Association of SLC11A1 3′UTR (GT)n Microsatellite Polymorphisms with Resistance to Paratuberculosis in Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Population and Sample Collection
2.2. DNA Extraction and Genotyping of the SLC11A1 3′UTR (GT)n Microsatellite
2.3. RNA Extraction and Gene Expression Analysis
2.4. Statistical Analysis
3. Results
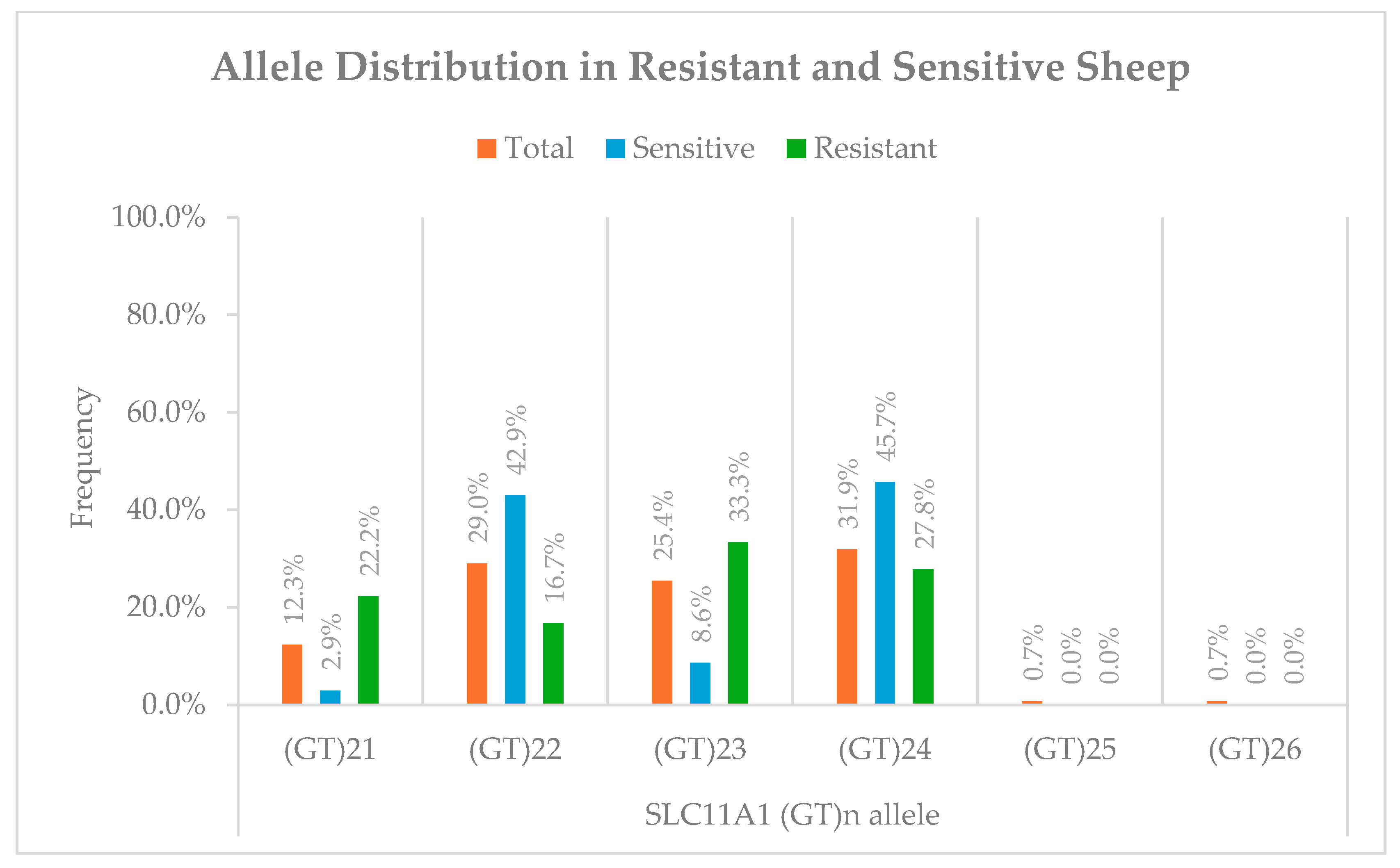
3.1. (GT)n Repeat Polymorphism Frequencies
3.2. Genotype–Phenotype Association
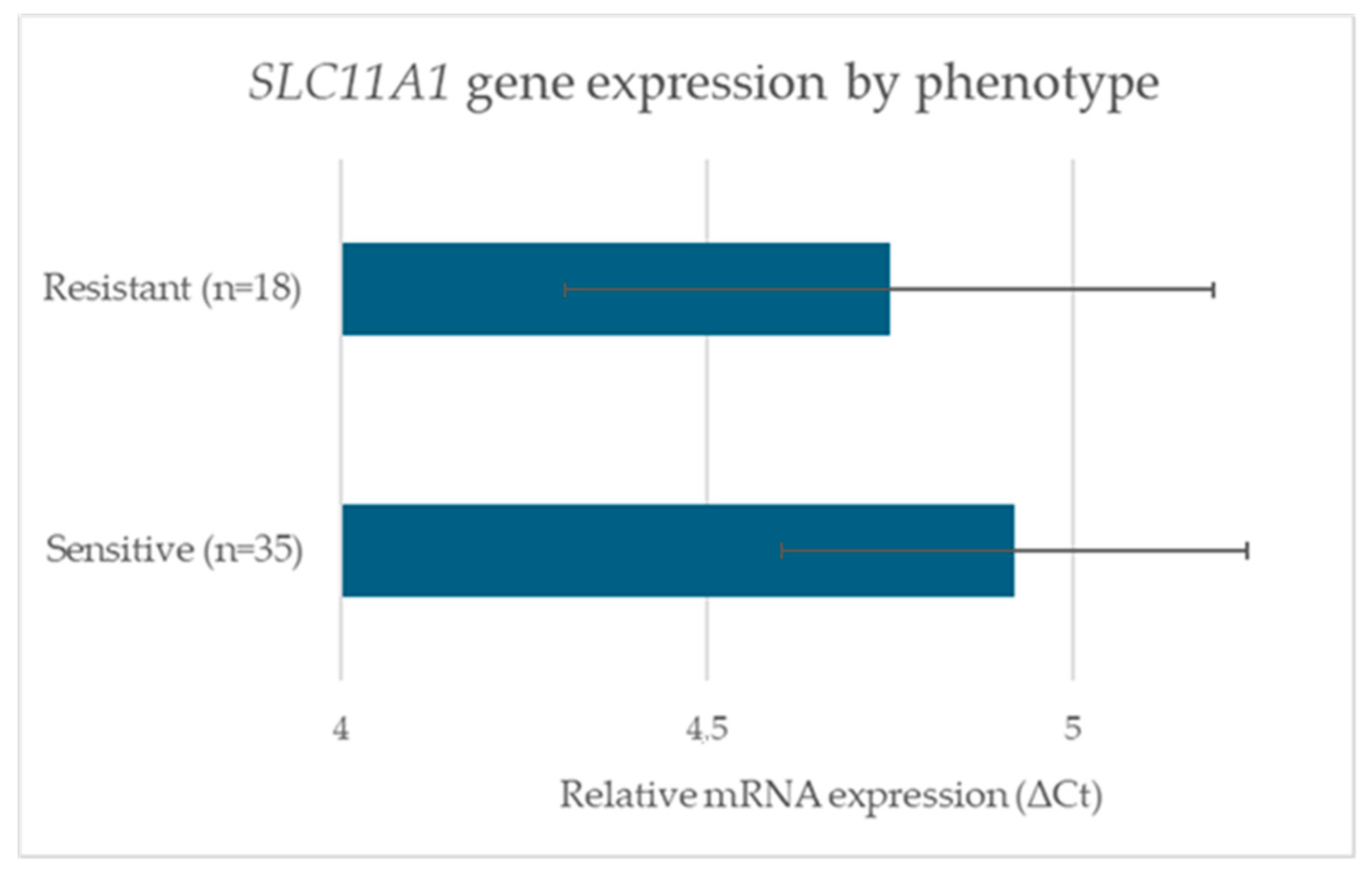
3.3. SLC11A1 Gene Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harris, N.B.; Barletta, R.G. Mycobacterium avium subsp. paratuberculosis in Veterinary Medicine. Clin. Microbiol. Rev. 2001, 14, 489–512. [Google Scholar] [CrossRef]
- Clarke, C.J. The pathology and pathogenesis of paratuberculosis in ruminants and other species. J. Comp. Pathol. 1997, 116, 217–261. [Google Scholar] [CrossRef] [PubMed]
- Bush, R.D.; Windsor, P.A.; Toribio, J.A.L.M.L. Losses of adult sheep due to ovine Johne’s disease in 12 infected flocks over a 3-year period. Aust. Vet. J. 2006, 84, 246–253. [Google Scholar] [CrossRef]
- Hasonova, L.; Pavlik, I. Economic impact of paratuberculosis in dairy cattle herds: A review. Vet. Med. 2006, 51, 193–211. [Google Scholar] [CrossRef]
- Raizman, E.A.; Fetrow, J.; Wells, S.J.; Godden, S.M.; Oakes, M.J.; Vazquez, G. The association between Mycobacterium avium subsp. paratuberculosis fecal shedding or clinical Johne’s disease and lactation performance on two Minnesota, USA dairy farms. Prev. Vet. Med. 2007, 78, 179–195. [Google Scholar] [CrossRef]
- Nielsen, S.S.; Toft, N. A review of prevalences of paratuberculosis in farmed animals in Europe. Prev. Vet. Med. 2009, 88, 1–14. [Google Scholar] [CrossRef]
- di Marco Lo Presti, V.; Ippolito, D.; Migliore, S.; Tolone, M.; Mignacca, S.A.; Marino, A.M.F.; Amato, B.; Calogero, R.; Vitale, M.; Vicari, D.; et al. Large-scale serological survey on Mycobacterium avium subsp. paratuberculosis infection in sheep and goat herds in Sicily, Southern Italy. Front. Vet. Sci. 2024, 11, 1334036. [Google Scholar] [CrossRef] [PubMed]
- Gautam, M.; Ridler, A.; Wilson, P.R.; Heuer, C. Control of clinical paratuberculosis in New Zealand pastoral livestock. N. Z. Vet. J. 2018, 66, 1–8. [Google Scholar] [CrossRef]
- Windsor, P.; Whittington, R. Ovine Paratuberculosis Control in Australia Revisited. Animals 2020, 10, 1623. [Google Scholar] [CrossRef] [PubMed]
- Koets, A.P.; Adugna, G.; Janss, L.L.G.; van Weering, H.J.; Kalis, C.H.J.; Wentink, G.H.; Rutten, V.P.M.G.; Schukken, Y.H. Genetic variation of susceptibility to Mycobacterium avium subsp. paratuberculosis infection in dairy cattle. J. Dairy Sci. 2000, 83, 2702–2708. [Google Scholar] [CrossRef]
