Pathogenicity and Genomic Characterization of Vibrio parahaemolyticus VSP1: A Pathogen Linked to Enteritis Outbreak in Shrimp (Penaeus vannamei)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Experiment Shrimp
2.2. Tissue Sectioning and H&E Staining
2.3. DNA Extraction and Full-Length 16S rRNA Gene Sequencing
2.4. Sequencing Data Analysis
2.5. Isolation and Identification of Potential Pathogens
2.6. Virulence Test with the Isolates
2.7. Growth Curve and Carbon Source Utilization of the Isolates
2.8. Genomic Analysis of the Isolates
2.9. Antimicrobial Susceptibility Testing
3. Results
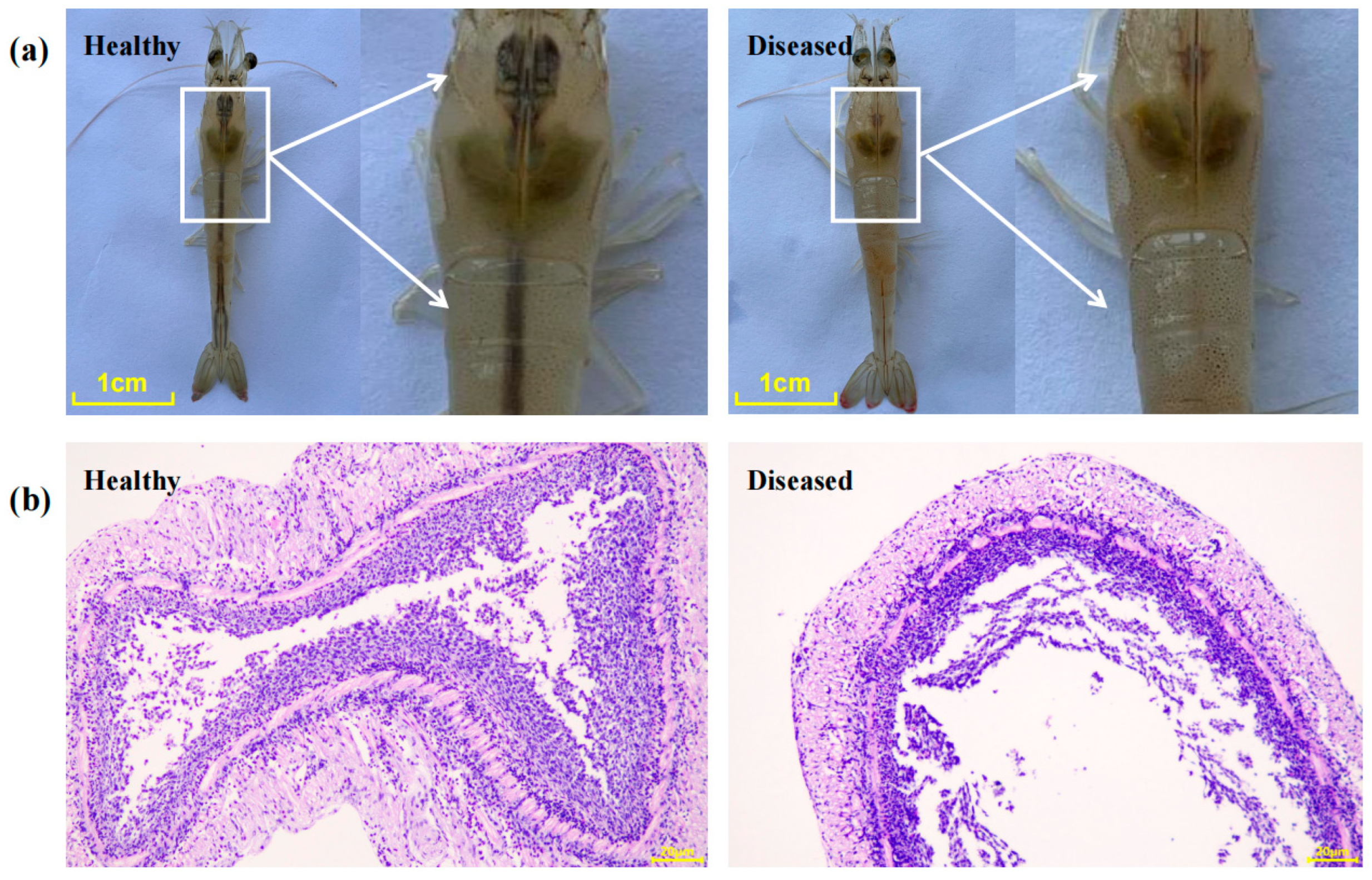
3.1. Clinical Signs and Histopathological Characterization of Diseased Shrimp
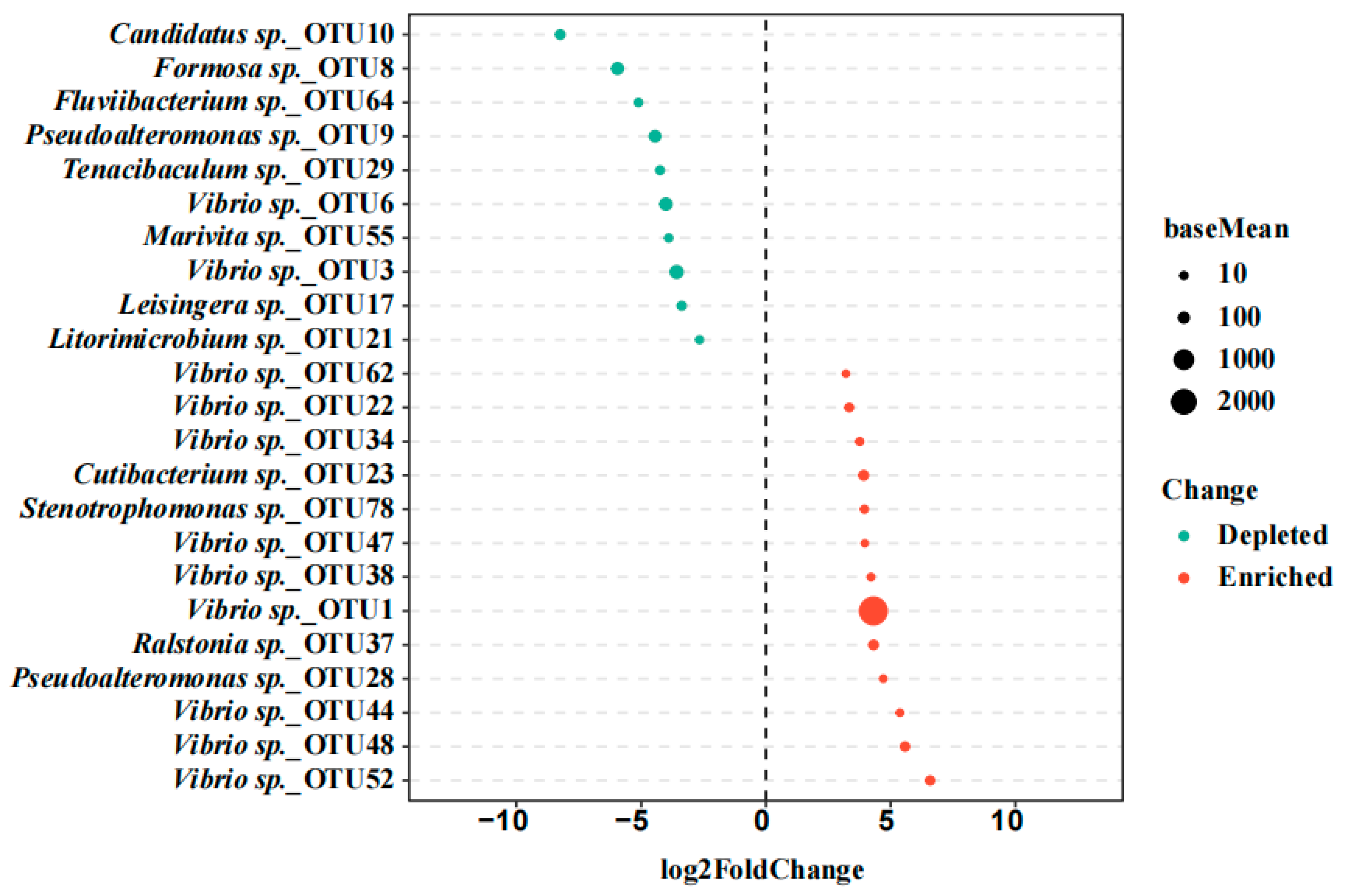
3.2. Screening of the Potential Pathogens Based on the Gut Bacterial Community Analysis
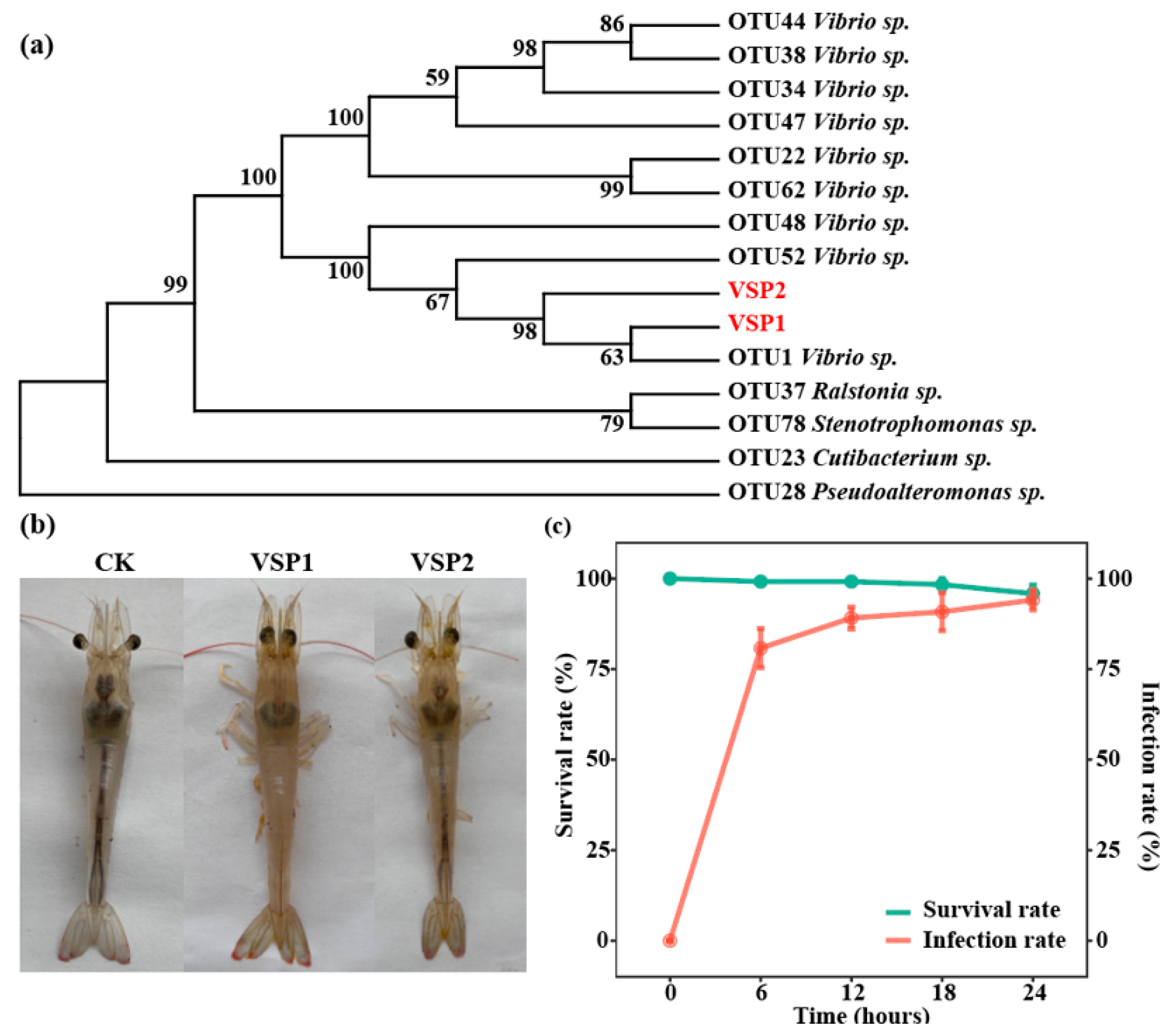
3.3. Targeted Isolation and Virulence Test of the Pathogen
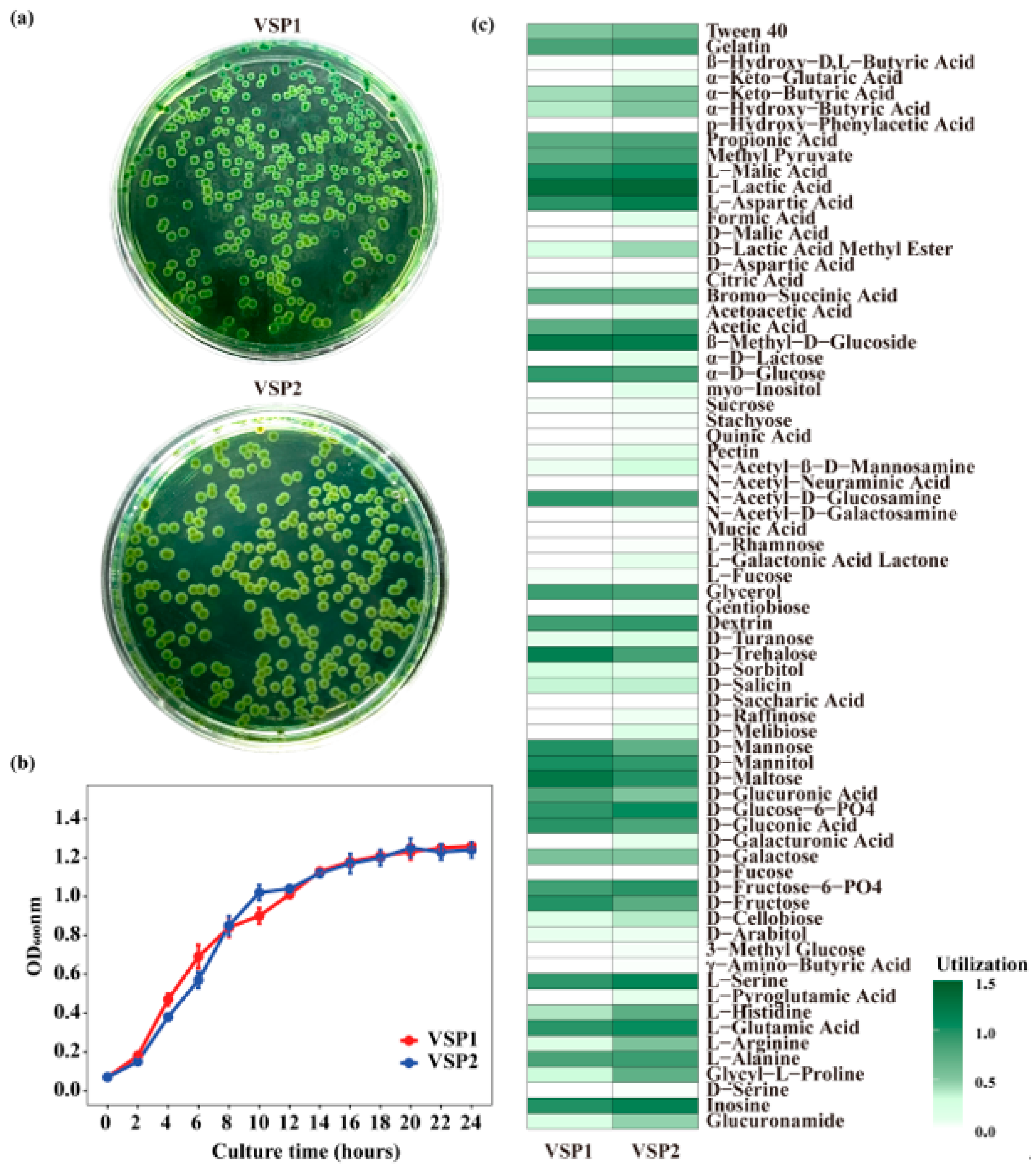
3.4. Biological Characteristics of the Isolated Strains
3.5. The Genome Characteristics of the Isolated Strains
3.6. Virulence and Antibiotic Resistance Genes of the Isolated Strains
4. Discussion
4.1. V. parahaemolyticus VSP1 Is the Pathogenic Bacterium Causing the Enteritis in Shrimp
4.2. VSP1 Has Multiple Virulence Factors, Supporting Its Infection, Colonization, and Damage to Shrimp
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2022. Towards Blue Transformation; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- Yu, Y.B.; Choi, J.H.; Kang, J.C.; Kim, H.J.; Kim, J.H. Shrimp bacterial and parasitic disease listed in the OIE: A review. Microb. Pathog. 2022, 166, 105545. [Google Scholar] [CrossRef]
- Jayasree, L.; Janakiram, P.; Madhavi, R. Characterization of Vibrio spp. Associated with Diseased Shrimp from Culture Ponds of Andhra Pradesh (India). J. World Aquac. Soc. 2006, 37, 523–532. [Google Scholar] [CrossRef]
- Kooloth Valappil, R.; Stentiford, G.D.; Bass, D. The rise of the syndrome—Sub-optimal growth disorders in farmed shrimp. Rev. Aquac. 2021, 13, 1888–1906. [Google Scholar] [CrossRef]
- Dalsgaard, A. The occurrence of human pathogenic Vibrio spp. and Salmonella in aquaculture*. Int. J. Food Sci. Technol. 2003, 33, 127–138. [Google Scholar] [CrossRef]
- Raszl, S.M.; Froelich, B.A.; Vieira, C.R.; Blackwood, A.D.; Noble, R.T. Vibrio parahaemolyticus and Vibrio vulnificus in South America: Water, seafood and human infections. J. Appl. Microbiol. 2016, 121, 1201–1222. [Google Scholar] [CrossRef] [PubMed]
