [CrossRef]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef]
- McMillan, E.A.; Jackson, C.R.; Frye, J.G. Transferable Plasmids of Salmonella enterica Associated with Antibiotic Resistance Genes. Front. Microbiol. 2020, 11, 562181. [Google Scholar] [CrossRef] [PubMed]
- Mazhar, S.H.; Li, X.; Rashid, A.; Su, J.; Xu, J.; Brejnrod, A.D.; Su, J.Q.; Wu, Y.; Zhu, Y.G.; Zhou, S.G.; et al. Co-selection of antibiotic resistance genes, and mobile genetic elements in the presence of heavy metals in poultry farm environments. Sci. Total Environ. 2021, 755, 142702. [Google Scholar] [CrossRef]
- Zhou, X.; Zhu, Y.; Yue, X.; Chen, S.; Li, J.; Yang, Y.; Liu, S.; Zhao, K.; Han, X.; Zou, L. Emergence of tigecycline-resistant Escherichia coli from swine feces and fertilized soil. Ecotoxicol. Environ. Saf. 2025, 304, 119136. [Google Scholar] [CrossRef]
- Prathan, R.; Bitrus, A.A.; Sinwat, N.; Angkititrakul, S.; Chuanchuen, R. Phylogenetic characterization of Salmonella enterica from pig production and humans in Thailand and Laos border provinces. Vet. World 2019, 12, 79–84. [Google Scholar] [CrossRef]
- Ananchaipattana, C.; Hosotani, Y.; Kawasaki, S.; Bari, M.L.; Yamaguchi, K.A.; Inatsu, Y. Serotyping, RAPD Grouping and Antibiotic Susceptibility Testing of <i>Salmonella Enterica</i> Isolated from Retail Foods in Thailand. Food Sci. Technol. Res. 2014, 20, 905–913. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, K.; Lin, R.; Xu, X.; Xu, F.; Lin, Q.; Hu, Y.; Zhang, H.; Zhang, J.; Liao, M.; et al. High Levels of Antibiotic Resistance in MDR-Strong Biofilm-Forming Salmonella Typhimurium ST34 in Southern China. Microorganisms 2023, 11, 2005. [Google Scholar] [CrossRef]
- Zhang, K.; Ge, H.; He, J.; Hu, M.; Xu, Z.; Jiao, X.; Chen, X. Salmonella Typhimurium ST34 Isolate Was More Resistant than the ST19 Isolate in China, 2007 - 2019. Foodborne Pathog. Dis. 2022, 19, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Ji, L.; Zha, Y.; Dong, F.; Xu, D. Antimicrobial Resistance and Genomic Characterization of Salmonella Serovars Typhimurium and 4,[5],12:i:- in Huzhou, China. Infect. Drug Resist. 2025, 18, 2765–2777. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, T.J.; Zhang, W.; Xie, C.; Yang, Y.; Chen, X.; Wang, Q.; Wang, H.N.; Lei, C.W. Prevalence and genetic diversity of multidrug-resistant Salmonella Typhimurium monophasic variant in a swine farm from China. Front. Microbiol. 2023, 14, 1200088. [Google Scholar] [CrossRef] [PubMed]
- Nambiar, R.B.; Elbediwi, M.; Ed-dra, A.; Wu, B.; Yue, M. Epidemiology and antimicrobial resistance of Salmonella serovars Typhimurium and 4,[5],12:i- recovered from hospitalized patients in China. Microbiol. Res. 2024, 282, 127631. [Google Scholar] [CrossRef]
- Gong, J.; Zhuang, L.; Zhang, D.; Zhang, P.; Dou, X.; Wang, C. Establishment of a Multiplex Loop-Mediated Isothermal Amplification Method for Rapid Detection of Sulfonamide Resistance Genes (sul1, sul2, sul3) in Clinical Enterobacteriaceae Isolates from Poultry. Foodborne Pathog. Dis. 2018, 15, 413–419. [Google Scholar] [CrossRef]
- Xie, X.; Wang, Z.; Zhang, K.; Li, Y.; Hu, Y.; Pan, Z.; Chen, X.; Li, Q.; Jiao, X. Pig as a reservoir of CRISPR type TST4 Salmonella enterica serovar Typhimurium monophasic variant during 2009-2017 in China. Emerg. Microbes Infect. 2020, 9, 1–4. [Google Scholar] [CrossRef]