- Reddacliff, L.A.; Beh, K.; McGregor, H.; Whittington, R.J. A preliminary study of possible genetic influences on the susceptibility of sheep to Johne’s disease. Aust. Vet. J. 2005, 83, 435–441. [Google Scholar] [CrossRef]
- Lugton, I.W. Cross-sectional study of risk factors for the clinical expression of ovine Johne’s disease on New South Wales farms. Aust. Vet. J. 2004, 82, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.M.; Searle, S. Genetic regulation of macrophage activation: Understanding the function of Nramp1 (=Ity/Lsh/Bcg). Immunol. Lett. 1999, 65, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Buschman, E.; Vidal, S.; Skamene, E. Nonspecific resistance to Mycobacteria: The role of the Nramp1 gene. Behring Inst. Mitteilungen 1997, 99, 51–57. [Google Scholar]
- Vidal, S.; Gros, P.; Skamene, E. Natural resistance to infection with intracellular parasites: Molecular genetics identifies Nramp1 as the Bcg/Ity/Lsh locus. J. Leukoc. Biol. 1995, 58, 382–390. [Google Scholar] [CrossRef]
- Wyllie, S.; Seu, P.; Goss, J.A. The natural resistance-associated macrophage protein 1 Slc11a1 (formerly Nramp1) and iron metabolism in macrophages. Microbes Infect. 2002, 4, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Thomas, N.; Joseph, S. Role of SLC11A1 gene in disease resistance. Biotechnol. Anim. Husb. 2012, 28, 99–106. [Google Scholar] [CrossRef][Green Version]
- Awomoyi, A.A.; Marchant, A.; Howson, J.M.M.; McAdam, K.P.W.J.; Blackwell, J.M.; Newport, M.J. Interleukin-10, polymorphism in SLC11A1 (formerly NRAMP1), and susceptibility to tuberculosis. J. Infect. Dis. 2002, 186, 1808–1814. [Google Scholar] [CrossRef]
- Meilang, Q.; Zhang, Y.; Zhang, J.; Zhao, Y.; Tian, C.; Huang, J.; Fan, H. Polymorphisms in the SLC11A1 gene and tuberculosis risk: A meta-analysis update. Int. J. Tuberc. Lung Dis. 2012, 16, 437–446. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, X.; Yang, Y.; Ding, Y.; Xue, W.; Meng, Y.; Zhu, W.; Yin, Z. Polymorphism, Expression of Natural Resistance-associated Macrophage Protein 1 Encoding Gene (NRAMP1) and Its Association with Immune Traits in Pigs. Asian-Australas. J. Anim. Sci. 2014, 27, 1189–1195. [Google Scholar] [CrossRef]
- Taka, S.; Gazouli, M.; Sotirakoglou, K.; Liandris, E.; Andreadou, M.; Triantaphyllopoulos, K.; Ikonomopoulos, J. Functional analysis of 3′UTR polymorphisms in the caprine SLC11A1 gene and its association with the Mycobacterium avium subsp. paratuberculosis infection. Vet. Immunol. Immunopathol. 2015, 167, 75–79. [Google Scholar] [CrossRef]