- Stentiford, G.D.; Neil, D.M.; Peeler, E.J.; Shields, J.D.; Small, H.J.; Flegel, T.W.; Vlak, J.M.; Jones, B.; Morado, F.; Moss, S.; et al. Disease will limit future food supply from the global crustacean fishery and aquaculture sectors. J. Invertebr. Pathol. 2012, 110, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Fu, X.; Li, F.; Li, Z.; Wang, A.; Jiang, S.; Liu, C.; Wang, H. Integrated microRNA-mRNA analysis reveals a possible molecular mechanism of enteritis susceptibility in Litopenaeus vannamei. Fish Shellfish Immunol. 2023, 136, 108699. [Google Scholar] [CrossRef]
- Battison, A.L.; Després, B.M.; Greenwood, S.J. Ulcerative enteritis in Homarus americanus: Case report and molecular characterization of intestinal aerobic bacteria of apparently healthy lobsters in live storage. J. Invertebr. Pathol. 2008, 99, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Chew, X.Z.; Gibson-Kueh, S.; Jerry, D.R.; Shen, X. Comparison of intestinal bacterial communities in asymptomatic and diseased Asian seabass (Lates calcarifer) with chronic enteritis and mixed bacterial infections. Aquaculture 2023, 572, 739516. [Google Scholar] [CrossRef]
- Segre, J.A. What does it take to satisfy Koch’s postulates two centuries later? Microbial genomics and Propionibacteria acnes. J. Investig. Dermatol. 2013, 133, 2141–2142. [Google Scholar] [CrossRef]
- Byrd, A.L.; Segre, J.A. Adapting Koch’s postulates. Science 2016, 351, 224–226. [Google Scholar] [CrossRef]
- Xiong, J.; Zhu, J.; Dai, W.; Dong, C.; Qiu, Q.; Li, C. Integrating gut microbiota immaturity and disease-discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environ. Microbiol. 2017, 19, 1490–1501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, M.; Wang, S.; Han, R.; Cao, Y.; Hua, W.; Mao, Y.; Zhang, X.; Pang, X.; Wei, C.; et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010, 4, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Lian, T.; Guo, G.; Gong, H.; Wu, C.; Han, P.; Weng, S.; He, J. Integration of microbiome and Koch’s postulates to reveal multiple bacterial pathogens of whitish muscle syndrome in mud crab, Scylla paramamosain. Microbiome 2023, 11, 155. [Google Scholar] [CrossRef]
- Li, L.; Meng, H.; Gu, D.; Li, Y.; Jia, M. Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol. Res. 2019, 222, 43–51. [Google Scholar] [CrossRef]
- Thirugnanasambandam, R.; Inbakandan, D.; Kumar, C.; Subashni, B.; Vasantharaja, R.; Stanley Abraham, L.; Ayyadurai, N.; Sriyutha Murthy, P.; Kirubagaran, R.; Ajmal Khan, S.; et al. Genomic insights of Vibrio harveyi RT-6 strain, from infected “Whiteleg shrimp” (Litopenaeus vannamei) using Illumina platform. Mol. Phylogenetics Evol. 2019, 130, 35–44. [Google Scholar] [CrossRef]