- Conte, D.; Mesa, D.; Krul, D.; Bail, L.; Ito, C.A.S.; Palmeiro, J.K.; Dalla-Costa, L.M. Comparative genomics of IncQ1 plasmids carrying bla(GES) variants from clinical and environmental sources in Brazil. Infect. Genet. Evol. 2024, 123, 105644. [Google Scholar] [CrossRef] [PubMed]
- Mourão, J.; Novais, C.; Machado, J.; Peixe, L.; Antunes, P. Metal tolerance in emerging clinically relevant multidrug-resistant Salmonella enterica serotype 4,[5],12:i:- clones circulating in Europe. Int. J. Antimicrob. Agents 2015, 45, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Visetnoi, S.; Nelles, W. Can Organic Pork Help Achieve Sustainable Development Goals in Thailand? Agriculture 2023, 13, 1822. [Google Scholar] [CrossRef]
- Elnekave, E.; Hong, S.L.; Lim, S.; Boxrud, D.; Rovira, A.; Mather, A.E.; Perez, A.; Alvarez, J. Transmission of Multidrug-Resistant Salmonella enterica Subspecies enterica 4,[5],12:i:- Sequence Type 34 between Europe and the United States. Emerg. Infect. Dis. 2020, 26, 3034–3038. [Google Scholar] [CrossRef]

| Stains ID | Serotype | Source | Years |
|---|---|---|---|
| T4 | S. Typhimurium | Carcass | 2024 |
| T5 | Monophasci S. Typhimurium | Carcass | 2024 |
| T8 | Monophasci S. Typhimurium | Carcass | 2024 |
| T140 | Monophasci S. Typhimurium | feces | 2024 |
| T146 | Monophasci S. Typhimurium | feces | 2024 |
| T220 | S. Typhimurium | Pork | 2024 |
| T268 | S. Typhimurium | Carcass | 2024 |
| T271 | S. Typhimurium | feces | 2024 |
| T272 | S. Typhimurium | feces | 2024 |
| Strain ID | T4 | T5 | T8 | T140 | T146 | T220 | T268 | T271 | T272 |
|---|---|---|---|---|---|---|---|---|---|
| Season | Summer | Summer | Summer | Raniy | Raniy | Raniy | Winter | Winter | Winter |
| Source | Carcass | Carcass | Carcass | feces | feces | Pork | Carcass | feces | feces |
| Reads (M) | 5,737,783 | 5,839,821 | 7,328,517 | 7,325,203 | 5,226,024 | 42,99,303 | 3,433,505 | 4,550,328 | 4,657,471 |
| Genome size (Mb) | 4,761,534 | 4,984,354 | 4,844,384 | 4,844,384 | 4,991,203 | 4,977,876 | 5,000,978 | 4,999,702 | 4,949,535 |
| GC (%) | 52.14% | 52.12% | 52.04% | 52.04% | 52.15% | 52.16% | 52.15% | 52.06% | 52.16% |
| No. of contigs | 26 | 58 | 32 | 32 | 74 | 71 | 63 | 61 | 73 |
| N50 (kb) | 531,293 | 280,191 | 594,705 | 594,705 | 224,050 | 223,414 | 236,456 | 224,050 | 223,414 |
| Strain ID | Replicon Types | oriT/Relaxase | T4SS | T4CP |
|---|---|---|---|---|
| T4 | IncQ1 | MobA_MobL | / | FtsK-like DNA translocase |
| T5 | IncQ1 | MOB_H | tfc19; tfc18; tfc22; tfc23; tfc24; tfc17; virb4; tfc15; tfc14; tfc13; tfc12; tfc11; tfc10; tfc9; tfc8; tfc7; tfc5; virB1; tfc3; tfc2 | TraD |
| T8 | / | / | / | FtsK-like DNA translocase |
| T140 | / | / | / | FtsK-like DNA translocase |
| T146 | IncQ1 | MOB_H | tfc2; tfc3; tfc5; tfc7; tfc8; tfc9; tfc10; tfc11; tfc12; tfc13; tfc14, tfc15; tfc17; tfc18; tfc19; tfc22; tfc23; tfc24; virB1; virb4 | TraD |
| T220 | Col(pHAD28) | MOB_H | tfc2; tfc3; tfc5; tfc7; tfc8; tfc9; tfc10; tfc11; tfc12; tfc13; tfc14; tfc15; tfc17; tfc18; tfc19; tfc22; tfc23; tfc24; virB1; virb4 | TraD |
| T268 | IncQ1 | MOB_H | tfc2; tfc3; tfc5; tfc7; tfc8; tfc9; tfc10; tfc11; tfc12; tfc13; tfc14; tfc15; tfc17; tfc18; tfc19; tfc22; tfc23; tfc24; virB1; virb4 | TraD |