- Taka, S.; Liandris, E.; Gazouli, M.; Sotirakoglou, K.; Theodoropoulos, G.; Bountouri, M.; Andreadou, M.; Ikonomopoulos, J. In vitro expression of the SLC11A1 gene in goat monocyte-derived macrophages challenged with Mycobacterium avium subsp. paratuberculosis. Infect. Genet. Evol. 2013, 17, 8–15. [Google Scholar] [CrossRef]
- Korou, L.M.; Liandris, E.; Gazouli, M.; Ikonomopoulos, J. Investigation of the association of the SLC11A1 gene with resistance/sensitivity of goats (Capra hircus) to paratuberculosis. Vet. Microbiol. 2010, 144, 353–358. [Google Scholar] [CrossRef]
- Abraham, A.; Naicy, T.; Raghavan, K.C.; Siju, J.; Aravindakshan, T. Evaluation of the association of SLC11A1 gene polymorphism with incidence of paratuberculosis in goats. J. Genet. 2017, 96, 641–646. [Google Scholar] [CrossRef]
- Balasubramaniam, S.; Kumar, S.; Sharma, A.; Mitra, A. Microsatellite (GT)n polymorphism at 3′UTR of SLC11A1 influences the expression of brucella LPS induced MCP1 mRNA in buffalo peripheral blood mononuclear cells. Vet. Immunol. Immunopathol. 2013, 152, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Larrañaga, O.; Garrido, J.M.; Manzano, C.; Iriondo, M.; Molina, E.; Gil, A.; Koets, A.P.; Rutten, V.P.M.G.; Juste, R.A.; Estonba, A. Identification of single nucleotide polymorphisms in the bovine solute carrier family 11 member 1 (SLC11A1) gene and their association with infection by Mycobacterium avium subspecies paratuberculosis. J. Dairy Sci. 2010, 93, 1713–1721. [Google Scholar] [CrossRef]
- Kravitz, A.; Liao, M.; Morota, G.; Tyler, R.; Cockrum, R.; Manohar, B.M.; Ronald, B.S.M.; Collins, M.T.; Sriranganathan, N. Retrospective Single Nucleotide Polymorphism Analysis of Host Resistance and Susceptibility to Ovine Johne’s Disease Using Restored FFPE DNA. Int. J. Mol. Sci. 2024, 25, 7748. [Google Scholar] [CrossRef]
- Bermingham, M.L.; Bishop, S.C.; Woolliams, J.A.; Pong-Wong, R.; Allen, A.R.; McBride, S.H.; Ryder, J.J.; Wright, D.M.; Skuce, R.A.; McDowell, S.W.; et al. Genome-wide association study identifies novel loci associated with resistance to bovine tuberculosis. Heredity 2014, 112, 543–551. [Google Scholar] [CrossRef]
- Gao, Y.; Jiang, J.; Yang, S.; Cao, J.; Han, B.; Wang, Y.; Zhang, Y.; Yu, Y.; Zhang, S.; Zhang, Q.; et al. Genome-wide association study of Mycobacterium avium subspecies Paratuberculosis infection in Chinese Holstein. BMC Genom. 2018, 19, 972. [Google Scholar] [CrossRef] [PubMed]
- Canive, M.; González-Recio, O.; Fernández, A.; Vázquez, P.; Badia-Bringué, G.; Lavín, J.L.; Garrido, J.M.; Juste, R.A.; Alonso-Hearn, M. Identification of loci associated with susceptibility to Mycobacterium avium subsp. paratuberculosis infection in Holstein cattle using combinations of diagnostic tests and imputed whole-genome sequence data. PLoS ONE 2021, 16, e0256091. [Google Scholar] [CrossRef] [PubMed]