- Goudenège, D.; Labreuche, Y.; Krin, E.; Ansquer, D.; Mangenot, S.; Calteau, A.; Médigue, C.; Mazel, D.; Polz, M.F.; Le Roux, F. Comparative genomics of pathogenic lineages of Vibrio nigripulchritudo identifies virulence-associated traits. ISME J. 2013, 7, 1985–1996. [Google Scholar] [CrossRef]
- Restrepo-Benavides, M.; Lozano-Arce, D.; Gonzalez-Garcia Laura, N.; Báez-Aguirre, F.; Ariza-Aranguren, G.; Faccini, D.; Zambrano María, M.; Jiménez, P.; Fernández-Bravo, A.; Restrepo, S.; et al. Unveiling potential virulence determinants in Vibrio isolates from Anadara tuberculosa through whole genome analyses. Microbiol. Spectr. 2024, 12, e02928-23. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Primers 2018, 4, 8. [Google Scholar] [CrossRef]
- Bertelli, C.; Greub, G. Rapid bacterial genome sequencing: Methods and applications in clinical microbiology. Clin. Microbiol. Infect. 2013, 19, 803–813. [Google Scholar] [CrossRef]
- Fu, S.; Wang, Q.; Wang, R.; Zhang, Y.; Lan, R.; He, F.; Yang, Q. Horizontal transfer of antibiotic resistance genes within the bacterial communities in aquacultural environment. Sci. Total Environ. 2022, 820, 153286. [Google Scholar] [CrossRef]
- Allard, M.W.; Bell, R.; Ferreira, C.M.; Gonzalez-Escalona, N.; Hoffmann, M.; Muruvanda, T.; Ottesen, A.; Ramachandran, P.; Reed, E.; Sharma, S.; et al. Genomics of foodborne pathogens for microbial food safety. Curr. Opin. Biotechnol. 2018, 49, 224–229. [Google Scholar] [CrossRef]
- Bentley, S.D.; Parkhill, J. Genomic perspectives on the evolution and spread of bacterial pathogens. Proc. Biol. Sci. 2015, 282, 20150488. [Google Scholar] [CrossRef]
- Kumar, V.; Bels, L.D.; Couck, L.; Baruah, K.; Bossier, P.; Broeck, W.V.d. PirABVP Toxin Binds to Epithelial Cells of the Digestive Tract and Produce Pathognomonic AHPND Lesions in Germ-Free Brine Shrimp. Toxins 2019, 11, 717. [Google Scholar] [CrossRef] [PubMed]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Guo, H.; Fu, X.; He, J.; Wang, R.; Yan, M.; Wang, J.; Dong, P.; Huang, L.; Zhang, D. Gut bacterial consortium enriched in a biofloc system protects shrimp against Vibrio parahaemolyticus infection. Microbiome 2023, 11, 230. [Google Scholar] [CrossRef]
- Fu, X.; He, J.; Wang, J.; Shen, F.; Qiu, J.; Chen, C.; Zhang, D.; Guo, H. Specific gut bacterial taxa inhabited in healthy shrimp (Penaeus vannamei) confer protection against Vibrio parahaemolyticus challenge. Aquaculture 2024, 579, 740192. [Google Scholar] [CrossRef]
- Spragge, F.; Bakkeren, E.; Jahn, M.T.; Araujo, E.B.N.; Pearson, C.F.; Wang, X.; Pankhurst, L.; Cunrath, O.; Foster, K.R. Microbiome diversity protects against pathogens by nutrient blocking. Science 2023, 382, eadj3502. [Google Scholar] [CrossRef]