| T271 | Col(BS512) IncQ1 IncX1 | MOB_H | tfc2; tfc3; tfc5; tfc7; tfc8; tfc9; tfc10; tfc11; tfc12; tfc13; tfc14; tfc15; tfc17; tfc18; tfc19; tfc22; tfc23; tfc24; virB1; virb4 | TraD |
| T272 | IncQ1 | MOB_H | tfc2; tfc3; tfc5; tfc7; tfc8; tfc9; tfc10; tfc11; tfc12; tfc13; tfc14; tfc15; tfc17; tfc18; tfc19; tfc22; tfc23; tfc24; virB1; virb4 | TraD |
| Strain ID | Cargo Genes | |||||
|---|---|---|---|---|---|---|
| ARG | VF | Metal Resistance | Degradation | Symbiosis | Anti-Crispr | |
| T4 | aac(6′)-Iaa tet(A) aph(6)-Id aph(3″)-Ib sul2 | SlrP; sopD2; msbA; nueA; ompA; pipB; sopB/sigD; csgG; csgF; csgE; csgD; csgB; csgA; csgC; flgB; flgC; flgE; flgF; flgG; flgH; flgI; flmH; sifA; spiC/ssaB; ssaC; ssaD; ssaE; sseA; sseB; sscA; sseC; sseD; sseE; sscB; sseF; sseG; ssaG; ssaH; ssaI; ssaJ; ssaK; ssaL; ssaM; ssaV; ssaN; ssaO; ssaP; ssaQ; ssaR; ssaS; ssaT; ssaU; sodB; steA; sifB; sseJ; steC; galU; kdsA; sopE2; flhA; flhB; cheZ; cheY; cheB; cheR; cheW; cheA; motB; motA; flhC; flhD; fliA; fliS; fliG; fliI; fliM; fliN; fliP; fliQ; fliR; rcsA; sopA; sopA; gogB; ugd; gnd; ddhB; ddhA; galF; wcaJ; manB; wcaG; gmd; wza; avrA; orgC; orgB/SctL; orgA/sctK; prgK; prgJ; prgI; prgH; sptP; sicP; sipA/sspA; sipD; sipC/sspC; sipB/sspB; sicA; spaS; spaR; spaQ; spaP; spaO/sctQ; invJ; invI; invC/sctN; invB; invA; invE; invG; invF; invH; sopD; AHA_3493; rfaE; wbbO; glf; entA; entB; entE; entC; fepB; entS; fepD; fepG; fepC; entF; fes; fepA; gtrA; gtrB; fimF; fimH; fimD; fimC; fimI; allD; allC; allB; allR; allA; allS; acrA; acrB; gmhA/lpcA; htpB; cheD; bcfA; bcfB; bcfC; bcfD; bcfD; bcfE; bcfF; bcfG; tufA; lpfE; lpfD; lpfC; lpfB; lpfA; rfaD; rfaF; misL; mgtB; mgtC; shdA; acrB; pla; pla; sseL; rcsB; lpxC; lpxA; lpxB; IlpA; iroB; iroC; iroD; iroN; pipB2; mig-14; luxS; algU; sinH; ratB; shdA; acrB; acrA; tufA; tufA | modE; modA; modB; modC; mntR; kdeA; nfsA; comR/ycfQ; bhsA/ycfR/comC; phoB; zinT/yodA; kpnO; kpnE; kpnF; kpnO; kpnO; kmrA; dsbB; znuA/yebL; znuC/yebM; znuB/yebI; ruvB; cutC; kpnO; arsA; mdtA; mdtB; mdtC; rcnB/yohN; yfeC; rcnR/yohL; rcnA/yohM; dsbC; zupT/ygiE; yqjH; zitB/ybgR; corC; cutE/lnt; cueR/ybbI; copA; acrD/yffA; golS; golT; zur/yjbK; actP/yjcG; cutA; mgtA; zntA/yhhO; nikR; cueP; acrD; mntH/yfeP; pmrG; kpnO; corD; cuiD; cutF/nlpE; oxyRkp; fieF/yiip; cpxR; cpxA; dsbA; corA; pstB; pstA; pstC; pstS; acrD/yffA; zraR/hydH; zraP; zntR/yhdM | bhpD; dcl | NoeL | AcrIF7; AcrIIA7; AcrIIA7; AcrIIC1 |
| T5 | aac(6′)-Iaa; tet(B); aph(6)-Id; aph(3″)-Ib; sul2; blaTEM-1B | sodCI; sseI/srfH; ompA; pipB; sopB/sigD; csgG; csgF; csgE; csgD; csgB; csgA; csgC; flgB; flgC; flgE; flgF; flgG; flgH; flgI; flmH; sifA; spiC/ssaB; ssaC; ssaD; ssaE; sseA; sseB; sscA; sseC; sseD; sseE; sscB; sseF; sseG; ssaG; ssaH; ssaI; ssaJ; ssaK; ssaL; ssaM; ssaV; ssaN; ssaO; ssaP; ssaQ; ssaR; ssaS; ssaT; ssaU; sodB; steA; sifB; sseJ; steC; galU; kdsA; sopE2; flhA; flhB; cheZ; cheY; cheB; cheR; cheW; cheA; motB; motA; flhC; flhD; fliA; iroB; iroC; iroD; iroN; pipB2; mig-14; luxS; avrA; orgC; orgB/SctL; orgA/sctK; prgK; prgJ; prgI; prgH; sptP; sicP; sipA/sspA; sipD; sipC/sspC; sipB/sspB; sicA; spaS; spaR; spaQ; spaP; spaO/sctQ; invJ; invI; invC/sctN; invB; invA; invE; invG; invF; invH; sopD; AHA_3493; rfaE; mgtC; mgtB; misL; rfaF; rfaD; lpfA; lpfB; lpfC; lpfD; lpfE; acrB; acrA; allS; allA; allR; allB; allC; allD; fimI; fimC; fimD; fimH; fimF; gtrB; gtrA; fepA; fes; entF; fepC; fepG; fepD; entS; fepB; entC; entE; entB; entA; glf; wbbO; bcfG; bcfF; bcfE; bcfD; bcfC; bcfB; bcfA; cheD; gtrA; gtrB; gmhA/lpcA; hsiC1/vipB; hsiB1/vipA; htpB; sseK2; wza; gmd; wcaG; manB; wcaJ; galF; ddhA; ddhB; gnd; ugd; sopA; IlpA; lpxB; lpxA; lpxC; rcsB; sseL; shdA; ratB; sinH; gogB; gtrB; sopD2; msbA; nueA; acrB; shdA; pla; fliS; fliG; fliI; fliM; fliN; fliP; fliQ; fliR; rcsA; acrB; acrA; slrP; tufA; tufA; sseK1; algU; sspH2; tufA | comR/ycfQ; bhsA/ycfR/comC; phoB; zinT/yodA; kpnO; kpnE; kpnF; kpnO; kpnO; kmrA; dsbB; znuA/yebL; znuC/yebM; znuB/yebI; ruvB; cutC; yfeC; rcnR/yohL; rcnA/yohM; dsbC; zupT/ygiE; yqjH; cueP; nikR; zntA/yhhO; acrD/yffA; copA; cueR/ybbI; cusS; cutE/lnt; corC; zitB/ybgR; mgtA; golS; golT; cutA; pcoE; pcoS; pcoR; pcoD; pcoC; pcoB; pcoA; silP; silA; silB; silF; silC; silR; silS; silE; arsC; arsB; arsA; arsD; arsR; actP/yjcG; rcnB/yohN; mdtC; mdtB; mdtA; cutF/nlpE; cuiD; corD; oxyRkp; fieF/yiip; cpxR; cpxA; dsbA; kpnO; pmrG; mntR; kdeA; nfsA; corA; pstS; pstC; pstA; pstB; zur/yjbK; acrD; mntH/yfeP; kpnO; acrD/yffA; modC; modB; modA; modE; zraP; zraR/hydH; zntR/yhdM; merR; merT; merP; merA; merD; merE | bhpD; dcl | NoeL | AcrIF7; AcrIIA7; AcrIIA7; AcrIIC1 |
| T8 | aac(6′)-Iaa; tet(A); aadA2; cmlA1; ant(3″)-Ia; sul3; aac(3)-IVa; blaTEM-1B | glf; wbbO; slrP; sopD2; msbA; nueA; ompA; pipB; sopB/sigD; csgG; csgF; csgE; csgD; csgB; csgA; csgC; flgB; flgC; flgE; flgF; flgG; flgH; flgI; flmH; sifA; spiC/ssaB; ssaC; ssaD; ssaE; sseA; sseB; sscA; sseC; sseD; sseE; sscB; sseF; sseG; ssaG; ssaH; ssaI; ssaJ; ssaK; ssaL; ssaM; ssaV; ssaN; ssaO; ssaP; ssaQ; ssaR; ssaS; ssaT; ssaU; sodB; steA; sifB; sseJ; steC; galU; IlpA; lpxB; lpxA; lpxD; lpxC; bcfG; bcfF; bcfE; bcfD; bcfC; bcfB; bcfA; cheD; htpB; gtrB; avrA; orgC; orgB/SctL; orgA/sctK; prgK; prgJ; prgI; prgH; sptP; sicP; sipA/sspA; sipD; sipC/sspC; sipB/sspB; sicA; spaS; spaR; spaQ; spaP; spaO/sctQ; invJ; invI; invC/sctN; invB; invA; invE; invG; invF; invH; sopD; AHA_3493; rfaE; acrA; acrB; mgtC; mgtB; misL; rfaF; rfaD; lpfA; lpfB; lpfC; lpfD; lpfE; rcsB; sseL; acrB; shdA; ratB; sinH; algU; hsiB1/vipA; hsiC1/vipB; gmhA/lpcA; acrB; acrA; allS; allA; allR; allB; allC; allD; fimI; fimC; fimD; fimH; fimF; gtrB; gtrA; fepA; fes; entF; fepC; fepG; fepD; entS; fepB; entC; entE; entB; entA; kdsA; sopE2; sspH2; flhA; flhB; cheZ; cheY; cheB; cheR; cheW; cheA; motB; motA; flhC; flhD; fliA; fliC; sseK2; wza; gmd; wcaG; manB; manB; gnd; ugd; sopA; rcsA; fliR; fliQ; fliP; fliN; fliM; fliI; fliG; fliF; fliS; wbtL; fliC; iroB; iroC; iroD; iroN; pipB2; mig-14; luxS; tufA; sseK1; manB; wcaG; gmd; galF; wcaJ; manB; tufA | cutE/lnt; corC; zitB/ybgR; modE; modA; modB; modC; mntR; kdeA; nfsA; comR/ycfQ; bhsA/ycfR/comC; phoB; zinT/yodA; kpnO; kpnE; kpnF; kpnO; kpnO; kmrA; cutF/nlpE; cuiD; corD; mgtA; cutA; actP/yjcG; zur/yjbK; yfeC; rcnR/yohL; rcnA/yohM; dsbC; zupT/ygiE; yqjH; acrD/yffA; cueP; nikR; zntA/yhhO; kpnO; pmrG; mntH/yfeP; acrD; golT; golS; acrD/yffA; copA; cueR/ybbI; cusS; dsbB; znuA/yebL; znuC/yebM; znuB/yebI; ruvB; cutC; rcnB/yohN; mdtC; mdtB; mdtA; oxyRkp; fieF/yiip; cpxR; cpxA; dsbA; arsA; kpnO; corA; pstS; pstC; pstA; pstB; zraP; zraR/hydH; zntR/yhdM; silE; silS; silR; silC; silF; silB; silA; silP; merR; merT; merP; merA; merD; merE | dcl; bhpD | NoeL; NoeL | AcrIF7; AcrIIA7; AcrIIA7; AcrIIC1 |