- Canive, M.; Badia-Bringué, G.; Vázquez, P.; Garrido, J.M.; Juste, R.A.; Fernandez, A.; González-Recio, O.; Alonso-Hearn, M. A Genome-Wide Association Study for Tolerance to Paratuberculosis Identifies Candidate Genes Involved in DNA Packaging, DNA Damage Repair, Innate Immunity, and Pathogen Persistence. Front. Immunol. 2022, 13, 820965. [Google Scholar] [CrossRef]
- Alonso-Hearn, M.; Badia-Bringué, G.; Canive, M. Genome-wide association studies for the identification of cattle susceptible and resilient to paratuberculosis. Front. Vet. Sci. 2022, 9, 935133. [Google Scholar] [CrossRef] [PubMed]
- Usai, M.G.; Casu, S.; Sechi, T.; Salaris, S.L.; Miari, S.; Mulas, G.; Cancedda, M.G.; Ligios, C.; Carta, A. Advances in understanding the genetic architecture of antibody response to paratuberculosis in sheep by heritability estimate and LDLA mapping analyses and investigation of candidate regions using sequence-based data. Genet. Sel. Evol. 2024, 56, 5. [Google Scholar] [CrossRef]
- Moioli, B.; D’Andrea, S.; de Grossi, L.; Sezzi, E.; de Sanctis, B.; Catillo, G.; Steri, R.; Valentini, A.; Pilla, F. Genomic scan for identifying candidate genes for paratuberculosis resistance in sheep. Anim. Prod. Sci. 2015, 56, 1046–1055. [Google Scholar] [CrossRef]
- Purdie, A.C.; Plain, K.M.; Begg, D.J.; de Silva, K.; Whittington, R.J. Candidate gene and genome-wide association studies of Mycobacterium avium subsp. paratuberculosis infection in cattle and sheep: A review. Comp. Immunol. Microbiol. Infect. Dis. 2011, 34, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Kravitz, A.; Pelzer, K.; Sriranganathan, N. The Paratuberculosis Paradigm Examined: A Review of Host Genetic Resistance and Innate Immune Fitness in Mycobacterium avium subsp. Paratuberculosis Infection. Front. Vet. Sci. 2021, 8, 721706. [Google Scholar] [CrossRef]
- Mataragka, A.; Sotirakoglou, K.; Gazouli, M.; Triantaphyllopoulos, K.A.; Ikonomopoulos, J. Parturition affects test-positivity in sheep with subclinical paratuberculosis; investigation following a preliminary analysis. J. King Saud Univ.-Sci. 2019, 31, 1399–1403. [Google Scholar] [CrossRef]
- National Research Council. Nutrient Requirements of Small Ruminants: Sheep, Goats, Cervids, and New World Camelids; The National Academies Press: Washington, DC, USA, 2007. [Google Scholar] [CrossRef]
- Kim, S.G.; Kim, E.H.; Lafferty, C.J.; Miller, L.J.; Koo, H.J.; Stehman, S.M.; Shin, S.J. Use of conventional and real-time polymerase chain reaction for confirmation of Mycobacterium avium subsp. paratuberculosis in a broth-based culture system ESP II. J. Vet. Diagn. Investig. 2004, 16, 448–453. [Google Scholar] [CrossRef]