- Li, R.; Zhu, H.; Ruan, J.; Qian, W.; Fang, X.; Shi, Z.; Li, Y.; Li, S.; Shan, G.; Kristiansen, K.; et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010, 20, 265–272. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Hudzicki, J. Kirby-Bauer disk diffusion susceptibility test protocol. Am. Soc. Microbiol. 2009, 15, 1–23. [Google Scholar]
- Hindler, J.; Richter, S.; Bernard, K.; Jones, S.; Castanheira, M.; Citron, D.; Couturier, M.; Fritsche, T.; Humphries, R.; Jorgensen, J. CLSI M45-Methods for An-Timicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Costa, R.; Araujo, R.; Sousa, O.; Vieira, R. Antibiotic-Resistant Vibrios in Farmed Shrimp. BioMed Res. Int. 2015, 2015, 505914. [Google Scholar] [CrossRef]
- Tang, K.F.J.; Bondad-Reantaso, M.G. Impacts of acute hepatopancreatic necrosis disease on commercial shrimp aquaculture. Rev. Sci. Tech. 2019, 38, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Wang, H.; Xie, G.; Zou, P.; Guo, C.; Liang, Y.; Huang, J. An isolate of Vibrio campbellii carrying the pir(VP) gene causes acute hepatopancreatic necrosis disease. Emerg. Microbes Infect. 2017, 6, e2. [Google Scholar] [CrossRef]
- de Souza Valente, C.; Wan, A.H.L. Vibrio and major commercially important vibriosis diseases in decapod crustaceans. J. Invertebr. Pathol. 2021, 181, 107527. [Google Scholar] [CrossRef]
- Flegel, T.W. Recent Research on Acute Hepatopancreatic Necrosis Disease (AHPND) and Enterocytozoon hepatopenaei in Thailand. Asian Fish. Sci. 2018, 31S, 257–269. [Google Scholar] [CrossRef]
- Aguilar-Rendon, K.G.; Lozano-Olvera, R.; Yanez-Rivera, B.; Soto-Rodriguez, S.A. Bacteriological and histopathological analysis of Penaeus vannamei experimentally infected with Vibrio parahaemolyticus-AHPND strains. Dis. Aquat. Organ. 2020, 140, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Khimmakthong, U.; Sukkarun, P. The spread of Vibrio parahaemolyticus in tissues of the Pacific white shrimp Litopenaeus vannamei analyzed by PCR and histopathology. Microb. Pathog. 2017, 113, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Salazar, G.J.; Galaviz-Silva, L.; Guzmán-Sáenz, F.M.; Hernández-Acosta, M.; Roy, L.A. Enteritis Hemocítica en Litopenaeus vannamei (Crustácea Decápoda) en cultivo de baja salinidad en Tamaulipas, México. Hidrobiológica 2015, 25, 139–145. [Google Scholar]
- Zorriehzahra, M.J. Early Mortality Syndrome (EMS) as new Emerging Threat in Shrimp Industry. Adv. Anim. Vet. Sci. 2015, 3, 64–72. [Google Scholar] [CrossRef]