| T140 | aac(6′)-Iaa; tet(A); aadA2; cmlA1; ant(3″)-Ia; sul3; aac(3)-IVa; blaTEM-1B | glf; wbbO; slrP; sopD2; msbA; nueA; ompA; pipB; sopB/sigD; csgG; csgF; csgE; csgD; csgB; csgA; csgC; flgB; flgC; flgE; flgF; flgG; flgH; flgI; flmH; sifA; spiC/ssaB; ssaC; ssaD; ssaE; sseA; sseB; sscA; sseC; sseD; sseE; sscB; sseF; sseG; ssaG; ssaH; ssaI; ssaJ; ssaK; ssaL; ssaM; ssaV; ssaN; ssaO; ssaP; ssaQ; ssaR; ssaS; ssaT; ssaU; sodB; steA; sifB; sseJ; steC; galU; IlpA; lpxB; lpxA; lpxD; lpxC; bcfG; bcfF; bcfE; bcfD; bcfC; bcfB; bcfA; cheD; htpB; gtrB; avrA; orgC; orgB/SctL; orgA/sctK; prgK; prgJ; prgI; prgH; sptP; sicP; sipA/sspA; sipD; sipC/sspC; sipB/sspB; sicA; spaS; spaR; spaQ; spaP; spaO/sctQ; invJ; invI; invC/sctN; invB; invA; invE; invG; invF; invH; sopD; AHA_3493; rfaE; acrA; acrB; mgtC; mgtB; misL; rfaF; rfaD; lpfA; lpfB; lpfC; lpfD; lpfE; rcsB; sseL; acrB; shdA; ratB; sinH; algU; hsiB1/vipA; hsiC1/vipB; gmhA/lpcA; acrB; acrA; allS; allA; allR; allB; allC; allD; fimI; fimC; fimD; fimH; fimF; gtrB; gtrA; fepA; fes; entF; fepC; fepG; fepD; entS; fepB; entC; entE; entB; entA; kdsA; sopE2; sspH2; flhA; flhB; cheZ; cheY; cheB; cheR; cheW; cheA; motB; motA; flhC; flhD; fliA; fliC; sseK2; wza; gmd; wcaG; manB; manB; gnd; ugd; sopA; rcsA; fliR; fliQ; fliP; fliN; fliM; fliI; fliG; fliF; fliS; wbtL; fliC; iroB; iroC; iroD; iroN; pipB2; mig-14; luxS; tufA; sseK1; manB; wcaG; gmd; galF; wcaJ; manB; tufA | cutE/lnt; corC; zitB/ybgR; modE; modA; modB; modC; mntR; kdeA; nfsA; comR/ycfQ; bhsA/ycfR/comC; phoB; zinT/yodA; kpnO; kpnE; kpnF; kpnO; kpnO; kmrA; cutF/nlpE; cuiD; corD; mgtA; cutA; actP/yjcG; zur/yjbK; yfeC; rcnR/yohL; rcnA/yohM; dsbC; zupT/ygiE; yqjH; acrD/yffA; cueP; nikR; zntA/yhhO; kpnO; pmrG; mntH/yfeP; acrD; golT; golS; acrD/yffA; copA; cueR/ybbI; cusS; dsbB; znuA/yebL; znuC/yebM; znuB/yebI; ruvB; cutC; rcnB/yohN; mdtC; mdtB; mdtA; oxyRkp; fieF/yiip; cpxR; cpxA; dsbA; arsA; kpnO; corA; pstS; pstC; pstA; pstB; zraP; zraR/hydH; zntR/yhdM; silE; silS; silR; silC; silF; silB; silA; silP; merR; merT; merP; merA; merD; merE | dcl; bhpD | NoeL; NoeL | AcrIF7; AcrIIA7; AcrIIA7; AcrIIC1 |
| T146 | aac(6′)-Iaa; aph(6)-Id; aph(3″)-Ib; sul2; blaTEM-1B | kdsA; galU; steC; sseJ; sifB; steA; sodB; ssaU; ssaT; ssaS; ssaR; ssaQ; ssaP; ssaO; ssaN; ssaV; ssaM; ssaL; ssaK; ssaJ; ssaI; ssaH; ssaG; sseG; sseF; sscB; sseE; sseD; sseC; sscA; sseB; sseA; ssaE; ssaD; ssaC; spiC/ssaB; sifA; flmH; flgI; flgH; flgG; flgF; flgE; flgC; flgB; csgC; csgA; csgB; csgD; csgE; csgF; csgG; sopB/sigD; pipB; ompA; sseI/srfH; sopD; AHA_3493; rfaE; lpfE; lpfD; lpfC; lpfB; lpfA; rfaD; rfaF; misL; wbbO; glf; entA; entB; entE; entC; fepB; entS; fepD; fepG; fepC; entF; fes; fepA; gtrA; gtrB; fimF; fimH; fimD; fimC; fimI; allD; allC; allB; allR; allA; allS; acrA; acrB; cheD; bcfA; bcfB; bcfC; bcfD; bcfE; bcfF; bcfG; htpB; IlpA; lpxB; lpxA; lpxC; sseK2; wza; gmd; wcaG; manB; wcaJ; galF; ddhA; ddhB; gnd; ugd; sopA; gtrA; gtrB; acrB; shdA; ratB; sinH; gogB; sseL; rcsB; luxS; avrA; orgC; orgB/SctL; orgA/sctK; prgK; prgJ; prgI; prgH; sptP; sicP; sipA/sspA; sipD; sipC/sspC; sipB/sspB; sicA; spaS; spaR; spaQ; spaP; spaO/sctQ; invJ; invI; invC/sctN; invB; invA; invE; invG; invF; invH; sopE2; flhA; flhB; cheZ; cheY; cheB; cheR; cheW; cheA; motB; motA; flhC; flhD; fliA; gtrB; sopD2; msbA; nueA; pla; fliS; fliG; fliI; fliM; fliN; fliP; fliQ; fliR; rcsA; hsiC1/vipB; hsiB1/vipA; mgtC; mgtB; mig-14; pipB2; iroN; iroD; iroC; iroB; acrA; acrB; slrP; tufA; sseK1; gmhA/lpcA; algU; sodCI; sspH2; tufA | dsbB; kmrA; kpnO; kpnO; kpnF; kpnE; kpnO; zinT/yodA; phoB; bhsA/ycfR/comC; comR/ycfQ; rcnR/yohL; rcnA/yohM; dsbC; zupT/ygiE; yqjH; zntA/yhhO; nikR; cueP; zitB/ybgR; corC; cutE/lnt; cusS; cueR/ybbI; copA; acrD/yffA; mgtA; actP/yjcG; arsR; arsD; arsA; arsB; arsC; silE; silS; silR; silC; silF; silB; silA; silP; pcoA; pcoB; pcoC; pcoD; pcoR; pcoS; pcoE; cutA; cutF/nlpE; cuiD; corD; rcnB/yohN; mdtC; mdtB; mdtA; golS; golT; acrD; dsbA; cpxA; cpxR; fieF/yiip; oxyRkp; pmrG; kpnO; yfeC; znuA/yebL; znuC/yebM; znuB/yebI; ruvB; cutC; corA; pstS; pstC; pstA; pstB; zur/yjbK; nfsA; kdeA; mntH/yfeP; kpnO; acrD/yffA; modE; modA; modB; modC; zraP; zraR/hydH; mntR; zntR/yhdM; merR; merT; merP; merA; merD; merE | bhpD; dcl | NoeL | AcrIF7; AcrIIA7; AcrIIC1; AcrIIA7 |
| T220 | aac(6′)-Iaa; tet(B); sul2; aph(3″)-Ib; aph(6)-Id; qnrB19; blaTEM-1B | aac(6′)-Iaa; tet(B); sul2; aph(3″)-Ib; aph(6)-Id; qnrB19; blaTEM-1B | dsbB; kmrA; kpnO; kpnO; kpnF; kpnE; kpnO; zinT/yodA; phoB; bhsA/ycfR/comC; comR/ycfQ; zntA/yhhO; nikR; cueP; zitB/ybgR; corC; cutE/lnt; cusS; cueR/ybbI; copA; acrD/yffA; actP/yjcG; arsR; arsD; arsA; arsB; arsC; silE; silS; silR; silC; silF; silB; silA; silP; pcoA; pcoB; pcoC; pcoD; pcoR; pcoS; pcoE; cutA; mgtA; yqjH; zupT/ygiE; rcnR/yohL; rcnA/yohM; dsbC; cutF/nlpE; cuiD; corD; rcnB/yohN; mdtC; mdtB; mdtA; golS; golT; acrD; oxyRkp; fieF/yiip; cpxR; cpxA; dsbA; znuA/yebL; znuC/yebM; znuB/yebI; ruvB; cutC; kpnO; pmrG; yfeC; corA; pstB; pstA; pstC; pstS; zur/yjbK; kdeA; nfsA; mntH/yfeP; kpnO; acrD/yffA; modC; modB; modA; modE; zraP; zraR/hydH; mntR; zntR/yhdM; merR; merT; merP; merA; merD; merE | bhpD; dcl | NoeL | AcrIF7; AcrIIC1; AcrIIA7 |
| T268 | aac(6′)-Iaa; aph(6)-Id; aph(3″)-Ib; sul2; blaTEM-1B | csgG; csgF; csgE; csgD; csgB; csgA; csgC; flgB; flgC; flgE; flgF; flgG; flgH; flgI; flmH; sifA; spiC/ssaB; ssaC; ssaD; ssaE; sseA; sseB; sscA; sseC; sseD; sseE; sscB; sseF; sseG; ssaG; ssaH; ssaI; ssaJ; ssaK; ssaL; ssaM; ssaV; ssaN; ssaO; ssaP; ssaQ; ssaR; ssaS; ssaT; ssaU; sodB; steA; sifB; sseJ; steC; galU; kdsA; rfaE; AHA_3493; sopD; invH; invF; invG; invE; invA; invB; invC/sctN; invI; invJ; spaO/sctQ; spaP; spaQ; spaR; spaS; sicA; sipB/sspB; sipC/sspC; sipD; sipA/sspA; sicP; sptP; prgH; prgI; prgJ; prgK; orgA/sctK; orgB/SctL; orgC; avrA; luxS; lpfE; lpfD; lpfC; lpfB; lpfA; rfaD; rfaF; misL; mgtB; mgtC; wbbO; glf; entA; entB; entE; entC; fepB; entS; fepD; fepG; fepC; entF; fes; fepA; gtrA; gtrB; fimF; fimH; fimD; fimC; fimI; allD; allC; allB; allR; allA; allS; acrA; acrB; bcfG; bcfF; bcfE; bcfD; bcfC; bcfB; bcfA; cheD; sseK2; wza; gmd; wcaG; manB; wcaJ; galF; ddhA; ddhB; gnd; ugd; sopA; htpB; lpxC; lpxA; lpxB; IlpA; gtrA; gtrB; rcsB; sseL; shdA; ratB; sinH; gogB; fliA; flhD; flhC; motA; motB; cheA; cheW; cheR; cheB; cheY; cheZ; flhB; flhA; sopE2; sodCI; sseI/srfH; ompA; pipB; sopB/sigD; acrB; shdA; gmhA/lpcA; hsiC1/vipB; hsiB1/vipA; gtrB; nueA; msbA; sopD2; pla; tufA; rcsA; fliR; fliQ; fliP; fliN; fliM; fliI; fliG; fliS; iroB; iroC; iroD; iroN; pipB2; mig-14; acrA; acrB; slrP; sseK1; tufA; algU; sspH2 | comR/ycfQ; bhsA/ycfR/comC; phoB; zinT/yodA; kpnO; kpnE; kpnF; kpnO; kpnO; kmrA; dsbB; yqjH; zupT/ygiE; dsbC; rcnA/yohM; rcnR/yohL; yfeC; zntA/yhhO; nikR; cueP; zitB/ybgR; corC; cutE/lnt; cusS; cueR/ybbI; copA; acrD/yffA; mgtA; rcnB/yohN; mdtC; mdtB; mdtA; cutA; pcoE; pcoS; pcoR; pcoD; pcoC; pcoB; pcoA; silP; silA; silB; silF; silC; silR; silS; silE; arsC; arsB; arsA; arsD; arsR; actP/yjcG; corD; cuiD; cutF/nlpE; golS; golT; oxyRkp; fieF/yiip; cpxR; cpxA; dsbA; kpnO; pmrG; mntR; kdeA; nfsA; cutC; ruvB; znuB/yebI; znuC/yebM; znuA/yebL; corA; acrD; pstB; pstA; pstC; pstS; zur/yjbK; mntH/yfeP; zntR/yhdM; kpnO; acrD/yffA; modC; modB; modA; modE; zraR/hydH; zraP; merE; merD; merA; merP; merT; merR | dcl; bhpD | NoeL | AcrIF7; AcrIIA7; AcrIIA7; AcrIIC1 |
| T271 | aac(6′)-Iaa; tet(B); aph(6)-Id; aph(3″)-Ib; sul2; blaTEM-1B | sifA; spiC/ssaB; ssaC; ssaD; ssaE; sseA; sseB; sscA; sseC; sseD; sseE; sscB; sseF; sseG; ssaG; ssaH; ssaI; ssaJ; ssaK; ssaL; ssaM; ssaV; ssaN; ssaO; ssaP; ssaQ; ssaR; ssaS; ssaT; ssaU; sodB; steA; sifB; sseJ; steC; galU; kdsA; sopE2; flhA; flhB; cheZ; cheY; cheB; cheR; cheW; cheA; motB; motA; flhC; flhD; fliA; AHA_3493; sopD; invH; invF; invG; invE; invA; invB; invC/sctN; invI; invJ; spaO/sctQ; spaP; spaQ; spaR; spaS; sicA; sipB/sspB; sipC/sspC; sipD; sipA/sspA; sicP; sptP; prgH; prgI; prgJ; prgK; orgA/sctK; orgB/SctL; orgC; avrA; luxS; mig-14; pipB2; iroN; iroD; iroC; iroB; misL; rfaF; rfaD; lpfA; lpfB; lpfC; lpfD; lpfE; cheD; bcfA; bcfB; bcfC; bcfD; bcfE; bcfF; bcfG; htpB; flmH; flgI; flgH; flgG; flgF; flgE; flgC; flgB; csgC; csgA; csgB; csgD; csgE; csgF; csgG; sopB/sigD; pipB; ompA; sseI/srfH; sodCI; nueA; msbA; sopD2; lpxC; lpxA; lpxB; IlpA; sopA; ugd; gnd; ddhB; ddhA; galF; wcaJ; manB; wcaG; gmd; wza; sseK2; wbbO; glf; entA; entB; entE; entC; fepB; entS; fepD; fepG; fepC; entF; fes; fepA; acrB; shdA; ratB; sinH; algU; sseL; rcsB; rfaE; gtrA; gtrB; fimF; fimH; fimD; fimC; fimI; allD; allC; allB; allR; allA; allS; acrA; acrB; gtrB; pla; fliS; fliG; fliI; fliM; fliN; fliP; fliQ; fliR; rcsA; hsiC1/vipB; hsiB1/vipA; acrB; acrA; slrP; sseK1; gmhA/lpcA; mgtC; mgtB; sspH2; tufA | phoB; zinT/yodA; kpnO; kpnE; kpnF; kpnO; kpnO; kmrA; dsbB; znuA/yebL; znuC/yebM; znuB/yebI; ruvB; cutC; dsbC; rcnA/yohM; rcnR/yohL; yfeC; cueP; nikR; zntA/yhhO; mgtA; cutA; pcoE; pcoS; pcoR; pcoD; pcoC; pcoB; pcoA; silP; silA; silB; silF; silC; silR; silS; silE; arsC; arsB; arsA; arsD; arsR; actP/yjcG; bhsA/ycfR/comC; comR/ycfQ; corD; cuiD; cutF/nlpE; mdtA; mdtB; mdtC; rcnB/yohN; zitB/ybgR; corC; cutE/lnt; cusS; acrD; golS; golT; dsbA; cpxA; cpxR; fieF/yiip; oxyRkp; pmrG; kpnO; yqjH; zupT/ygiE; cueR/ybbI; copA; acrD/yffA; corA; pstB; pstA; pstC; pstS; zur/yjbK; nfsA; kdeA; mntH/yfeP; kpnO; acrD/yffA; modC; modB; modA; modE; zraP; zraR/hydH; mntR; zntR/yhdM; merE; merD; merA; merP; merT; merR | bhpD; dcl | NoeL | AcrIIA7; AcrIF7; AcrIIC1 |