- Dowling, R.J.O.; Bienzle, D. Gene-expression changes induced by Feline immunodeficiency virus infection differ in epithelial cells and lymphocytes. J. Gen. Virol. 2005, 86, 2239–2248. [Google Scholar] [CrossRef]
- Taylor, D.L.; Zhong, L.; Begg, D.J.; de Silva, K.; Whittington, R.J. Toll-like receptor genes are differentially expressed at the sites of infection during the progression of Johne’s disease in outbred sheep. Vet. Immunol. Immunopathol. 2008, 124, 132–151. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pinedo, P.J.; Buergelt, C.D.; Donovan, G.A.; Melendez, P.; Morel, L.; Wu, R.; Langaee, T.Y.; Rae, D.O. Candidate gene polymorphisms (BoIFNG, TLR4, SLC11A1) as risk factors for paratuberculosis infection in cattle. Prev. Vet. Med. 2009, 91, 189–196. [Google Scholar] [CrossRef]
- Canive, M.; Casais, R.; Jimenez, J.A.; Blanco-Vazquez, C.; Amado, J.; Garrido, J.M.; Juste, R.A.; Alonso-Hearn, M. Correlations between single nucleotide polymorphisms in bovine CD209, SLC11A1, SP110 and TLR2 genes and estimated breeding values for several traits in Spanish Holstein cattle. Heliyon 2020, 6, e04254. [Google Scholar] [CrossRef]
- Okuni, J.B.; Afayoa, M.; Ojok, L. Survey of Candidate Single-Nucleotide Polymorphisms in SLC11A1, TLR4, NOD2, PGLYRP1, and IFNγ in Ankole Longhorn Cattle in Central Region of Uganda to Determine Their Role in Mycobacterium avium Subspecies paratuberculosis Infection Outcome. Front. Vet. Sci. 2021, 8, 614518. [Google Scholar] [CrossRef]
- Gopi, B.; Vir Singh, R.; Kumar, S.; Kumar, S.; Chauhan, A.; Sonwane, A.; Kumar, A.; Bharati, J.; Vir Singh, S. Effect of selected single nucleotide polymorphisms in SLC11A1, ANKRA2, IFNG and PGLYRP1 genes on host susceptibility to Mycobacterium avium subspecies paratuberculosis infection in Indian cattle. Vet. Res. Commun. 2022, 46, 209–221. [Google Scholar] [CrossRef]
- Wu, H.; Cheng, D.; Wang, L. Association of polymorphisms of Nramp1 gene with immune function and production performance of large white pig. J. Genet. Genom. 2008, 35, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Sechi, L.A.; Dow, C.T. Mycobacterium avium ss. paratuberculosis Zoonosis—The Hundred Year War—Beyond Crohn’s Disease. Front. Immunol. 2015, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Dukkipati, V.S.R.; Blair, H.T.; Garrick, D.J.; Lopez-Villalobos, N.; Whittington, R.J.; Reddacliff, L.A.; Eppleston, J.; Windsor, P.; Murray, A. Association of microsatellite polymorphisms with immune responses to a killed Mycobacterium avium subsp. Paratuberculosis Vaccine Merino sheep. N. Z. Vet. J. 2010, 58, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Whittington, R.J.; Begg, D.J.; de Silva, K.; Plain, K.M.; Purdie, A.C. Comparative immunological and microbiological aspects of paratuberculosis as a model mycobacterial infection. Vet. Immunol. Immunopathol. 2012, 148, 29–47. [Google Scholar] [CrossRef]