- Tiruvayipati, S.; Bhassu, S. Host, pathogen and the environment: The case of Macrobrachium rosenbergii, Vibrio parahaemolyticus and magnesium. Gut Pathog. 2016, 8, 15. [Google Scholar] [CrossRef]
- Varadi, L.; Luo, J.L.; Hibbs, D.E.; Perry, J.D.; Anderson, R.J.; Orenga, S.; Groundwater, P.W. Methods for the detection and identification of pathogenic bacteria: Past, present, and future. Chem. Soc. Rev. 2017, 46, 4818–4832. [Google Scholar] [CrossRef]
- Siddique, A.B.; Moniruzzaman, M.; Ali, S.; Dewan, M.N.; Islam, M.R.; Islam, M.S.; Amin, M.B.; Mondal, D.; Parvez, A.K.; Mahmud, Z.H. Characterization of Pathogenic Vibrio parahaemolyticus Isolated From Fish Aquaculture of the Southwest Coastal Area of Bangladesh. Front. Microbiol. 2021, 12, 635539. [Google Scholar] [CrossRef]
- Hammerl, J.A.; Gollner, C.; Jackel, C.; Swidan, F.; Gutmann, H.; Strauch, E. The Acquisition of the scr Gene Cluster Encoding Sucrose Metabolization Enzymes Enables Strains of Vibrio parahaemolyticus and Vibrio vulnificus to Utilize Sucrose as Carbon Source. Front. Microbiol. 2021, 12, 754464. [Google Scholar] [CrossRef]
- Williams, S.L.; Jensen, R.V.; Kuhn, D.D.; Stevens, A.M. Analyzing the metabolic capabilities of a Vibrio parahaemolyticus strain that causes Early Mortality Syndrome in shrimp. Aquaculture 2017, 476, 44–48. [Google Scholar] [CrossRef]
- Khan, F.; Tabassum, N.; Anand, R.; Kim, Y.M. Motility of Vibrio spp.: Regulation and controlling strategies. Appl. Microbiol. Biotechnol. 2020, 104, 8187–8208. [Google Scholar] [CrossRef]
- Shime-Hattori, A.; Iida, T.; Arita, M.; Park, K.S.; Kodama, T.; Honda, T. Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol. Lett. 2006, 264, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Watnick Paula, I.; Fullner Karla, J.; Kolter, R. A Role for the Mannose-Sensitive Hemagglutinin in Biofilm Formation by Vibrio cholerae El Tor. J. Bacteriol. 1999, 181, 3606–3609. [Google Scholar] [CrossRef]
- Manning, P.A. The tcp gene cluster of Vibrio cholerae. Gene 1997, 192, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Floyd, K.A.; Lee, C.K.; Xian, W.; Nametalla, M.; Valentine, A.; Crair, B.; Zhu, S.; Hughes, H.Q.; Chlebek, J.L.; Wu, D.C.; et al. c-di-GMP modulates type IV MSHA pilus retraction and surface attachment in Vibrio cholerae. Nat. Commun. 2020, 11, 1549. [Google Scholar] [CrossRef]
- Zhu, J.; Miller, M.B.; Vance, R.E.; Dziejman, M.; Bassler, B.L.; Mekalanos, J.J. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2002, 99, 3129–3134. [Google Scholar] [CrossRef]
- Higgins, D.A.; Pomianek, M.E.; Kraml, C.M.; Taylor, R.K.; Semmelhack, M.F.; Bassler, B.L. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 2007, 450, 883–886. [Google Scholar] [CrossRef]