| T272 | aac(6′)-Iaa; tet(B); sul2; aph(3″)-Ib; aph(6)-Id; blaTEM-1B | gogB; sinH; ratB; shdA; acrB; csgG; csgF; csgE; csgD; csgB; csgA; csgC; flgB; flgC; flgE; flgF; flgG; flgH; flgI; flmH; sifA; spiC/ssaB; ssaC; ssaD; ssaE; sseA; sseB; sscA; sseC; sseD; sseE; sscB; sseF; sseG; ssaG; ssaH; ssaI; ssaJ; ssaK; ssaL; ssaM; ssaV; ssaN; ssaO; ssaP; ssaQ; ssaR; ssaS; ssaT; ssaU; sodB; steA; sifB; sseJ; steC; galU; kdsA; sopE2; flhA; flhB; cheZ; cheY; cheB; cheR; cheW; cheA; motB; motA; flhC; flhD; fliA; iroB; iroC; iroD; iroN; pipB2; mig-14; luxS; avrA; orgC; orgB/SctL; orgA/sctK; prgK; prgJ; prgI; prgH; sptP; sicP; sipA/sspA; sipD; sipC/sspC; sipB/sspB; sicA; spaS; spaR; spaQ; spaP; spaO/sctQ; invJ; invI; invC/sctN; invB; invA; invE; invG; invF; invH; sopD; AHA_3493; lpfE; lpfD; lpfC; lpfB; lpfA; rfaD; rfaF; misL; cheD; bcfA; bcfB; bcfC; bcfD; bcfE; bcfF; bcfG; htpB; lpxC; lpxA; lpxB; IlpA; sopA; ugd; gnd; ddhB; ddhA; galF; wcaJ; manB; wcaG; gmd; wza; sseK2; wbbO; glf; entA; entB; entE; entC; fepB; entS; fepD; fepG; fepC; entF; fes; fepA; rfaE; gmhA/lpcA; sseL; rcsB; gtrA; gtrB; fimF; fimH; fimD; fimC; fimI; allD; allC; allB; allR; allA; allS; acrA; acrB; gtrB; nueA; msbA; sopD2; pla; hsiB1/vipA; hsiC1/vipB; mgtC; mgtB; acrB; acrA; sopB/sigD; pipB; ompA; sseI/srfH; slrP; fliS; fliG; fliI; fliM; fliN; fliP; fliQ; fliR; rcsA; tufA; sseK1; algU; sodCI; sspH2; sspH2; tufA | acrD; comR/ycfQ; bhsA/ycfR/comC; phoB; zinT/yodA; kpnO; kpnE; kpnF; kpnO; kpnO; kmrA; dsbB; znuA/yebL; znuC/yebM; znuB/yebI; ruvB; cutC; yfeC; rcnR/yohL; rcnA/yohM; dsbC; zntA/yhhO; nikR; cueP; mgtA; actP/yjcG; arsR; arsD; arsA; arsB; arsC; silE; silS; silR; silC; silF; silB; silA; silP; pcoA; pcoB; pcoC; pcoD; pcoR; pcoS; pcoE; cutA; corD; cuiD; cutF/nlpE; mdtA; mdtB; mdtC; rcnB/yohN; zitB/ybgR; corC; cutE/lnt; yqjH; zupT/ygiE; golS; golT; oxyRkp; fieF/yiip; cpxR; cpxA; dsbA; pmrG; kpnO; cusS; cueR/ybbI; copA; acrD/yffA; corA; pstS; pstC; pstA; pstB; zur/yjbK; nfsA; kdeA; mntH/yfeP; acrD/yffA; modE; modA; modB; modC; kpnO; zraP; zraR/hydH; mntR; zntR/yhdM; merR; merT; merP; merA; merD; merE | bhpD; dcl | NoeL | AcrIIC1; AcrIF7; AcrIIA7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Wang, N.; Angkititrakul, S.; Li, W.; Luo, Z.; Hou, M.; Wu, Y.; Shi, Y.; Wang, Y.; Li, F.; et al. Genomic Evidence for the Rise of Salmonella Typhimurium ST34 with Increased Plasmid-Mediated Resistance in the Thailand Pork Chain. Pathogens 2025, 14, 1190. https://doi.org/10.3390/pathogens14121190
Liu H, Wang N, Angkititrakul S, Li W, Luo Z, Hou M, Wu Y, Shi Y, Wang Y, Li F, et al. Genomic Evidence for the Rise of Salmonella Typhimurium ST34 with Increased Plasmid-Mediated Resistance in the Thailand Pork Chain. Pathogens. 2025; 14(12):1190. https://doi.org/10.3390/pathogens14121190
Chicago/Turabian StyleLiu, Hongmei, Ning Wang, Sunpetch Angkititrakul, Wengui Li, Zhongyang Luo, Mingpeng Hou, Yi Wu, Yubo Shi, Yuelin Wang, Fengyun Li, and et al. 2025. "Genomic Evidence for the Rise of Salmonella Typhimurium ST34 with Increased Plasmid-Mediated Resistance in the Thailand Pork Chain" Pathogens 14, no. 12: 1190. https://doi.org/10.3390/pathogens14121190
APA StyleLiu, H., Wang, N., Angkititrakul, S., Li, W., Luo, Z., Hou, M., Wu, Y., Shi, Y., Wang, Y., Li, F., Liu, Y., Wu, X., & Suksawat, F. (2025). Genomic Evidence for the Rise of Salmonella Typhimurium ST34 with Increased Plasmid-Mediated Resistance in the Thailand Pork Chain. Pathogens, 14(12), 1190. https://doi.org/10.3390/pathogens14121190