- Begg, D.J.; Whittington, R.J. Experimental animal infection models for Johne’s disease, an infectious enteropathy caused by Mycobacterium avium subsp. paratuberculosis. Vet. J. 2008, 176, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Larrañaga, O.; Langa, J.; Rendo, F.; Manzano, C.; Iriondo, M.; Estonba, A. Genomic selection signatures in sheep from the Western Pyrenees. Genet. Sel. Evol. 2018, 50, 9. [Google Scholar] [CrossRef] [PubMed]
| Target | Primers (5′-3′) | Size | Thermal Profile | Reference |
|---|---|---|---|---|
| IS900 | F 1: AATGACGGTTACGGAGGTGGT R 2: GCAGTAATGGTCGGCCTTACC Pr 3: TCCACGCCCGCCCAGACAGG | 76 bp | 95 °C for 3 min; 40 cycles of 95 °C for 3 s, 60 °C for 20 s, 72 °C for 1 s; 43 °C for 30 s | [39] |
| 3′UTR SLC11A1 | F: ACCTGGTCTGGACCTGTCTCATCA R: CATTGCAAGGTAGGTGTCCCCAT | 346 bp | 95 °C for 3 min; 35 cycles of 95 °C for 30 s, 59 °C for 30 s, 72 °C for 20 s; 72 °C for 3 min | [23] |
| GAPDH | F: TTCCAGTATGATTCCACCCATG R: GCCTTTCCATTGATGACGAG | 80 bp | 42 °C for 5 min; 95 °C for 15 s; 40 cycles of 95 °C for 5 s, 52 °C for 20 s, 72 °C for 1 s; 40 °C for 30 s | [41] |
| SLC11A1 mRNA | F: GGCTGTGGCTGGATTCAAAC R: ATGGTCAGCCAGAGGAGAATG | 168 bp | 42 °C for 5 min; 95 °C for 15 s; 40 cycles of 95 °C for 5 s, 57 °C for 20 s, 72 °C for 1 s; 40 °C for 30 s | [23] |
| β-actin | F: TGTCTCTGTACGCTTCTGG R: GTGGTGGTGAAACTGTAGC | 190 bp | 95 °C for 3 min; 40 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s; 72 °C for 3 min | [40] |
| Species | Variant/Region Analyzed | Association with Resistance/Susceptibility | Notes | References |
|---|---|---|---|---|
| Sheep | Genetic influences (preliminary, candidate-based) | Suggested possible genetic effect on Johne’s disease susceptibility | Early evidence, not locus-specific | [11] |
| Sheep | Retrospective SNP analysis | Identified associations near SLC11A1 with MAP resistance | Based on FFPE DNA, SNP focus | [27] |
| Sheep | GWAS (antibody response to MAP) | Regions linked to immune response; SLC11A1 implicated | High-resolution genomic mapping | [33] |
| Sheep | GWAS (SNPs across genome) | Regions associated with MAP resistance; included SLC11A1 | SNP-based, no microsatellite resolution | [34] |
| Sheep | 3′UTR (GT)n microsatellite | (GT)21 and (GT)23 associated with resistance; (GT)22 and (GT)24 with susceptibility | Association found despite no difference in basal expression | [This study] |