- Bridges, A.A.; Bassler, B.L. The intragenus and interspecies quorum-sensing autoinducers exert distinct control over Vibrio cholerae biofilm formation and dispersal. PLoS Biol. 2019, 17, e3000429. [Google Scholar] [CrossRef] [PubMed]
- Burdette, D.L.; Yarbrough, M.L.; Orvedahl, A.; Gilpin, C.J.; Orth, K. Vibrio parahaemolyticus orchestrates a multifaceted host cell infection by induction of autophagy, cell rounding, and then cell lysis. Proc. Natl. Acad. Sci. USA 2008, 105, 12497–12502. [Google Scholar] [CrossRef]
- Makino, K.; Oshima, K.; Kurokawa, K.; Yokoyama, K.; Uda, T.; Tagomori, K.; Iijima, Y.; Najima, M.; Nakano, M.; Yamashita, A.; et al. Genome sequence of Vibrio parahaemolyticus: A pathogenic mechanism distinct from that of V cholerae. Lancet 2003, 361, 743–749. [Google Scholar] [CrossRef]
- Park, K.-S.; Ono, T.; Rokuda, M.; Jang, M.-H.; Okada, K.; Iida, T.; Honda, T. Functional Characterization of Two Type III Secretion Systems of Vibrio parahaemolyticus. Infect. Immun. 2004, 72, 6659–6665. [Google Scholar] [CrossRef]
- Calder, T.; de Souza Santos, M.; Attah, V.; Klimko, J.; Fernandez, J.; Salomon, D.; Krachler, A.M.; Orth, K. Structural and regulatory mutations in Vibrio parahaemolyticus type III secretion systems display variable effects on virulence. FEMS Microbiol. Lett. 2014, 361, 107–114. [Google Scholar] [CrossRef]
- Johnson, T.L.; Fong, J.C.; Rule, C.; Rogers, A.; Yildiz, F.H.; Sandkvist, M. The Type II secretion system delivers matrix proteins for biofilm formation by Vibrio cholerae. J. Bacteriol. 2014, 196, 4245–4252. [Google Scholar] [CrossRef]
- Wang, R.; Sun, L.; Wang, Y.; Deng, Y.; Fang, Z.; Liu, Y.; Liu, Y.; Sun, D.; Deng, Q.; Gooneratne, R. Growth and Hemolysin Production Behavior of Vibrio parahaemolyticus in Different Food Matrices. J. Food Prot. 2018, 81, 246–253. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Stockley, L.; Rangdale, R.; Martinez-Urtaza, J. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European perspective. Environ. Microbiol. Rep. 2010, 2, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, J.M.; Rui, H.; Zhou, X.; Iida, T.; Kodoma, T.; Ito, S.; Davis, B.M.; Bronson, R.T.; Waldor, M.K. Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 2012, 8, e1002593. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Krachler, A.M.; Broberg, C.A.; Li, Y.; Mirzaei, H.; Gilpin, C.J.; Orth, K. Type III effector VopC mediates invasion for Vibrio species. Cell Rep. 2012, 1, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Gewurz, B.E.; Ritchie, J.M.; Takasaki, K.; Greenfeld, H.; Kieff, E.; Davis, B.M.; Waldor, M.K. A Vibrio parahaemolyticus T3SS effector mediates pathogenesis by independently enabling intestinal colonization and inhibiting TAK1 activation. Cell Rep. 2013, 3, 1690–1702. [Google Scholar] [CrossRef]