| Goats | Functional analysis, 3′UTR microsatellite | Variants affected inducible expression under MAP challenge | Demonstrated functional mechanism | [21] |
| Goats | 3′UTR (GT)n microsatellite | Shorter alleles enriched in resistant goats | Consistent with ovine findings | [23] |
| Goats | 3′UTR microsatellite | Specific alleles associated with reduced paratuberculosis incidence | Validated earlier results | [24] |
| Cattle | SNPs in SLC11A1 | Associated with MAP infection risk | Consistent across populations | [26] |
| Cattle | Candidate gene SNPs (SLC11A1, TLR4, IFNG) | Associations with MAP susceptibility | Population-specific variation | [43] |
| Cattle | SNPs in SLC11A1 and others | Linked with breeding values for MAP traits | Large-scale genomic approach | [44] |
| Cattle | SNPs in SLC11A1 | No association with MAP infection | SNPs polymorphic variants showed no allele/genotype differences between cattle | [45] |
| Cattle | SLC11A1 SNP rs109453173 | Associated with resistance (GG genotype/G allele protective; CC/CG linked to susceptibility) | Case–control study; suggests potential resistance marker | [46] |
| Buffalo | 3′UTR microsatellite | Allelic variation influenced MCP1 mRNA after Brucella challenge | Functional immune effects | [25] |
| Pigs | SLC11A1 polymorphisms | Associated with immune traits | Cross-species evidence of functional role | [47] |
| Humans | SLC11A1 SNPs and promoter variants | Associated with tuberculosis susceptibility | Strong parallels with livestock | [18,48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mataragka, A.; Klavdianos Papastathis, A.; Ikonomopoulos, J. Association of SLC11A1 3′UTR (GT)n Microsatellite Polymorphisms with Resistance to Paratuberculosis in Sheep. Pathogens 2025, 14, 1150. https://doi.org/10.3390/pathogens14111150
Mataragka A, Klavdianos Papastathis A, Ikonomopoulos J. Association of SLC11A1 3′UTR (GT)n Microsatellite Polymorphisms with Resistance to Paratuberculosis in Sheep. Pathogens. 2025; 14(11):1150. https://doi.org/10.3390/pathogens14111150
Chicago/Turabian StyleMataragka, Antonia, Anastasios Klavdianos Papastathis, and John Ikonomopoulos. 2025. "Association of SLC11A1 3′UTR (GT)n Microsatellite Polymorphisms with Resistance to Paratuberculosis in Sheep" Pathogens 14, no. 11: 1150. https://doi.org/10.3390/pathogens14111150
APA StyleMataragka, A., Klavdianos Papastathis, A., & Ikonomopoulos, J. (2025). Association of SLC11A1 3′UTR (GT)n Microsatellite Polymorphisms with Resistance to Paratuberculosis in Sheep. Pathogens, 14(11), 1150. https://doi.org/10.3390/pathogens14111150