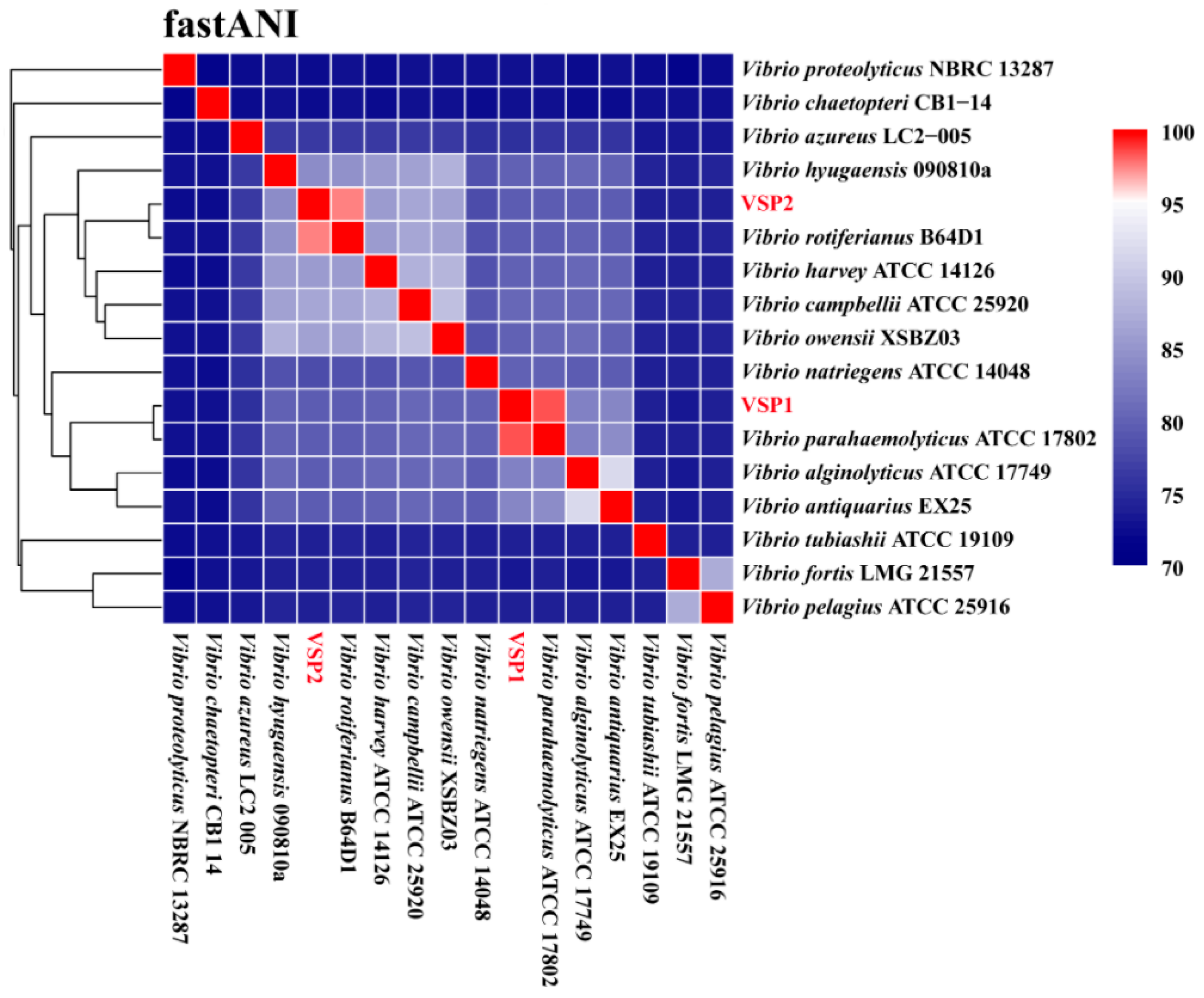
| VSP1 | VSP2 | |
|---|---|---|
| Assembly statistics | ||
| Genome completeness (%) | 100 | 100 |
| Genome contamination (%) | 0.08 | 2.12 |
| Genome size (bp) | 5,366,644 | 5,603,652 |
| GC content (%) | 45.0 | 45.0 |
| Prediction and annotation | ||
| Coding sequences (CDSs) | 4829 | 5037 |
| Average gene length (bp) | 320 | 322 |
| Annotated genes (Bakta) | 4276 | 4471 |
| Pathogenicity-related genes | ||
| Genes annotated in VFDB | 156 | 153 |
| Genes annotated in CARD | 6 | 6 |
| Prophage elements | 25 | 17 |
| Virulence Factors | Gene Number | ||
|---|---|---|---|
| V. parahaemolyticus ATCC 17802 | VSP1 | VSP2 | |
| Adherence | |||
| Mannose-sensitive hemagglutinin (MSHA type IV pilus) | 14 | 15 | 17 |
| Type IV pilus | 4 | 4 | 0 |
| Type IV pili (Yersinia) | 0 | 1 | 0 |
| Antiphagocytosis | |||
| Capsular polysaccharide | 16 | 15 | 15 |
| Motility | |||
| Flagella | 56 | 57 | 87 |
| Iron acquisition system | |||
| Enterobactin receptors | 2 | 2 | 0 |
| Heme receptors | 2 | 2 | 1 |
| Periplasmic binding protein-dependent ABC transport systems | 4 | 4 | 6 |
| Acinetobactin (Acinetobacter) | 0 | 0 | 5 |
| Quorum Sensing Factors | |||
| Autoinducer-2 | 1 | 1 | 1 |
| Cholerae autoinducer-1 | 1 | 1 | 1 |
| Secretion system | |||
| EPS type II secretion system | 12 | 12 | 12 |
| T3SS1 | 34 | 34 | 0 |
| T3SS1 secreted effectors | 4 | 4 | 0 |
| T3SS2 | 10 | 0 | 0 |
| T3SS2 secreted effectors | 1 | 0 | 0 |
| AAI/SCI-II T6SS (Escherichia) | 0 | 0 | 2 |
| Toxin | |||
| Thermolabile hemolysin | 1 | 1 | 1 |
| Thermostable direct hemolysin | 1 | 0 | 0 |
| Alpha-hemolysin (Escherichia) | 3 | 0 | 0 |
| Phytotoxin phaseolotoxin (Pseudomonas) | 0 | 0 | 1 |
| Endotoxin | |||
| LOS (Haemophilus) | 0 | 1 | 0 |
| Acid resistance | |||
| Urease (Helicobacter) | 2 | 0 | 3 |
| Glycosylation system | |||
| O-linked flagellar glycosylation (Campylobacter) | 1 | 0 | 0 |
| Immune evasion | |||
| Capsule (Acinetobacter) | 1 | 2 | 0 |
| LPS (Francisella) | 0 | 0 | 1 |
| LPS glucosylation (Shigella) | 1 | 0 | 0 |
| Others | |||
| O-antigen (Yersinia) | 1 | 0 | 0 |
| Strain | Predicted ARG | Identity (%) | Drug Class | Resistance Mechanism |
|---|---|---|---|---|
| VSP1 | CARB-18 | 100 | β-lactam | Antibiotic inactivation |
| CRP | 95.24 | Macrolide, Fluoroquinolone, β-lactam | Antibiotic efflux | |
| TxR | 86.29 | Tetracycline | Antibiotic efflux | |
| parE | 78.98 | Fluoroquinolone | Target alteration | |
| adeF | 42.68 | Fluoroquinolone, Tetracycline | Antibiotic efflux | |
| vanT | 33.06 | Glycopeptide | Target alteration | |
| VSP2 | CRP | 95.24 | Macrolide, Fluoroquinolone, β-lactam | Antibiotic efflux |
| parE | 79.14 | Fluoroquinolone | Target alteration | |
| tet(B) | 69.59 | Tetracycline | Antibiotic efflux | |
| PBP3 | 45.25 | Cephalosporin, Penicillin | Target alteration | |
| adeF | 43.15 | Fluoroquinolone, Tetracycline | Antibiotic efflux | |
| vanT | 33.06 | Glycopeptide | Target alteration |
| Antibiotic Class | Antibiotics | Concentration (µg) | Diameter of Inhibition Zone (mm)/ Antibiotic Sensitivity | |
|---|---|---|---|---|
| VSP1 | VSP2 | |||
| β-lactam | Ampicillin (AMP) | 10 | 0 R | 0 R |
| Piperacillin (PIP) | 100 | 0 R | 0 R | |
| Cefotaxime (CTX) | 30 | 22 R | 22 R | |
| Cefoxitin (FOX) | 30 | 20 S | 20 S | |
| Cefepime (FEP) | 30 | 19 I | 18 R | |
| Imipenem (IPM) | 10 | 16 R | 16 R | |
| Ceftazidime (CAZ) | 30 | 19 I | 19 I | |
| Fluoroquinolones | Levofloxacin (LEV) | 5 | 13 R | 15 I |
| Quinolones | Ciprofloxacin (CIP) | 5 | 15 R | 14 R |
| Tetracyclines | Tetracycline (TE) | 30 | 0 R | 12 R |
| Aminoglycosides | Gentamicin (CEN) | 10 | 12 R | 10 R |
| Chloramphenicols | Chloramphenicol (CMP) | 30 | 32 S | 29 S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Shen, F.; Tian, M.; Zeng, F.; Huang, L.; Yao, J.; Zong, C.; Chen, J.; Zhang, D.; Guo, H. Pathogenicity and Genomic Characterization of Vibrio parahaemolyticus VSP1: A Pathogen Linked to Enteritis Outbreak in Shrimp (Penaeus vannamei). Pathogens 2025, 14, 1188. https://doi.org/10.3390/pathogens14111188
Wang J, Shen F, Tian M, Zeng F, Huang L, Yao J, Zong C, Chen J, Zhang D, Guo H. Pathogenicity and Genomic Characterization of Vibrio parahaemolyticus VSP1: A Pathogen Linked to Enteritis Outbreak in Shrimp (Penaeus vannamei). Pathogens. 2025; 14(11):1188. https://doi.org/10.3390/pathogens14111188
Chicago/Turabian StyleWang, Jing, Fengguang Shen, Meng Tian, Fanqi Zeng, Lei Huang, Jiayun Yao, Can Zong, Jiong Chen, Demin Zhang, and Haipeng Guo. 2025. "Pathogenicity and Genomic Characterization of Vibrio parahaemolyticus VSP1: A Pathogen Linked to Enteritis Outbreak in Shrimp (Penaeus vannamei)" Pathogens 14, no. 11: 1188. https://doi.org/10.3390/pathogens14111188
APA StyleWang, J., Shen, F., Tian, M., Zeng, F., Huang, L., Yao, J., Zong, C., Chen, J., Zhang, D., & Guo, H. (2025). Pathogenicity and Genomic Characterization of Vibrio parahaemolyticus VSP1: A Pathogen Linked to Enteritis Outbreak in Shrimp (Penaeus vannamei). Pathogens, 14(11), 1188. https://doi.org/10.3390/pathogens14111188






